Significance
The high-mobility group box 1 (HMGB1) protein is abundantly expressed in the nucleus where it regulates chromatin function. More recently, it was found to also function in the cytoplasm and extracellular milieu for the regulation of immunity and inflammation. However, the in vivo study of HMGB1 has been hampered by the fact that HMGB1-deficient mice die soon after birth. In this study, we successfully generated Hmgb1-floxed mice to achieve conditional inactivation of the gene in a cell- and tissue-specific manner. We demonstrate that cytosolic HMGB1 in myeloid cells is critical for the protection of the host from endotoxemia and bacterial infection by inducing autophagy, a cellular response critical for maintaining cellular viability in the setting of various stresses including infection.
Keywords: LPS, IL-1β, IL-18
Abstract
High-mobility group box 1 (HMGB1) is a DNA-binding protein abundantly expressed in the nucleus that has gained much attention for its regulation of immunity and inflammation. Despite this, whether and how HMGB1 contributes to protective and/or pathological responses in vivo is unclear. In this study, we constructed Hmgb1-floxed (Hmgb1f/f) mice to achieve the conditional inactivation of the gene in a cell- and tissue-specific manner by crossing these mice with an appropriate Cre recombinase transgenic strain. Interestingly, although mice with HMGB1 ablation in myeloid cells apparently develop normally, they are more sensitive to endotoxin shock compared with control mice, which is accompanied by massive macrophage cell death. Furthermore, these mice also show an increased sensitivity to Listeria monocytogenes infection. We also provide evidence that the loss of HMGB1 in macrophages results in the suppression of autophagy, which is commonly induced by lipopolysaccharide stimulation or L. monocytogenes infection. Thus, intracellular HMGB1 contributes to the protection of mice from endotoxemia and bacterial infection by mediating autophagy in macrophages. These newly generated HMGB1 conditional knockout mice will serve a useful tool with which to study further the in vivo role of this protein in various pathological conditions.
Of the four members of the high-mobility group box (HMGB) family, HMGB1 is the best studied, given its versatile functions in various aspects of cellular responses (1–5). Ubiquitously expressed in all cells, HMGB1 is found en masse in the nucleus and is supposedly released into the extracellular fluid through an endoplasmic reticulum–Golgi pathway-independent mechanism from immune cells such as monocytes or macrophages after stimulation with lipopolysaccharide (LPS), proinflammatory cytokines, or nitric oxide (1, 6). The release of HMGB1 is also regulated by the inflammasome, a multiprotein oligomer that activates caspase-1 to promote the maturation of inflammatory cytokines, interleukin-1β (IL-1β) and IL-18, and by dying cells, typically those undergoing necrosis (7–10). Secreted or released, HMGB1 is known to participate in the activation of cell surface innate immune receptors, typically Toll-like receptors (TLRs), thereby affecting many aspects of the host’s inflammatory responses upon infection or noxious stresses (1–5). Perhaps most notably is the crucial role of HMGB1 in LPS-induced endotoxemia, whereby administration of an anti-HMGB1 antibody significantly protects mice from lethality (1, 11). The study of released HMGB1 is complicated by a number of complex posttranslational modifications made to the protein, including acetylation and redox modifications that may regulate HMGB1 function (12–14).
HMGB1 can regulate immune reactions in several ways. Cytosolic HMGB1, together with the other members of the family, function as universal sentinels or chaperones for immunogenic nucleic acids by facilitating the recognition of nucleic acids by more discriminative, nucleic acid-sensing innate receptors (15–17). In addition, HMGB1 regulates autophagy, a cellular response that functions in clearing long-lived proteins and dysfunctional organelles to generate substrates for adenosine triphosphate (ATP) production during periods of starvation and other types of cellular stress events (13, 18–20). This mechanism contributes to antimicrobial responses against invading microorganisms (21, 22). Indeed, microorganisms can induce autophagy by stimulating innate immune receptors, such as TLRs, by a process in which bacteria are captured by phagocytosis but remain within intact vacuoles, an autophagic process termed microtubule-associated protein light chain 3 (LC3)-associated phagocytosis (LAP), which promotes the maturation of autophagosomes into autolysosomes (23, 24).
Collectively, these studies place HMGB1 in the center of immunological events where it uniquely functions intracellularly and extracellularly as a mediator of immune and inflammatory responses. The biological and clinical importance of HMGB1 is underscored by the dysregulation of this protein in a number of pathological conditions, including sepsis, ischemia–reperfusion injury, arthritis, and cancer (1, 3–5). Nonetheless, in vivo validation of the versatile functions described above is lacking due to the lethality of the Hmgb1-deficient mice, thought to cause lethal hypoglycemia in newborn mice (25). In the present study, we describe the generation of Hmgb1-floxed (Hmgb1f/f) mice that enabled the cell- and tissue-specific deletion of the gene when crossed with an appropriate Cre recombinase transgenic strain. We demonstrate in this study a protective role of intracellular HMGB1 in macrophages where it serves as a crucial regulator of autophagosome formation in the context of LPS stimulation or bacterial infection in vivo. Finally, we will discuss the future prospects of HMGB1 research using these newly generated mutant mice.
Results
Generation of Hmgb1f/f Mice.
To study the function of HMGB1 in distinct cells and tissues, we generated mice with a conditional knockout of the Hmgb1 gene by using the Cre-loxP system. As depicted in Fig. S1A, the targeting vector was constructed to cause the deletion of exons 2–4 upon expression of Cre protein. A neomycin resistance (neo) gene flanked by two loxP sites was introduced into intron 1 of the gene, whereas a third loxP site was generated downstream of exon 4. Mouse embryonic stem cells (ES) were electroporated with this vector, selected in the presence of G418 and then homologous recombinant clones identified by PCR and confirmed by Southern blot analysis (Fig. S1B). Cre protein was then transiently expressed in the targeted ES clones to delete the loxP-flanked neo gene (Fig. S1C). The resulting ES clones, carrying the loxP-flanked (floxed) Hmgb1 gene, were used to generate chimeric mice that successfully transmitted the gene in the germ line. Mice homozygous for the floxed Hmgb1 gene (Hmgb1f/f) were born at the expected Mendelian ratios and presented with no obvious abnormalities. When these mice were crossed with mice transgenic for a CAG promoter-driven cre gene in which cre recombinase expression is constitutively and broadly driven by the cytomegalovirus early enhancer element and chicken β-actin promoter (26), mice died soon after birth, which is consistent with a previous report (25) (Fig. S2).
Ablation of the Hmgb1 Gene in Myeloid Cells.
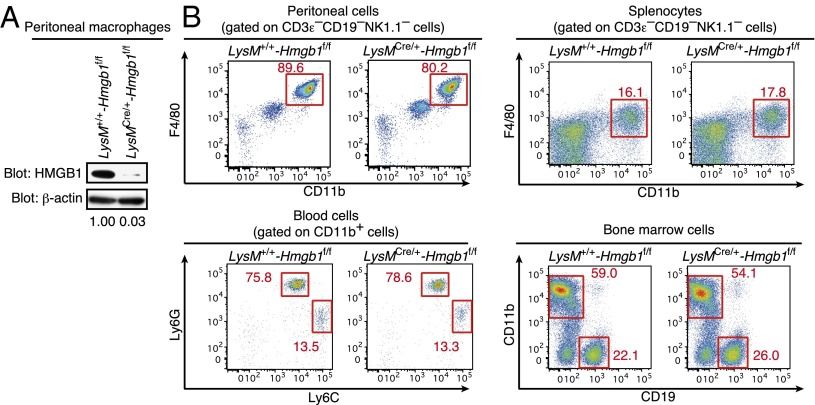
In cells of myeloid lineage, in particular macrophages, HMGB1 is released following stimulation by TLR ligands or other noxious agents (1, 6), supporting the concept that HMGB1 plays a role in inflammatory responses. To examine the function of HMGB1 in these cells, we crossed Hmgb1f/f mice and mice with the cre gene inserted into the endogenous M lysozyme (LysM) locus. Mice carrying Hmgb1f/f and the LysM cre (LysMCre/+-Hmgb1f/f) were born and developed normally. As shown in Fig. 1A, HMGB1 expression was barely detectable in peritoneal macrophages of these mice, whereas HMGB1 was expressed normally in other cells such as T and B lymphocytes (Fig. S3A). We further examined myeloid cell populations in the peritoneal cavity, spleen, blood, and bone marrow of these mice. As shown in Fig. 1B, no overt difference was found in the cellularity of the myeloid lineage, indicating that the loss of HMGB1 does not affect the development of these cells. Further, we observed no difference in the in vitro differentiation of M1 or M2 macrophages in LysMCre/+-Hmgb1f/f mice (Fig. S3B).
Fig. 1.
HMGB1 expression and myeloid cell population of LysMCre/+-Hmgb1f/f mice. (A) Peritoneal macrophages were obtained from LysM+/+-Hmgb1f/f and LysMCre/+-Hmgb1f/f mice. Whole cell extracts (20 μg) were prepared and subjected to immunoblot analysis to detect HMGB1 protein. HMGB1 band density is presented relative to β-actin band density. (B) Single cell suspensions were prepared from the peritoneal cavity, spleen, peripheral blood, and bone marrow from LysM+/+-Hmgb1f/f or LysMCre/+-Hmgb1f/f mice and stained with the indicated combination of the following fluorochrome-conjugated antibodies: anti-F4/80 PerCP-Cy5, anti-CD11b APC, anti-Ly6G PE-Cy7, anti-Ly6C FITC, anti-CD19 PB, anti-NK1.1 PB, and anti-CD3ε PB antibodies. CD3ε−CD19−NK1.1− cells (Upper) or CD11b+ cells (Lower Left) were gated and shown. The numbers represent the percentage of cells contained in each region.
Susceptibility of LysMCre/+-Hmgb1f/f Mice to LPS-Induced Endotoxemia.
There has been a particular focus on HMGB1 in the context of LPS-induced endotoxemia, wherein HMGB1 released by myeloid cells is reported to be capable of activating various innate receptors, especially TLR4, and thereby exacerbating pathogenic inflammatory responses (11, 27). We therefore first examined whether and how LysMCre/+-Hmgb1f/f mice respond to LPS. We observed that LysMCre/+-Hmgb1f/f mice were more vulnerable to i.p. injection of LPS and were accompanied by a massive amount of tissue destruction in the lungs compared with LysM+/+-Hmgb1f/f mice (Fig. 2A). This observation indicates there is a protective role of HMGB1, which seemingly contrasts with the prevailing notion that it exacerbates endotoxemia (1, 2, 11).
Fig. 2.
Role of HMGB1 in myeloid cells in LPS-induced shock. (A) LysM+/+-Hmgb1f/f (n = 8) or LysMCre/+-Hmgb1f/f (n = 6) mice were intraperitoneally injected with LPS (17.5 mg/kg). Mice survival was monitored (Left). Histological analysis of lungs from LysM+/+-Hmgb1f/f or LysMCre/+-Hmgb1f/f mice injected with LPS, assessed by microscopy of sections stained with hematoxylin and eosin (Right). Original magnification, 40×. (Scale bars, 50 μm.) (B–D) Cytokine production in sera from LysM+/+-Hmgb1f/f or LysMCre/+-Hmgb1f/f mice injected with LPS as in A. TNF-α, IL-6, and IL-12p40 (B) and IL-1β and IL-18 (C) protein levels were measured by ELISA. (D) HMGB1 levels in plasma 8 h after LPS injection were also measured. Data are shown as means ± SD; *P < 0.05. (E) Peritoneal macrophages from LysM+/+-Hmgb1f/f or LysMCre/+-Hmgb1f/f mice were preincubated with ultrapure LPS (50 ng/mL) for 4 h followed by ATP stimulation (750 µM) for 1 h. IL-1β (Left) and IL-18 (Right) protein levels were measured by ELISA. We also observed that Il1b and Il18 mRNAs were normally induced by LPS stimulation in peritoneal macrophages from LysMCre/+-Hmgb1f/f mice. Data are shown as means ± SD of triplicate determinants. **P < 0.01.
We then examined serum levels of proinflammatory cytokines thought to participate in the pathogenic response to LPS and found that levels of TNF-α, IL-6, and IL-12p40 in LysMCre/+-Hmgb1f/f mice were similar to those found in LysM+/+-Hmgb1f/f mice (Fig. 2B). Interestingly, however, there was a notable increase of IL-1β and IL-18 in the serum of the LysMCre/+-Hmgb1f/f mice, indicating a hyperactivation of an inflammasome pathway(s) (Fig. 2C). In this experimental setting, serum HMGB1 levels were only marginally lower compared with those of LysM+/+-Hmgb1f/f mice (Fig. 2D), demonstrating that myeloid cells are not the main source of LPS-induced serum HMGB1 and that the increased vulnerability to LPS reflects a function of intracellular HMGB1 vis-à-vis extracellular HMGB1.
The elevated IL-1β and IL-18 expression in the serum of the LPS-stimulated LysMCre/+-Hmgb1f/f mice prompted us to examine the activation of the inflammasome in the mutant macrophages. We stimulated macrophages from LysMCre/+-Hmgb1f/f mice with LPS and ATP and then examined IL-1β and IL-18 production, a hallmark of inflammasome activation. As shown in Fig. 2E, a marked increase of both IL-1β and IL-18 was observed in the LysMCre/+-Hmgb1f/f macrophages. In view of a previous report showing that caspase-1–deficient mice are highly resistant to LPS-induced endotoxemia (28), our observation indicates that intracellular HMGB1 in macrophages functions as a negative regulator of the inflammasome activation pathway, which may account for our observed LPS sensitivity in the LysMCre/+-Hmgb1f/f mice.
Critical Contribution of HMGB1 at the Interface Between Autophagy and Cell Death.
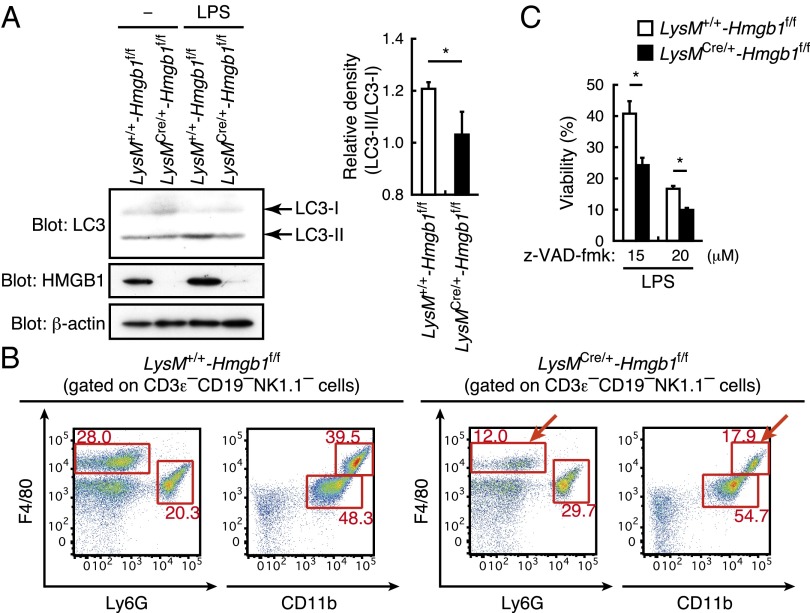
It has been shown that the inflammasome pathway is regulated by autophagy which, as described above, has multiple immunological functions and influences immune responses to infection (13, 18, 19, 21–23). Because LPS can activate autophagy through the TLR4/Toll/IL-1 receptor domain–containing adapter-inducing interferon-β (TRIF)/p38 MAPK signaling pathway and autophagic proteins regulate caspase-1–mediated innate immune responses (29–31), we wished to examine whether enhanced inflammasome activation in the absence of HMGB1 (Fig. 2E) is a consequence of an unregulated autophagic process. We examined the status of microtubule-associated protein light chain 3 (LC3), a well-known marker of autophagosomes (32). As shown in Fig. 3A, LPS stimulation of LysMCre/+-Hmgb1f/f macrophages resulted in a significant decrease in the formation of two cleaved forms of LC3, LC3-I and LC3-II, which typically represent the induction of autophagosomes (32, 33), compared with LysM+/+-Hmgb1f/f macrophages. These data indicate that HMGB1 is required for the autophagic process.
Fig. 3.
Role of HMGB1 in myeloid cells to autophagy and cell death. (A) Bone marrow macrophages were stimulated with LPS (1 µg/mL) for 1 h. Whole cell lysates were prepared and subjected to immunoblot analysis. LC3-I/II, HMGB1, and β-actin were detected (Left). LC3-II/LC3-I relative band density (n = 3) is also shown (Right). *P < 0.05. (B) Single cell suspensions were prepared from the peritoneal cavity of LysM+/+-Hmgb1f/f or LysMCre/+-Hmgb1f/f mice 12 h after i.p. LPS injection (17.5 mg/kg) and stained with the indicated combination of the following fluorochrome-conjugated antibodies: anti-F4/80 PerCP-Cy5, anti-CD11b APC, anti-Ly6G PE-Cy7, anti-CD19 PB, anti-NK1.1 PB, and anti-CD3ε PB antibodies. CD3ε−CD19−NK1.1− cells were gated and shown. The numbers represent the percentage of cells contained in each region. Arrows represent macrophage population. (C) Bone marrow macrophages were stimulated with LPS (500 ng/mL) for 24 h with z-VAD-fmk (15 or 20 µM), and cell viability was measured by the crystal violet staining method. *P < 0.05.
We also examined the in vivo regulation of macrophage fate following LPS administration in these mice. As shown in Fig. 3B, we observed a marked decrease of macrophage populations in LysMCre/+-Hmgb1f/f mice compared with LysM+/+-Hmgb1f/f mice. Because suppression of autophagy results in cell death, this observation is consistent with the above in vitro results. Although TLR4 stimulation alone cannot induce cell death, it can if the pan-caspase inhibitor z-VAD-fmk is present (34, 35). Under this in vitro experimental setting, a marked enhancement of macrophage cell death was observed in the absence of HMGB1 (Fig. 3C). Thus, these observations in toto indicate that HMGB1 critically regulates autophagy and cell death in macrophages following TLR4 activation.
Because autophagy functions in a wide variety of cell types, we asked whether this observation can also be made in dendritic cells or hepatocytes, by respectively expressing the cre recombinase gene under the promoter of CD11c or albumin (Cd11cCre/+-Hmgb1f/f or AlbCre/+-Hmgb1f/f mice). In both cases, we observed a phenotype similar to that seen in LysMCre/+-Hmgb1f/f mice, namely, an elevation of serum IL-1β and IL-18 upon LPS stimulation, but no change in serum TNF-α, IL-6, or IL-12p40 levels (Fig. S4 A and B). Thus, HMGB1-mediated autophagy is likely to be a cell-type–independent mechanism.
Requirement of Macrophage HMGB1 in Antibacterial Responses.
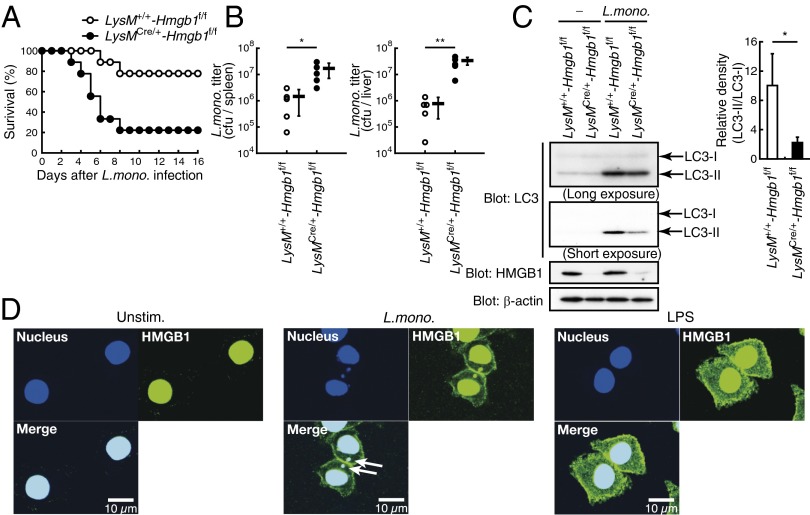
Because autophagy also participates in the host’s response against infection by various pathogens, particularly the clearance of intracellular bacteria, we next examined LysMCre/+-Hmgb1f/f mice in response to a pathogenic bacterium Listeria monocytogenes. As shown in Fig. 4A, these mice showed a marked vulnerability to the bacterial infection compared with LysM+/+-Hmgb1f/f mice. Expectedly, there was a marked elevation of bacterial burden seen in the spleen and liver of LysMCre/+-Hmgb1f/f mice (Fig. 4B), although mRNA levels of inflammatory cytokines remained essentially unchanged (Fig. S5).
Fig. 4.
Hmgb1-deficient mice in myeloid cells are vulnerable to L. monocytogenes infection. (A) LysM+/+-Hmgb1f/f (n = 9) and LysMCre/+-Hmgb1f/f (n = 9) mice were infected with L. monocytogenes (5 × 105 cfu) intraperitoneally. Mice survival was monitored every 24 h. (B) L. monocytogenes titers in the spleen (Left) or liver (Right) from LysM+/+-Hmgb1f/f (n = 5) and LysMCre/+-Hmgb1f/f (n = 5) mice infected for 3 d as in A were measured. Each symbol represents an individual mouse; long horizontal lines indicate the mean. **P < 0.01 and *P < 0.05. (C) Peritoneal macrophages were infected with L. monocytogenes [multiplicity of infection (MOI) of 5] for 1 h. Whole cell lysates were prepared and subjected to immunoblot analysis. LC3-I/II, HMGB1, and β-actin were detected (Left). LC3-II/LC3-I relative band density (n = 3) is also shown (Right). *P < 0.05. (D) Translocation of HMGB1 from nucleus to cytosol upon L. monocytogenes infection and LPS stimulation. RAW264.7 cells were untreated (Left), infected with L. monocytogenes (MOI of 1; Center), or stimulated with LPS (1 μg/mL; Right) for 1 h. Cells were then fixed and stained with anti-HMGB1 antibody and DAPI. Confocal images of HMGB1 (green) and nuclei (blue) are shown. (Scale bars, 10 μm.) Arrows show colocalization of HMGB1 and L. monocytogenes.
Interestingly, the generation of LC3-II was significantly suppressed in peritoneal macrophages of infected LysMCre/+-Hmgb1f/f mice compared with those from LysM+/+-Hmgb1f/f mice, indicating a critical role for HMGB1 in bacteria-induced autophagy in these cells (Fig. 4C). We also observed that upon bacterial infection of macrophages in vitro, there was a noticeable accumulation of HMGB1 in the cytosol, which also occurs upon LPS stimulation (36, 37) (Fig. 4D). Of note, cytosolic HMGB1 merged with bacterial DNA, suggesting an association of this protein with autophagosomes for autophagy-mediated bacterial eradication (Fig. 4D). Collectively, these results support the notion that HMGB1, released from the nucleus to the cytosol upon infection, participates in bacteria-induced autophagy, given that the absence of this protein severely affects bacterial clearance.
Discussion
In this study, we generated conditional knockout mice of HMGB1 to gain insight into its biological roles, specifically as it relates to immune responses, of this protein in vivo. These conditional knockout mice permitted the ablation of HMGB1 in a tissue- and cell-type–specific manner. Before our study, HMGB1 was identified as a multifunctional mediator of immune and inflammatory responses by virture of its role as danger-associated molecular patterns (DAMPs) or alarmins upon its release from the cell, which occurs typically by cell death or a still poorly characterized mechanism (1–5). Intracellular HMGB1 is also an immune regulator, particularly in the cytoplasm where it functions as sensor and/or chaperone for immunogenic nucleic acids and mediates autophagy (13, 15, 17, 20). This study illustrates the in vivo validation of HMGB1 in myeloid cells, showing its protective role in the cytoplasm against LPS-induced endotoxemia and infection by L. monocytogenes.
Interestingly, previous studies have shown that HMGB1 functions as a crucial instigator of LPS-induced endotoxemia (1, 2, 11). Our present study by no means excludes this pathological aspect of HMGB1. However, we demonstrate that cells of myeloid lineage are not the main source of proinflammatory HMGB1, as serum HMGB1 levels, induced by LPS, were only marginally affected in LysMCre/+-Hmgb1f/f mice (Fig. 2D). Thus, it will be of great interest to clarify which cells are responsible for the proinflammatory HMGB1 as well as how it instigates inflammatory responses. In a similar context, the inflammatory function of hepatocyte-derived HMGB1 has been reported in ischemia–reperfusion injury. We therefore examined AlbCre/+-Hmgb1f/f mice but basically did not observe any overt differences compared with HMGB1-intact mice, suggesting that the hepatocyte-derived HMGB1 plays little, if any, role in this inflammatory pathogenesis (Fig. S6). Thus, the role and source of HMGB1 in this ischemia–reperfusion injury model also needs further clarification.
The antimicrobial functions of autophagy are known to serve as a series of barriers against invading microorganisms, wherein the antimicrobial function of LAP, which involves the engagement of the autophagic machinery while the bacterium is confined in the nascent and presumably intact phagosome, is well characterized. Our results indicate a critical role of HMGB1 in this process as revealed by an increased vulnerability to L. monocytogenes infection and the suppression of LC3-II generation in macrophages, the hallmark of LAP (Fig. 4C). Thus, this study reveals a unique, antibacterial function of HMGB1.
Our study also corroborates other reports that indicate the involvement of HMGB1 in the autophagic process (13, 20). It has been reported that stimuli that enhance reactive oxygen species promote cytosolic translocation of HMGB1 from the nucleus where it directly interacts with the autophagy-related (ATG) protein Beclin-1, a Bcl-2–homology (BH)-3 domain-only protein that is essential for autophagy induction (13, 20). Whether and/or how the suppression of autophagy in the absence of HMGB1 upon LPS stimulation or bacterial infection, revealed in this study, integrates to Beclin-1 and/or other molecules involved in autophagy remains to be determined further.
In summary, this study offers in vivo evidence for the protective aspect of cytosolic HMGB1 against endotoxemia and bacterial infection. The availability of the mice for conditional ablation of HMGB1 will provide new avenues of research for the investigation of this protein, which apparently shuttles from nucleus to cytoplasm to the extracellular environment. In particular, in view of the fact that the main source of serum HMGB1 upon LPS stimulation is not cells of myeloid cell lineage, further investigation by generation of additional Cre-loxP mutant mice will be an aide in the examination of the function of extracellular HMGB1 during endotoxemia.
It has been conventional wisdom that effector molecules are newly synthesized and released during immune or inflammatory responses typically starting at the level of transcriptional activation of the corresponding genes (23, 38, 39). The present findings together with previous reports may point to the existence of an additional mechanism, that is, the nucleus, wherein HMGB1 is abundantly accumulated without cellular stimulation, functions as “arsenal” for this and other effector molecules to be released into the cytosol and extracellular milieu so as to ensure prompt and effective cellular responses. Indeed, the newly generated Hmgb1f/f mice will allow further investigation to fully identify HMGB1’s multiple functions during homeostasis and pathogenesis.
Materials and Methods
Mice.
C57BL/6 mice were purchased from CLEA Japan, Inc. LysM-Cre knockin mice, Alb-Cre transgenic mice, CAG-Cre transgenic mice, and Cd11c-Cre transgenic mice were obtained from The Jackson Laboratory. All animal care and experiments conformed to the guidelines for animal experiments of the University of Tokyo, and were approved by the animal research committee of the University of Tokyo.
Construction of the Targeting Vector.
A targeting vector was constructed to delete exons 2–4, which includes the translation initiation site of the Hmgb1 gene. Hmgb1 genomic DNA was amplified by PCR by using specific primers as follows: sense, 5′-actagtgcttgtctgtttcacagttttcgttac-3′ and antisense, 5′-gtcgacagttatcaagtataatcccctaacactgg-3′. The fragment was inserted between two loxP sites of the vector pKSTKNEOloxP, which has herpes simplex virus thymidine kinase and loxP-flanked pGK-neo. Then the 10.7-kb genomic fragment 5′ upstream of exon 2–4 and 3′ downstream 5-kb genomic fragment was inserted.
Reagents and Cells.
LPS O55:B4 was purchased from Sigma. Ultrapure LPS and ATP were purchased from InvivoGen. z-VAD-fmk was purchased from Sigma. The mouse macrophage cell line RAW264.7 cells were cultured and maintained as described (13, 15, 17, 20). For preparing peritoneal macrophages, mice were injected with 2 mL of 4% (wt/vol) thioglycollate solution intraperitoneally. The peritoneal cavity was washed by PBS, containing 2% (vol/vol) FCS and 1 mM EDTA to collect peritoneal exudate cells 4 d after the injection, and cells were plated on Petri dishes for 2 h in RPMI, supplemented with 10% (vol/vol) FCS. After incubation, cells were recollected and seeded for each assay. Bone marrow cells were extracted from each mouse as described previously (17, 40), then seeded in 15-cm cell culture dishes and cultured in RPMI supplemented with 10% (vol/vol) FCS and M-CSF (100 ng/mL; Peprotech) for 6 d. M-CSF cultured bone-marrow–derived macrophages (BMMs) were replated for each assay. CD3ε+ T cells and CD19+ B cells were prepared by using a CD3ε MicroBead kit (Miltenyi Biotec) and a CD19 MicroBead kit (Miltenyi Biotec), respectively, according to the manufacturer’s protocol.
LPS-Induced Lethal Shock.
Mice were injected intraperitoneally with LPS (17.5 mg/kg weight) and monitored every 12 h. Whole blood samples were taken sequentially from tails, and sera were prepared by centrifugation for 5 min at 4 °C at 8,000 × g after incubation at room temperature for 30 min. For plasma sample preparation, blood samples were treated with 5 mM EDTA.
Statistical Analysis.
Differences between control and experimental groups were evaluated with a Student t test.
Additional information can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank K. J. Tracey, N. Mizushima, T. Nishimura, and A. Kuma for their invaluable advice and M. Taniguchi, H. Tanabe, R. Fujii, K. Adachi, and H. Ueki for their technical assistance. This work was supported in part by the Core Research for Evolutional Science and Technology of the Japan Science and Technology Agency; a Grant-In-Aid for Scientific Research in Innovative Areas from the Ministry of Education, Culture, Sports, and Science; and the Uehara Memorial Foundation. A.M. is a research fellow of the Japan Society for the Promotion of Science. The Department of Molecular Immunology is supported by BONAC Corporation and Kyowa Hakko Kirin Co., Ltd.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320808110/-/DCSupplemental.
References
- 1.Andersson U, Tracey KJ. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol. 2011;29:139–162. doi: 10.1146/annurev-immunol-030409-101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianchi ME. HMGB1 loves company. J Leukoc Biol. 2009;86(3):573–576. doi: 10.1189/jlb.1008585. [DOI] [PubMed] [Google Scholar]
- 3.Harris HE, Andersson U, Pisetsky DS. HMGB1: A multifunctional alarmin driving autoimmune and inflammatory disease. Nat Rev Rheumatol. 2012;8(4):195–202. doi: 10.1038/nrrheum.2011.222. [DOI] [PubMed] [Google Scholar]
- 4.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): Nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5(4):331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 5.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 6.Gardella S, et al. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep. 2002;3(10):995–1001. doi: 10.1093/embo-reports/kvf198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franchi L, Núñez G. Immunology. Orchestrating inflammasomes. Science. 2012;337(6100):1299–1300. doi: 10.1126/science.1229010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu B, Wang H, Andersson U, Tracey KJ. Regulation of HMGB1 release by inflammasomes. Protein Cell. 2013;4(3):163–167. doi: 10.1007/s13238-012-2118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nyström S, et al. TLR activation regulates damage-associated molecular pattern isoforms released during pyroptosis. EMBO J. 2013;32(1):86–99. doi: 10.1038/emboj.2012.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481(7381):278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285(5425):248–251. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- 12.Sterner R, Vidali G, Allfrey VG. Studies of acetylation and deacetylation in high mobility group proteins. Identification of the sites of acetylation in HMG-1. J Biol Chem. 1979;254(22):11577–11583. [PubMed] [Google Scholar]
- 13.Tang D, Billiar TR, Lotze MT. A Janus tale of two active high mobility group box 1 (HMGB1) redox states. Mol Med. 2012;18:1360–1362. doi: 10.2119/molmed.2012.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venereau E, et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J Exp Med. 2012;209(9):1519–1528. doi: 10.1084/jem.20120189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanov S, et al. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood. 2007;110(6):1970–1981. doi: 10.1182/blood-2006-09-044776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian J, et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8(5):487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 17.Yanai H, et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462(7269):99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- 18.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8(9):741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 20.Tang D, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190(5):881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240(1):92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogawa M, Mimuro H, Yoshikawa Y, Ashida H, Sasakawa C. Manipulation of autophagy by bacteria for their own benefit. Microbiol Immunol. 2011;55(7):459–471. doi: 10.1111/j.1348-0421.2011.00343.x. [DOI] [PubMed] [Google Scholar]
- 23.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13(10):722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez J, et al. Microtubule-associated protein 1 light chain 3 α (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci USA. 2011;108(42):17396–17401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calogero S, et al. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22(3):276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 26.Sakai K, Miyazaki Ji. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun. 1997;237(2):318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- 27.El Mezayen R, et al. Endogenous signals released from necrotic cells augment inflammatory responses to bacterial endotoxin. Immunol Lett. 2007;111(1):36–44. doi: 10.1016/j.imlet.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuida K, et al. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;267(5206):2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 29.Nakahira K, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456(7219):264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y, et al. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27(1):135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3(6):542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 33.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SO, Han J. Pan-caspase inhibitor zVAD enhances cell death in RAW246.7 macrophages. J Endotoxin Res. 2001;7(4):292–296. doi: 10.1179/096805101101532873. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y, Kim SO, Li Y, Han J. Autophagy contributes to caspase-independent macrophage cell death. J Biol Chem. 2006;281(28):19179–19187. doi: 10.1074/jbc.M513377200. [DOI] [PubMed] [Google Scholar]
- 36.Li W, et al. A hepatic protein, fetuin-A, occupies a protective role in lethal systemic inflammation. PLoS ONE. 2011;6(2):e16945. doi: 10.1371/journal.pone.0016945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu CX, Sun H, Liu Q, Guo H, Gong JP. LPS induces HMGB1 relocation and release by activating the NF-κB-CBP signal transduction pathway in the murine macrophage-like cell line RAW264.7. J Surg Res. 2012;175(1):88–100. doi: 10.1016/j.jss.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 38.Denes A, Lopez-Castejon G, Brough D. Caspase-1: Is IL-1 just the tip of the ICEberg? Cell Death Dis. 2012;3:e338. doi: 10.1038/cddis.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinon F, Burns K, Tschopp J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol Cell. 2002;10(2):417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 40.Negishi H, et al. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc Natl Acad Sci USA. 2005;102(44):15989–15994. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.