Significance
Defects in the enzyme β-galactocerebrosidase (GALC) result in the devastating neurodegenerative disorder Krabbe disease. GALC is responsible for the degradation and recycling of glycosphingolipids that form the primary lipid component of the myelin sheath that insulates nerve cells. A detailed understanding of how GALC processes substrate will facilitate the development of new drug therapies for Krabbe disease. This study reveals a series of structural snapshots of GALC captured during different steps of the catalytic cycle. These structures identify specific residues within the active site that undergo significant movements during substrate cleavage, providing key insight into the catalytic mechanism of GALC.
Keywords: β-galactosylceramidase, lysosomal storage disease, glycosyl hydrolase, pharmacological chaperone therapy
Abstract
Glycosphingolipids are ubiquitous components of mammalian cell membranes, and defects in their catabolism by lysosomal enzymes cause a diverse array of diseases. Deficiencies in the enzyme β-galactocerebrosidase (GALC) cause Krabbe disease, a devastating genetic disorder characterized by widespread demyelination and rapid, fatal neurodegeneration. Here, we present a series of high-resolution crystal structures that illustrate key steps in the catalytic cycle of GALC. We have captured a snapshot of the short-lived enzyme–substrate complex illustrating how wild-type GALC binds a bona fide substrate. We have extensively characterized the enzyme kinetics of GALC with this substrate and shown that the enzyme is active in crystallo by determining the structure of the enzyme–product complex following extended soaking of the crystals with this same substrate. We have also determined the structure of a covalent intermediate that, together with the enzyme–substrate and enzyme–product complexes, reveals conformational changes accompanying the catalytic steps and provides key mechanistic insights, laying the foundation for future design of pharmacological chaperones.
The recycling and degradation of eukaryotic membrane components occurs in the lysosome and is essential for cellular maintenance. The molecular mechanisms of lysosomal lipid degradation are primarily informed by the study of a class of human diseases, sphingolipidoses, which are caused by inherited defects in glycosphingolipid catabolism. Krabbe disease is a devastating neurodegenerative disorder that is caused by deficiencies in the lysosomal enzyme β-galactocerebrosidase (GALC) (enzyme commission 3.2.1.46). It is essential for the catabolism of galactosphingolipids, including the principal lipid component of myelin, β-d-galactocerebroside (Fig. 1A) (1). GALC function has also been implicated in cancer cell metabolism, primary open-angle glaucoma and the maintenance of a hematopoietic stem cell niche (2–4).
Fig. 1.
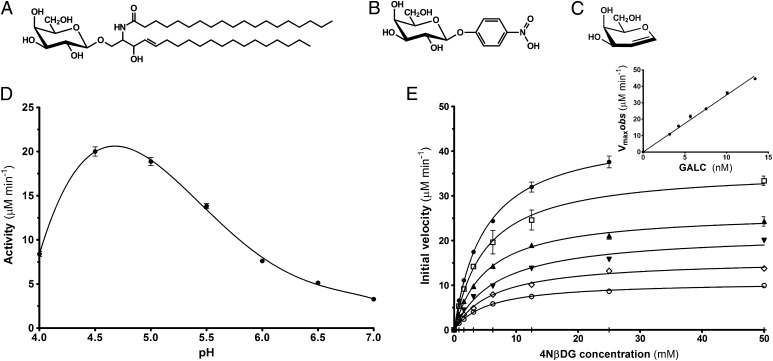
Enzyme kinetic analysis of GALC. (A–C) Schematic diagram of the natural substrate β-d-galactocerebroside (A), the chromogenic substrate 4NβDG (B), and the inhibitor d-galactal (C). (D) pH profile of enzyme activity for GALC. SEM error bars are shown. (E) Michaelis–Menten plots of initial velocity vs. substrate concentration at GALC concentrations of 3.18 nM (○), 4.24 nM, (◇), 5.66 nM, (▼), 7.54 nM, (▲), 10.1 nM, (□), and 13.4 nM (●). (Inset) Plot of Vmaxobs vs. GALC concentration showing kcat (57.8 s−1) as the gradient. SEM error bars are shown.
GALC catalyzes the hydrolysis of β-d-galactocerebroside to β-d-galactose and ceramide, as well as the breakdown of psychosine to β-d-galactose and sphingosine. In both cases, removal of the galactosyl moiety is thought to occur via a retaining two-step glycosidic bond hydrolysis reaction (5, 6). Our recent structure of murine GALC identified two active site glutamate residues geometrically consistent with this mechanism (7). In the first step, the carboxylate group of E258 is hypothesized to perform a nucleophilic attack at ring position C1, forming an enzyme–substrate intermediate, releasing the first product (ceramide or sphingosine) as the leaving group. In the second step, E182 is thought to act as a general acid/base to deprotonate a water molecule, which then attacks the ring, releasing the enzyme and the second product (galactose).
Defects in GALC lead to the accumulation of cytotoxic metabolites that elicit complex, and still only partially understood, cellular events resulting in apoptosis of myelin-forming oligodendrocytes and Schwann cells of the central and peripheral nervous system (8–10). This leads to the characteristic demyelination and neurodegeneration observed in Krabbe disease patients (11, 12). A wide range of disease-causing missense mutations have been identified throughout the GALC gene (13–21). Some of these mutations will result in catalytically inactive protein, whereas other mutations may destabilize the protein, compromising lysosomal delivery by causing retention in the endoplasmic reticulum (ER). For such mutants with residual catalytic activity, pharmacological chaperone therapy (PCT) may be an appropriate treatment strategy (22–24). This approach involves the use of small molecules that bind to and stabilize the defective enzyme in the ER and facilitate its delivery to the site of action, such as the lysosome. PCT is currently under investigation for the related lysosomal storage disorders Gaucher disease (25–27) and Fabry disease (28, 29), with a number of candidate ligands now in clinical trials (23).
Here, we present a comprehensive enzymatic characterization of GALC accompanied by crystal structures of wild-type GALC in complex with substrate, product, and covalent intermediate, illustrating key stable species along the catalytic pathway. These structures not only reveal the short-lived enzyme–substrate complex captured using hydrolysable substrate and wild-type enzyme but also highlight unexpected conformational changes of active site residues. We confirm the hypothesized mechanism, including the involvement of E258 and E182, but also identify key roles for additional active site residues.
Results
Enzyme Kinetics.
Accurate measurement of the GALC kinetic parameters has been hampered by the low expression levels of the enzyme in tissues and the difficulty in purifying the enzyme because of its hydrophobicity (30, 31). We have previously determined the structure of mouse GALC by purifying the enzyme from stably expressing HEK 293T cells: the murine enzyme shares 83% sequence identity with the human enzyme, and all active site residues are conserved (7). To characterize the enzyme kinetics of GALC, we used the chromogenic substrate 4-nitrophenyl-β-d-galactopyranoside (4NβDG) (Fig. 1B) because of its high solubility at a range of pH values. We confirm that the optimum pH of GALC activity is between pH 4.5 and 5.0 consistent with its lysosomal localization (Fig. 1D). Michaelis–Menten plots were used to determine Km and Vmax parameters at several enzyme concentrations (Fig. 1E and Fig. S1). At pH 4.6, the Km for 4NβDG is 5.1 ± 0.3 mM and the Vmax is 46.6 ± 1.6 nmol⋅min−1⋅µg−1, giving a kcat of 57.8 s−1. These values are comparable to those of another lysosomal hydrolase, human α-galactosidase, determined using a similar substrate (Km, 8.3 mM; kcat, 63.5 s−1) (32) and the hydrolysis of lactose and allolactose by Escherichia coli β-galactosidase (Km, ∼1 mM; kcat, ∼60 s−1) (33, 34).
Enzyme–Substrate Complex.
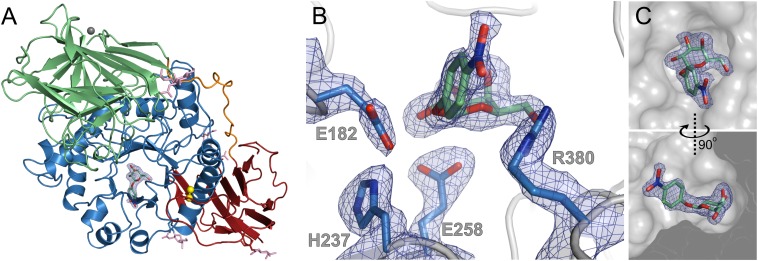
Typically enzyme–substrate complexes can only be formed by using catalytically inactive mutant forms of an enzyme or nonhydrolysable substrate analogs. The combination of small crystal size (5.4 × 10−5 mm3), high solvent content (61.9%), and reduced catalytic activity of GALC at pH 6.8, the pH at which crystals were grown, made GALC crystals excellent candidates for diffusion-trapping experiments (Fig. S2) (35, 36). Rapid soaking (20–40 s) and cryocooling of crystals allowed us to capture the substrate 4NβDG in the active site of the wild-type enzyme (Fig. 2A). Electron-density maps revealed unambiguously the presence of the unhydrolysed substrate in the active site, and the enzyme–substrate complex structure was refined at 2.1-Å resolution (Fig. 2B, Fig. S3A, and Table 1).
Fig. 2.
Structure of the wild-type GALC enzyme in complex with substrate. (A) Ribbon diagram showing the overall structure of GALC with the substrate 4NβDG bound in the active site. The electron density (2FO-FC contoured at 0.25 e−/Å3, blue) is shown for uncleaved substrate bound in the active site pocket of GALC. Surface glycans (pink sticks) are shown. (B) Detail of the GALC active site with bound, uncleaved substrate showing active site residues (sticks) and electron density (as above). (C) Surface representation of the GALC active site (gray) with the substrate and electron density shown (as above).
Table 1.
Data collection and refinement statistics
| Statistic | GALC-4NβDG | GALC-d-galactal | GALC-galactose |
| Data collection | |||
| Space group | R 3 2 | R 3 2 | R 3 2 |
| Cell dimensions | |||
| a, b, c (Å) | 250.0, 250.0, 77.8 | 249.5, 249.5, 77.7 | 248.8, 248.8, 77.6 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 56.38–2.09 (2.15–2.09)* | 47.46–1.97 (2.02–1.97)* | 47.01–2.06 (2.12–2.06)* |
| Rmerge | 0.153 (1.287) | 0.138 (1.040) | 0.143 (1.402) |
| <I/σI> | 13.5 (2.5) | 10.6 (2.2) | 13.9 (2.4) |
| CC1/2 | 0.999 (0.579) | 0.998 (0.598) | 0.998 (0.618) |
| Completeness (%) | 99.9 (99.4) | 99.8 (100.0) | 99.9 (100.0) |
| Redundancy | 19.0 (14.8) | 9.7 (9.2) | 13.5 (13.8) |
| Refinement | |||
| Resolution (Å) | 56.38–2.09 (2.12–2.09) | 47.45–1.97 (1.99–1.97) | 44.25–2.06 (2.09–2.06) |
| No. reflections | 54646 (2561) | 64813 (2683) | 56378 (2673) |
| Rwork/Rfree | 0.187/0.224 | 0.170/0.198 | 0.172/0.206 |
| Ramachandran favored region (%) | 95.29 | 95.76 | 95.44 |
| Ramachandran outliers (%) | 0.47 | 0.47 | 0.47 |
| No. of atoms | |||
| Protein | 5119 | 5127 | 5119 |
| Carbohydrate | 98 | 98 | 98 |
| Ligand | 21 | 10 | 12 |
| Ca2+ ion | 1 | 1 | 1 |
| Water | 272 | 267 | 206 |
| B-factors | |||
| Protein | 55.1 | 36.1 | 45.3 |
| Carbohydrate | 88.1 | 70.2 | 85.4 |
| Ligand | 54.9 | 22.3 | 43.5 |
| Ca2+ ion | 72.2 | 36.4 | 59.7 |
| Water | 50.5 | 35.7 | 40.0 |
| r.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | 0.007 | 0.007 |
| Bond angles (°) | 1.12 | 1.07 | 1.05 |
Values in parentheses are for highest-resolution shell. One crystal was used per structure.
The galactosyl hydroxyl groups make multiple specific contacts with the enzyme (Fig. S4A) but do not require significant conformational change of active site loops, indicating that the substrate binding pocket of GALC is highly preorganized. This is in contrast to the considerable conformational changes that occur in loops around the active site of the related enzyme β-glucocerebrosidase (25, 37). The scissile bond and 4-nitrophenyl aglycone project from the surface of the enzyme and contribute very little to substrate specificity or recognition (Fig. 2C). This is consistent with the requirement for lipid-binding saposin proteins in order for GALC to cleave the natural, lipidated substrates, and it is likely that the specificity of binding is conferred by the saposin proteins rather than the hydrophobic lipid tails of the substrate molecules (38, 39).
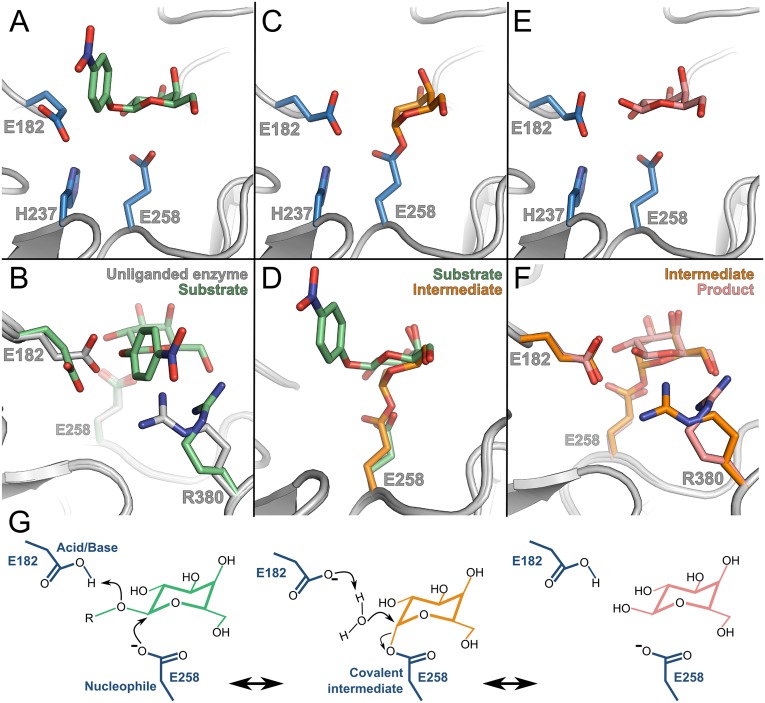
Although there is not substantial movement of active site loops upon substrate binding, to accommodate substrate in the active site pocket, the side chain conformations of R380 and the catalytic acid/base residue E182 are significantly altered (Figs. 3 A and B). The R380 side chain changes conformation to form hydrogen bonds with the galactosyl 6-hydroxyl, whereas the movement of the E182 side chain is necessary to accommodate the scissile bond of the substrate. Despite this movement, E182 maintains the hydrogen bond with H237 seen in the absence of substrate. In the enzyme–substrate complex, the Oε2 atom of the E182 side chain is ∼3.1 Å from the leaving group oxygen, ready to donate a proton during departure of 4-nitrophenol. Unlike several other retaining β-glycosidases where alignment of the anomeric carbon with the syn lone pair of the nucleophile is achieved by distorting the substrate into 1S3 geometry, in the GALC enzyme–substrate complex, the substrate binds in an undistorted low-energy 4C1 conformation similar to that seen for E. coli β-galactosidase (33, 40).
Fig. 3.
Structures of the GALC active site illustrating conformational changes along the reaction coordinate. (A) Wild-type enzyme in complex with substrate 4NβDG (green). (B) Overlay of the GALC active site residues in the absence (gray) and presence (green) of substrate. (C) Covalent intermediate structure illustrating d-galactal (orange) covalently attached to the catalytic nucleophile. (D) Movement of E258 and the pyranose ring between substrate binding (green) and covalent linkage with the inhibitor d-galactal (orange). (E) Enzyme–product complex formed following extended incubation with substrate illustrating catalytic activity of GALC in crystallo. (F) The two different conformations of the R380 side chain in the covalent intermediate (orange) and product (pink) complexes. (G) Schematic representation of the proposed retaining two-step glycosidic bond hydrolysis reaction.
Enzyme–Intermediate Complex.
To capture a structural snapshot of the covalent enzyme–intermediate complex, we incubated GALC crystals with 1,2-dideoxy-d-lyxo-hex-1-enopyranose (d-galactal) (Fig. 1C). d-galactal is a slow-binding inhibitor of E. coli β-galactosidase and can form an intermediate with a half-life sufficiently long to allow soaking and cryocooling of crystals (33, 41). We showed that d-galactal is a potent inhibitor of GALC (Ki = 32 ± 1.8 μM; Fig. S5) and were able to trap the intermediate complex for structure determination. The enzyme acts as a catalyst for the hydration of the double bond of d-galactal to form 2-deoxygalactose via a 2-deoxygalactosyl–galactocerebrosidase intermediate (Fig. 3C). The covalent linkage (∼1.3 Å) between the C1 atom of d-galactal and the Oε2 atom of E258 is unambiguous and only requires a minor conformational change of E258 (Fig. 3D and Fig. S3B). To form the covalent linkage, there is a significant movement of the C1 atom of the pyranose ring compared with the enzyme–substrate complex, about 1.3 Å deeper into the active site pocket (Fig. 3D). The pyranose ring retains an undistorted 4C1 chair conformation and is stabilized by two new hydrogen bonds to Y238 (Fig. S4B). This movement upon intermediate formation is much more subtle than that seen for E. coli β-galactosidase (33) and human α-galactosidase (32), more closely resembling that seen for the covalent intermediate of hen egg white lysozyme captured using a catalytically inactivated mutant enzyme (42).
The crucial role of E258 in the catalytic mechanism was tested by generating a mutant of GALC, where this residue was changed to glutamine (E258Q), resulting in the change of a single nonhydrogen atom (Oε2 to N). At pH 4.6, this mutant enzyme has a melting temperature (Tm) similar to the wild-type enzyme and is correctly folded, and yet all activity is lost, confirming the role of E258 as the catalytic nucleophile (Fig. S6).
Enzyme–Product Complex.
Extended soaking (10 min) of GALC crystals with substrate revealed only the product galactose in the active site, demonstrating that GALC is active in crystallo (Fig. 3E and Figs. S3C and S4C). This structure confirms that we have crystallized a catalytically competent conformation of the enzyme and, in combination with the enzyme–substrate and covalent intermediate structures, provides a series of snapshots illustrating key stages of the GALC catalytic cycle (Fig. 3G and Movie S1).
Discussion
Movement of Active Site Residues and the Role of R380.
The structural snapshots captured here reveal that during catalysis the side chain of the catalytic acid/base residue E182 can adopt two distinct conformations (Fig. 3 A and C). The reversible movement of catalytic side chains has been observed before for other enzymes (43, 44). In addition, we observe that the noncatalytic substrate-binding residue R380 also alternates between two different conformations throughout the catalytic cycle (Fig. 3 B and F). Upon binding of substrate, the side chain of R380 moves to accommodate the substrate and forms a hydrogen bond with the galactosyl 6-hydroxyl. Upon formation of the covalent intermediate, R380 moves back to a conformation similar to that seen in the unliganded enzyme. However, in this complex, two ordered water molecules that were not present in the unliganded structure appear in the active site, hydrogen bonded to both R380 and E182 (Figs. S4B and S7). To complete the catalytic cycle, E182 activates one of these waters to attack the enzyme-bound intermediate. In the enzyme–product complex, the side chain of R380 is again hydrogen bonded to the galactose 6-hydroxyl, as seen in the enzyme–substrate complex. The importance of R380 for GALC activity is highlighted by the severe infantile Krabbe disease that arises from its mutation to tryptophan or leucine (17, 45).
Substrate Distortion.
Based on studies with hen egg white lysozyme, a general catalytic mechanism has been proposed for all retaining β-glycosidases involving initial substrate distortion and the electrophilic migration of the anomeric carbon along the reaction coordinate to form a covalent intermediate, typically from 1S3 to 4C1 via a 4H3 oxocarbenium ion-like transition state (40, 42). In this mechanism, accumulating positive charge at the anomeric center is stabilized by the p-like lone pair of the endocyclic oxygen when the C1, C2, C5, and O atoms are coplanar. Although this has been validated in several subsequent Michaelis complexes (46, 47), here we observe that in all stable species along the reaction coordinate, the galactosyl moiety remains in the lowest-energy 4C1 conformation. This does not preclude distortion as part of the reaction coordinate but is in agreement with the observed lack of substrate distortion seen for the E. coli β-galactosidase (33). It has been proposed that for polymeric substrates the distortion of one sugar is compensated for by the binding interactions of others (46). The shallow substrate-binding pocket present in GALC means that there are very few specific interactions other than those to the galactosyl moiety, suggesting that, for this substrate, distortion may not be energetically stable.
Toward PCT.
Currently, the only available treatment for early infantile Krabbe disease is hematopoietic stem cell transplantation. Although other related lysosomal storage diseases (LSDs) can be treated by enzyme-replacement therapy, for Krabbe disease this strategy is unsuitable because it is primarily a disease of the central nervous system: the administered enzyme will not cross the blood brain barrier and thus will not reach the site of action. For LSDs where the mutant enzyme possesses some residual catalytic activity, PCT is a feasible alternative treatment approach because even partial restoration of trafficking to the lysosome can provide sufficient enzyme activity to prevent disease (48). High-throughput screening of small-molecule libraries for PCT candidates for Krabbe disease has so far been unable to identify any small molecules that significantly increase GALC activity (49). One potential PCT candidate for Krabbe disease is α-lobeline, which was shown in tissue culture to increase the activity of the hyperglycosylated mutant D528N form of GALC (50). However, this molecule was not effective with other mutated forms of GALC and, because of its chemical structure, is not likely to bind the active site of GALC, potentially limiting its specificity.
The elucidation of glycosyl hydrolase structures and mechanisms has driven the recent development of candidate PCT molecules that specifically bind the enzyme active site to stabilize partially defective enzymes implicated in a range of human diseases (26, 51, 52). Glycan-analog PCT candidates have been identified for other LSDs. One example is the iminosugar 1-deoxynojirimycin, which has been shown to stabilize α-galactosidase A by binding to the active site (28) and is currently in phase III clinical trials for the treatment of Fabry disease (29). Three related molecules, isofagomine (25), N-butyl-, and N-nonyl-deoxynojirimycin (26) have been shown to bind the acid β-glucosidase active site and increase enzymatic activity in cell lines and patient fibroblasts expressing clinically relevant mutations responsible for Gaucher disease (27, 53). The structures of GALC described here provide the atomic detail necessary to aid the design of small molecules that specifically bind the active site of GALC, facilitating the development of pharmacological chaperone therapies for Krabbe disease.
To summarize, we have shown how a bona fide substrate binds in the active site pocket of the wild-type GALC enzyme. Structural snapshots of the covalent intermediate and product complexes of GALC provide a comprehensive illustration of the catalytic cycle of this medically important enzyme. The insights gained from this series of structures into enzyme–ligand interactions and active site conformational dynamics provide an atomic framework for the rational design of small molecule inhibitors and pharmacological chaperones.
Materials and Methods
Protein Expression and Purification.
His6-tagged murine wild-type and E258Q GALC was recombinantly expressed by stably transfected HEK 293T cell lines and purified from conditioned medium using nickel-affinity chromatography. When stored at 4 °C, purified GALC was stable and retained full enzymatic activity for at least 2 wk. Detailed methods are provided in SI Materials and Methods.
Crystallization and Small-Molecule Soaks.
GALC protein was concentrated to 2.5 mg/mL in 150 mM NaCl, 10 mM Hepes (pH 7.4). Crystals were grown by sitting-drop vapor diffusion with microseeding (54) against a reservoir of 0.2 M sodium acetate, 0.1 M sodium cacodylate (pH 6.8), and 34% wt/vol polyethylene glycol 8000. For enzyme–substrate and enzyme–product complexes, crystals were soaked with 20 mM 4NβDG for 20–40 s and 10 min, respectively. For the covalent intermediate complex, crystals were soaked with 20 mM d-galactal for 2 h. Immediately after soaking, crystals were cryoprotected with perfluoropolyether oil before flash-cooling in liquid nitrogen. Diffraction data were recorded at the Diamond Light Source beamline I04-1. Final refinement statistics for all structures are shown in Table 1, and the refined models plus structure factors have been deposited in the Protein Data Bank. Detailed data collection and refinement methods are provided in SI Materials and Methods.
Enzyme Activity Assays.
Endpoint assays were conducted with chromogenic substrate 4-nitrophenyl-β-d-galactopyranoside (4NβDG). In all experiments, formation of product 4-nitrophenol was monitored spectrophotometrically by terminating reactions with stopping buffer (360 mM NaOH, 280 mM glycine, pH 10.6) and measuring A410. All experiments were performed at 37 °C with shaking. pH-profile experiments were conducted with 10 mM 4NβDG in a range of citrate/phosphate buffers (pH values: 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, and 7.0) supplemented with 50 mM NaCl. Steady-state kinetic experiments were performed in 20 mM sodium acetate, 150 mM NaCl, 0.1% vol/vol Nonidet P-40 (pH 4.6) with 4NβDG concentrations of 50, 25, 12.5, 6.25, 3.13, 1.56, and 0.78 mM and GALC concentrations of 13.4, 10.1, 7.54, 5.66, 4.24, and 3.18 nM. Experiments were performed in triplicate. Kinetic parameters were obtained by curve-fitting using Prism 5 (GraphPad). Details of procedures and analyses are provided in SI Materials and Methods. Validation of steady-state conditions is provided in Fig. S1.
Supplementary Material
Acknowledgments
We thank the staff of beamline I04-1 at the Diamond Light Source. We thank Begoña Cachón-González for the murine β-galactocerebrosidase expression construct. C.H.H. is funded by a Wellcome Trust PhD studentship; S.C.G. is supported by a Sir Henry Dale fellowship, jointly funded by the Wellcome Trust and The Royal Society (Grant 098406/Z/12/Z); R.J.R. is supported by a Principal Research Fellowship funded by the Wellcome Trust (Grant 082961/Z/07/Z); and J.E.D. is supported by a Royal Society University Research Fellowship (UF100371). The Cambridge Institute for Medical Research is supported by Wellcome Trust Strategic Award 100140.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4CCC (GALC-4NβDG), 4CCD (GALC-d-galactal), and 4CCE (GALC-galactose)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311990110/-/DCSupplemental.
References
- 1.Nagano S, et al. Expression and processing of recombinant human galactosylceramidase. Clin Chim Acta. 1998;276(1):53–61. doi: 10.1016/s0009-8981(98)00095-3. [DOI] [PubMed] [Google Scholar]
- 2.Beier UH, Görögh T. Implications of galactocerebrosidase and galactosylcerebroside metabolism in cancer cells. Int J Cancer. 2005;115(1):6–10. doi: 10.1002/ijc.20851. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, et al. GALC deletions increase the risk of primary open-angle glaucoma: The role of Mendelian variants in complex disease. PLoS ONE. 2011;6(11):e27134. doi: 10.1371/journal.pone.0027134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visigalli I, et al. The galactocerebrosidase enzyme contributes to the maintenance of a functional hematopoietic stem cell niche. Blood. 2010;116(11):1857–1866. doi: 10.1182/blood-2009-12-256461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandhoff K, Kolter T. Topology of glycosphingolipid degradation. Trends Cell Biol. 1996;6(3):98–103. doi: 10.1016/0962-8924(96)80999-8. [DOI] [PubMed] [Google Scholar]
- 6.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3(9):853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 7.Deane JE, et al. Insights into Krabbe disease from structures of galactocerebrosidase. Proc Natl Acad Sci USA. 2011;108(37):15169–15173. doi: 10.1073/pnas.1105639108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanazawa T, et al. Inhibition of cytokinesis by a lipid metabolite, psychosine. J Cell Biol. 2000;149(4):943–950. doi: 10.1083/jcb.149.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giri S, Khan M, Rattan R, Singh I, Singh AK. Krabbe disease: Psychosine-mediated activation of phospholipase A2 in oligodendrocyte cell death. J Lipid Res. 2006;47(7):1478–1492. doi: 10.1194/jlr.M600084-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Formichi P, et al. Psychosine-induced apoptosis and cytokine activation in immune peripheral cells of Krabbe patients. J Cell Physiol. 2007;212(3):737–743. doi: 10.1002/jcp.21070. [DOI] [PubMed] [Google Scholar]
- 11.D’Agostino AN, Sayre GP, Hayles AB. Krabbe’s disease. Globoid cell type of leukodystrophy. Arch Neurol. 1963;8:82–96. doi: 10.1001/archneur.1963.00460010098012. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka K, Nagara H, Kobayashi T, Goto I. The twitcher mouse: Accumulation of galactosylsphingosine and pathology of the sciatic nerve. Brain Res. 1988;454(1-2):340–346. doi: 10.1016/0006-8993(88)90835-9. [DOI] [PubMed] [Google Scholar]
- 13.Rafi MA, Luzi P, Zlotogora J, Wenger DA. Two different mutations are responsible for Krabbe disease in the Druze and Moslem Arab populations in Israel. Hum Genet. 1996;97(3):304–308. doi: 10.1007/BF02185759. [DOI] [PubMed] [Google Scholar]
- 14.Tappino B, et al. Identification and characterization of 15 novel GALC gene mutations causing Krabbe disease. Hum Mutat. 2010;31(12):E1894–E1914. doi: 10.1002/humu.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiumara A, et al. Krabbe leukodystrophy in a selected population with high rate of late onset forms: Longer survival linked to c.121G>A (p.Gly41Ser) mutation. Clin Genet. 2011;80(5):452–458. [Google Scholar]
- 16.De Gasperi R, et al. Molecular heterogeneity of late-onset forms of globoid-cell leukodystrophy. Am J Hum Genet. 1996;59(6):1233–1242. [PMC free article] [PubMed] [Google Scholar]
- 17.Wenger DA, Rafi MA, Luzi P. Molecular genetics of Krabbe disease (globoid cell leukodystrophy): Diagnostic and clinical implications. Hum Mutat. 1997;10(4):268–279. doi: 10.1002/(SICI)1098-1004(1997)10:4<268::AID-HUMU2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 18.Xu C, Sakai N, Taniike M, Inui K, Ozono K. Six novel mutations detected in the GALC gene in 17 Japanese patients with Krabbe disease, and new genotype-phenotype correlation. J Hum Genet. 2006;51(6):548–554. doi: 10.1007/s10038-006-0396-3. [DOI] [PubMed] [Google Scholar]
- 19.Lissens W, et al. A single mutation in the GALC gene is responsible for the majority of late onset Krabbe disease patients in the Catania (Sicily, Italy) region. Hum Mutat. 2007;28(7):742. doi: 10.1002/humu.9500. [DOI] [PubMed] [Google Scholar]
- 20.Fu L, et al. Molecular heterogeneity of Krabbe disease. J Inherit Metab Dis. 1999;22(2):155–162. doi: 10.1023/a:1005449919660. [DOI] [PubMed] [Google Scholar]
- 21.Furuya H, et al. Adult onset globoid cell leukodystrophy (Krabbe disease): Analysis of galactosylceramidase cDNA from four Japanese patients. Hum Genet. 1997;100(3-4):450–456. doi: 10.1007/s004390050532. [DOI] [PubMed] [Google Scholar]
- 22.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426(6968):905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 23.Parenti G. Treating lysosomal storage diseases with pharmacological chaperones: From concept to clinics. EMBO Mol Med. 2009;1(5):268–279. doi: 10.1002/emmm.200900036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan JQ. A contradictory treatment for lysosomal storage disorders: Inhibitors enhance mutant enzyme activity. Trends Pharmacol Sci. 2003;24(7):355–360. doi: 10.1016/S0165-6147(03)00158-5. [DOI] [PubMed] [Google Scholar]
- 25.Lieberman RL, et al. Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease. Nat Chem Biol. 2007;3(2):101–107. doi: 10.1038/nchembio850. [DOI] [PubMed] [Google Scholar]
- 26.Brumshtein B, et al. Crystal structures of complexes of N-butyl- and N-nonyl-deoxynojirimycin bound to acid beta-glucosidase: Insights into the mechanism of chemical chaperone action in Gaucher disease. J Biol Chem. 2007;282(39):29052–29058. doi: 10.1074/jbc.M705005200. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y, et al. Ex vivo and in vivo effects of isofagomine on acid β-glucosidase variants and substrate levels in Gaucher disease. J Biol Chem. 2012;287(6):4275–4287. doi: 10.1074/jbc.M111.280016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guce AI, Clark NE, Rogich JJ, Garman SC. The molecular basis of pharmacological chaperoning in human α-galactosidase. Chem Biol. 2011;18(12):1521–1526. doi: 10.1016/j.chembiol.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asano N, et al. In vitro inhibition and intracellular enhancement of lysosomal alpha-galactosidase A activity in Fabry lymphoblasts by 1-deoxygalactonojirimycin and its derivatives. Eur J Biochem. 2000;267(13):4179–4186. doi: 10.1046/j.1432-1327.2000.01457.x. [DOI] [PubMed] [Google Scholar]
- 30.Chen YQ, Wenger DA. Galactocerebrosidase from human urine: Purification and partial characterization. Biochim Biophys Acta. 1993;1170(1):53–61. doi: 10.1016/0005-2760(93)90175-9. [DOI] [PubMed] [Google Scholar]
- 31.Sakai N, et al. Purification and characterization of galactocerebrosidase from human lymphocytes. J Biochem. 1994;116(3):615–620. doi: 10.1093/oxfordjournals.jbchem.a124569. [DOI] [PubMed] [Google Scholar]
- 32.Guce AI, et al. Catalytic mechanism of human alpha-galactosidase. J Biol Chem. 2010;285(6):3625–3632. doi: 10.1074/jbc.M109.060145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juers DH, et al. A structural view of the action of Escherichia coli (lacZ) beta-galactosidase. Biochemistry. 2001;40(49):14781–14794. doi: 10.1021/bi011727i. [DOI] [PubMed] [Google Scholar]
- 34.Huber RE, Wallenfels K, Kurz G. The action of beta-galactosidase (Escherichia coli) on allolactose. Can J Biochem. 1975;53(9):1035–1038. doi: 10.1139/o75-142. [DOI] [PubMed] [Google Scholar]
- 35.Hajdu J, et al. Analyzing protein functions in four dimensions. Nat Struct Biol. 2000;7(11):1006–1012. doi: 10.1038/80911. [DOI] [PubMed] [Google Scholar]
- 36.Hajdu J. Fast crystallography and time-resolved structures. Annu Rev Biophys Biomol Struct. 1993;22:467–498. doi: 10.1146/annurev.bb.22.060193.002343. [DOI] [PubMed] [Google Scholar]
- 37.Wei RR, et al. X-ray and biochemical analysis of N370S mutant human acid β-glucosidase. J Biol Chem. 2011;286(1):299–308. doi: 10.1074/jbc.M110.150433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandhoff K, Kolter T. Biosynthesis and degradation of mammalian glycosphingolipids. Philos Trans R Soc Lond B Biol Sci. 2003;358(1433):847–861. doi: 10.1098/rstb.2003.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harzer K, et al. Saposins (sap) A and C activate the degradation of galactosylceramide in living cells. FEBS Lett. 1997;417(3):270–274. doi: 10.1016/s0014-5793(97)01302-1. [DOI] [PubMed] [Google Scholar]
- 40.Vocadlo DJ, Davies GJ. Mechanistic insights into glycosidase chemistry. Curr Opin Chem Biol. 2008;12(5):539–555. doi: 10.1016/j.cbpa.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 41.Wentworth DF, Wolfenden R. Slow binding of D-galactal, a “reversible” inhibitor of bacterial beta-galactosidase. Biochemistry. 1974;13(23):4715–4720. doi: 10.1021/bi00720a006. [DOI] [PubMed] [Google Scholar]
- 42.Vocadlo DJ, Davies GJ, Laine R, Withers SG. Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature. 2001;412(6849):835–838. doi: 10.1038/35090602. [DOI] [PubMed] [Google Scholar]
- 43.Millard CB, et al. Reaction products of acetylcholinesterase and VX reveal a mobile histidine in the catalytic triad. J Am Chem Soc. 1999;121(42):9883–9884. [Google Scholar]
- 44.Barabas O, et al. Catalytic mechanism of alpha-phosphate attack in dUTPase is revealed by X-ray crystallographic snapshots of distinct intermediates, 31P-NMR spectroscopy and reaction path modelling. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selleri S, et al. Deletion of exons 11-17 and novel mutations of the galactocerebrosidase gene in adult- and early-onset patients with Krabbe disease. J Neurol. 2000;247(11):875–877. doi: 10.1007/s004150070076. [DOI] [PubMed] [Google Scholar]
- 46.Sulzenbacher G, Driguez H, Henrissat B, Schülein M, Davies GJ. Structure of the Fusarium oxysporum endoglucanase I with a nonhydrolyzable substrate analogue: Substrate distortion gives rise to the preferred axial orientation for the leaving group. Biochemistry. 1996;35(48):15280–15287. doi: 10.1021/bi961946h. [DOI] [PubMed] [Google Scholar]
- 47.Tews I, et al. Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat Struct Biol. 1996;3(7):638–648. doi: 10.1038/nsb0796-638. [DOI] [PubMed] [Google Scholar]
- 48.Schueler UH, et al. Correlation between enzyme activity and substrate storage in a cell culture model system for Gaucher disease. J Inherit Metab Dis. 2004;27(5):649–658. doi: 10.1023/b:boli.0000042959.44318.7c. [DOI] [PubMed] [Google Scholar]
- 49.Ribbens J, et al. A high-throughput screening assay using Krabbe disease patient cells. Anal Biochem. 2013;434(1):15–25. doi: 10.1016/j.ab.2012.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee WC, et al. Molecular characterization of mutations that cause globoid cell leukodystrophy and pharmacological rescue using small molecule chemical chaperones. J Neurosci. 2010;30(16):5489–5497. doi: 10.1523/JNEUROSCI.6383-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuzwa SA, et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat Chem Biol. 2008;4(8):483–490. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- 52.Ficko-Blean E, Stubbs KA, Nemirovsky O, Vocadlo DJ, Boraston AB. Structural and mechanistic insight into the basis of mucopolysaccharidosis IIIB. Proc Natl Acad Sci USA. 2008;105(18):6560–6565. doi: 10.1073/pnas.0711491105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sánchez-Ollé G, et al. Promising results of the chaperone effect caused by imino sugars and aminocyclitol derivatives on mutant glucocerebrosidases causing Gaucher disease. Blood Cells Mol Dis. 2009;42(2):159–166. doi: 10.1016/j.bcmd.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 54.Walter TS, et al. Semi-automated microseeding of nanolitre crystallization experiments. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64(Pt 1):14–18. doi: 10.1107/S1744309107057260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.