Significance
We developed an approach to fine-map large genomic regions that play important roles for malaria parasite resistance in the vector mosquito, Anopheles gambiae. Direct-association studies in wild mosquitoes from malaria-endemic areas in Africa identified three mutations in three genes that are associated significantly with Plasmodium falciparum infection. We confirmed the involvement of these genes in parasite infection by silencing them. The discovered genes will help us understand interactions between the mosquitoes and parasites. The identified single-nucleotide polymorphisms could be used to develop a malaria risk-assessment tool and may help eliminate parasite-susceptible mosquitoes.
Keywords: synteny, genomics, traits, chromosomal domains, gene expression
Abstract
The malaria parasite-resistance island (PRI) of the African mosquito vector, Anopheles gambiae, was mapped to five genomic regions containing 80 genes, using coexpression patterns of genomic blocks. High-throughput sequencing identified 347 nonsynonymous single-nucleotide polymorphisms within these genes in mosquitoes from malaria-endemic areas in Kenya. Direct association studies between nonsynonymous single-nucleotide polymorphisms and Plasmodium falciparum infection identified three naturally occurring genetic variations in each of three genes (An. gambiae adenosine deaminase, fibrinogen-related protein 30, and fibrinogen-related protein 1) that were associated significantly with parasite infection. A role for these genes in the resistance phenotype was confirmed by RNA interference knockdown assays. Silencing fibrinogen-related protein 30 increased parasite infection significantly, whereas ablation of fibrinogen-related protein 1 transcripts resulted in mosquitoes nearly free of parasites. The discovered genes and single-nucleotide polymorphisms are anticipated to be useful in the development of tools for malaria control in endemic areas in Africa.
Malaria parasites cause more than 225 million clinical cases and >781,000 deaths per year (1). Anopheles gambiae is one of the most important vectors in Africa transmitting Plasmodium falciparum (2). Parasites have to evade the innate immune system before mosquitoes can infect humans. Identification of naturally occurring parasite resistance genes and their functions are of fundamental interest and may be used to develop malaria control strategies.
Many genes related to innate immunity have been reported (3–5). Among these are pattern recognition receptors, including leucine-rich-repeats proteins, fibrinogen-related protein family members (FREPs or FBNs), thioester-containing proteins (TEPs), and C-type lectins (CTLs). Some gene products inhibit the development of oocysts; for example, Tep1, Anopheles Plasmodium-responsive leucine-rich repeat 1 C (APL1C), and leucine-rich repeat immune protein 1 (LRIM1) form a complex that activates the mosquito complement-like system in hemolymph to kill parasites (5, 6). Other mosquito gene products facilitate parasite development by protecting them from the innate immune attack. The C-type lectins CTL4 and CTLM2 can protect Plasmodium ookinetes from melanization (7). Moreover, it has been proposed that parasite invasion is mediated by specific proteins, and this hypothesis is supported by experiments that demonstrate the preferential adherence of ookinetes to midguts and the inhibition of parasite invasion by a peptide, salivary gland and midgut peptide 1 (SM1) (8).
The parasite resistance phenotype is a multigenic trait in natural An. gambiae populations; previous genetic mapping studies identified several contributing loci. A major parasite resistance island (PRI) was identified on the basis of segregation in two parental strains, with differences in vector competence originating in Eastern and Western Africa (4, 9). However, identification of the malaria resistance genes within the PRI in wild-derived mosquitoes is a significant challenge because the locus is large (>10 Mb) and it is difficult to acquire sufficient samples for direct-association studies. Synchronizing large numbers of same-age (3–5-d-old) wild adult mosquitoes is difficult, and low blood-feeding rates (<10%) on membrane feeders make it a logistical challenge to obtain enough samples for genome-wide direct-association studies. Thus, it is necessary to narrow the size range of the PRI for further analysis.
We used chromosomal domain expression to find coexpression patterns of candidate immune genomic blocks that could include resistance genes. The approach is based on the conservation of innate immunity from flies to humans (10, 11). The innate immune genes also are conserved across mosquito species and clustered in their genomes as coexpressed chromosomal domains (12–14). We compared the An. gambiae genome with Culex quinquefasciatus, a distantly related species that is estimated to have last shared a common ancestor ∼145 million years ago (15, 16), to find conserved chromosomal domains. Our approach differs from previous studies in vector competence, in which candidate genes were identified on the basis of changes in transcription profiles after parasite challenges. We used chromosomal domains as expression modules (17, 18) and a number of known infection-related genes as a training set to find expression profiles that were enriched significantly with the training genes. Five regions, containing 80 genes in total, were identified within the PRI that have characteristic coexpression patterns. Further association studies and RNAi experiments discovered and confirmed three genes whose products affect Plasmodium parasite infection of An. gambiae.
Results
Syntenic Blocks in An. gambiae and C. quinquefasciatus.
We compared C. quinquefasciatus supercontigs with the An. gambiae reference genome on the basis of translated protein sequences. Neighboring conserved regions were joined to define the boundaries of syntenic blocks. For example, comparison of C. quinquefasciatus contig3.1 and An. gambiae chromosome 2L identified nine conserved syntenic blocks (Fig. 1A). These steps were repeated between 3,171 C. quinquefasciatus supercontigs and the An. gambiae chromosomal arms, resulting in a total of 1,242 syntenic blocks detected in the latter species (Fig. 1B). The chromosome arms 2L, 2R, 3L, 3R, and X contain 237, 348, 197, 228, and 232 syntenic blocks, respectively, which is similar to a previous report (n = 1,514) that was based on ortholog analysis (Table S1; ref. 16). The median size of the syntenic blocks in An. gambiae was ∼37.7 kb in length, with ∼12 predicted protein-coding genes. Most individual C. quinquefasciatus supercontigs mapped to a single chromosome arm in the An. gambiae genome, confirming a previous conclusion that few translocation events had occurred between different chromosomes (Fig. 1B; ref. 16). There were 1,252 nonconserved genomic regions in the An. gambiae genome. The major malaria resistance locus, PRI, has 56 conserved syntenic blocks with a median length of ∼45 kb. The PRI was detected previously in mosquitoes with a chromosomal inversion between 20,528,089 and 42,165,532 bp (4, 9, 19). However, our map is based on the pink-eye standard (PEST) strain, which lacks this inversion, being used as the reference genome. This results in the PRI localizing in our map to two noncontiguous domains.
Fig. 1.
Synteny between An. gambiae and C. quinquefasciatus. (A) Example of synteny detection between An. gambiae and C. quinquefasciatus. The C. quinquefasciatus suppercontig3.1 was mapped to nine conserved syntenic regions on the An. gambiae chromosome 2L. The red and blue dots represent genes transcribed from the same or a complementary strand, respectively. (B) Synteny between the An. gambiae genome and the C. quinquefasciatus supercontigs. The pink blocks indicate the positions of the PRI. The PRI was detected originally in mosquitoes with a chromosomal inversion that is absent in the reference genome strain (4, 9, 19). This results in two noncontiguous PRI domains in the map shown here.
Expression Profiles of Syntenic Blocks for Innate Immunity.
Approximately 186,344 probes on the Affymetrix microarray chip were aligned, based on their sequences, to the current An. gambiae reference genome. Among these probes, 147,549 were mapped uniquely to chromosomes 2, 3, and the X. Detailed information on probe sequence, location, and signals can be downloaded in text format from http://omics.ou.edu/download/.ExpressionPattern/OligoSignalAndPosition.tab.
The hierarchical expression tree of all genomic blocks constructed on the basis of the probe signal shows that the mRNA abundance resulting from transcription in many genomic regions was lower at 24, 48, and 72 h after blood feeding when comparing young (3 d postemergence), naive (NBF), and 3 h post-blood-feeding mosquitoes (BF3h) (Fig. 2A). These observations are consistent with a physiological switch from host-seeking activity to egg production after a blood meal. In addition, expression in most genomic blocks was different between younger (≤4 d after adult emergence) and older (14 d postemergence) animals, which is consistent with previous observations (20). Therefore, the genomic block expression tree is consistent with the overall transcript abundance profiles in different conditions. A detailed image of the genomic block expression tree is available in Fig. S1.
Fig. 2.
Immune genomic blocks are enriched significantly in three branches of the expression tree. (A) The expression tree of conserved syntenic blocks and nonconserved genomic regions from 14 expression samples. The red and green coloration indicate above- and below-average z-scores, respectively. NBM, no-blood meal; PBMxxh, various hours post-blood meal; MBM, multiple blood feeding. The letter “d” in PBM15d and NBM18d stands for days. Fig. S1 provides a detailed expression tree. (B) Enrichment of genomic blocks that contain the trained immune genes. Enrichment scores were transformed from P value with the equation of score = −ln(p), where p is the binomial probability. The enrichment score profile was smoothed using the Ramer-Douglas-Peucker line smooth algorithm with a window size of 40 leaves. α, β, and χ are characteristic profiles that were enriched significantly with training genes. The detailed genomic regions in the three profiles are shown in Fig. S2.
Quantitative trait loci (QTL) discovery is based on the segregation of genetic variations and traits in pedigree analyses. The underlying genetic variations may not alter the expression of that gene, and it may not be regulated differentially by variable conditions such as infection. Therefore, we used known infection-related genes as a training set to identify enrichments of characteristic expression patterns. This strategy differs from approaches that determine candidate genes on the basis of the differentially expressed genes under parasite challenge. Seventy known genes were used to label genomic blocks on the expression tree to detect enrichment profiles related to parasite infection (Fig. 2B; Tables S2 and S3). Three branches were enriched significantly with immunity blocks (Fig. 2B). Profile α contains 15 nonconserved and 25 conserved genomic blocks (Fig. S2). Three paper clip-like domain-containing (CLIP)-proteins, CLIPB15, CLIPC2, and CLIPB5, related to immune modulation in insects (21) are within this branch, and a fourth, inhibitor of apoptosis 5, is an inhibitor of apoptosis. The presence of these genes supports the hypothesis that genomic blocks in profile α might function as immune modulators. Profile β contains 44 nonconserved and 90 conserved genomic blocks with 13 known immune genes. Five conserved genomic blocks, designated sequentially PRI-a, PRI-b, PRI-c, PRI-d, and PRI-e, are located within the PRI at 2L (∼15 Mb to 20,528,089 bp and ∼35 Mb to 42,165,532 bp) (Table 1; ref. 4). Interestingly, the products of several of the 13 immune genes, including APL1C, CTLs, serpins, and caspases, are reported to be involved in the defense against parasite infection (4, 22–24). Therefore, we further analyzed the profile β conserved genomic blocks within the PRI for possible roles in immunity against Plasmodium parasites. Profile χ contains nine nonconserved and 19 conserved genomic blocks. Four of five known profile χ immune genes are Toll-like proteins and peptidoglycan recognition proteins. Toll-pathway and peptidoglycan recognition proteins are used for defense against bacteria and fungi (25, 26), and therefore the profile χ expression pattern and associated genomic blocks might be related to immunity against these microbiota. Nonconserved genomic regions were underrepresented significantly in all three profiles (P = 7 × 10−5).
Table 1.
Syntenic blocks within PRI in the An. gambiae genome that shared the profile β expression pattern
| Block name | Start position, bp | End position, bp | Length, bp | Number of protein-encoding genes |
| PRI-a | 18,536,947 | 18,697,902 | 160,956 | 10 |
| PRI-b | 37,924,199 | 38,048,948 | 124,750 | 10 |
| PRI-c | 39,757,958 | 39,994,106 | 236,149 | 12 |
| PRI-d | 40,901,916 | 41,287,903 | 385,988 | 29 |
| PRI-e | 41,830,390 | 42,164,453 | 334,064 | 19 |
Genetic Variation in Wild-Derived Mosquitoes from Malaria-Endemic Areas in Kenya.
More than 10,000 mosquito larvae were collected independently from different habitats in Kenya and reared to adults under laboratory conditions. Approximately 1,000 morphologically confirmed An. gambiae, 3 to 5 d old, were fed on P. falciparum-infected human blood, with ∼10% of them becoming engorged.
We used whole-genome sequencing to detect genetic variation in the wild An. gambiae populations. According to previous reports (4, 9), the frequency of genetic alleles for PRI is ∼0.17; thus, >8 mosquitoes are required to gain 95% confidence of not missing the candidate genetic alleles. The genomic DNAs of nine confirmed An. gambiae (five uninfected, with no oocysts in midguts, and four infected with two, eight, eight, and nine oocysts, respectively) that fed on the same P. falciparum-infected human blood were sequenced to detect genetic variation. The length of each sequence read was 100 bp, and the whole-genome coverage for each individual was >36. More than 1.6 × 106 SNPs were detected genome-wide in each individual mosquito (Table 2). This high SNP frequency supports a previous report of ∼1 SNP per 200 bp (2). Furthermore, 347 SNPs encoded nonsynonymous changes in genes within the candidate genomic regions and were present in at least two mosquitoes (Table S4). These SNPs were verified manually, using the GBrowse graphics user interface (27), and the results indicated a low error rate (<1%) caused by sequencing or detection.
Table 2.
Summary of resequencing nine individual mosquitoes
| Individual mosquito ID | Total reads | Coverage, fold | SNPs |
| 541 | 186,329,422 | 66.9 | 1,927,849 |
| 544 | 127,341,586 | 45.8 | 1,709,907 |
| 545 | 100,603,962 | 36.1 | 1,604,083 |
| 551 | 135,849,298 | 48.9 | 1,651,555 |
| 553 | 153,042,132 | 55.1 | 1,781,589 |
| 564 | 179,647,640 | 64.6 | 1,888,508 |
| 565 | 206,448,504 | 74.3 | 1,971,906 |
| 566 | 208,407,812 | 74.9 | 1,954,330 |
| 567 | 181,056,110 | 65.2 | 1,937,986 |
Fibrinogen-Related Protein 1, Fibrinogen-Related Protein 30, and An. gambiae Adenosine Deaminase Genes Were Identified by the Association Between Genetic Variation and P. falciparum Infection in Wild An. gambiae.
Eighty candidate genes within PRIs a–e were prioritized by the relationship between the 347 SNPs and the P. falciparum infection in the nine mosquitoes (Table S4). Five SNPs within five genes associated with mean intensity of infection (P value < 0.05), as well as infection prevalence (P value <0.12) (Table 3), were chosen for further analysis.
Table 3.
Direct-association analyses between five SNPs in five candidate genes and P. falciparum infection in wild-derived An. gambiae
| Block | Position on 2L, bp | Gene name (VectorBase ID) | SNP | Amino acid | Prevalence P value* | Intensity P value* | Prevalence P value† | Intensity P value† |
| PRI-c | 39,852,810 | AgADA (AGAP006906) | T/C | C→R | 0.119 | 0.0496 | 0.0046 | 0.0021 |
| PRI-c | 39,966,795 | FBN30 (AGAP006914) | T/C | F→L | 0.048 | 4.16e-06 | 0.030 | 0.0091 |
| PRI-d | 41,165,983 | FREP1 (AGAP007031) | A/T | L→Q | 0.119 | 0.0496 | 0.226 | 0.018 |
| PRI-d | 41,246,582 | BRCA2 (AGAP007032) | A/C | K→Q | 0.119 | 0.0496 | 0.221 | 0.502 |
| PRI-e | 42,069,973 | LRRx (AGAP007060) | A/T | S→T | 0.119 | 0.0496 | 0.740 | 0.161 |
Bold indicates associated significantly (P < 0.05). BRCA2, breast cancer 2 susceptibility protein-like; LRRx, leucine-rich repeat protein x.
P values were calculated on the basis of nine wild-derived individual mosquitoes sequenced by the Illumina paired-end approach.
P values were calculated based on 22 wild-derived individual mosquitoes.
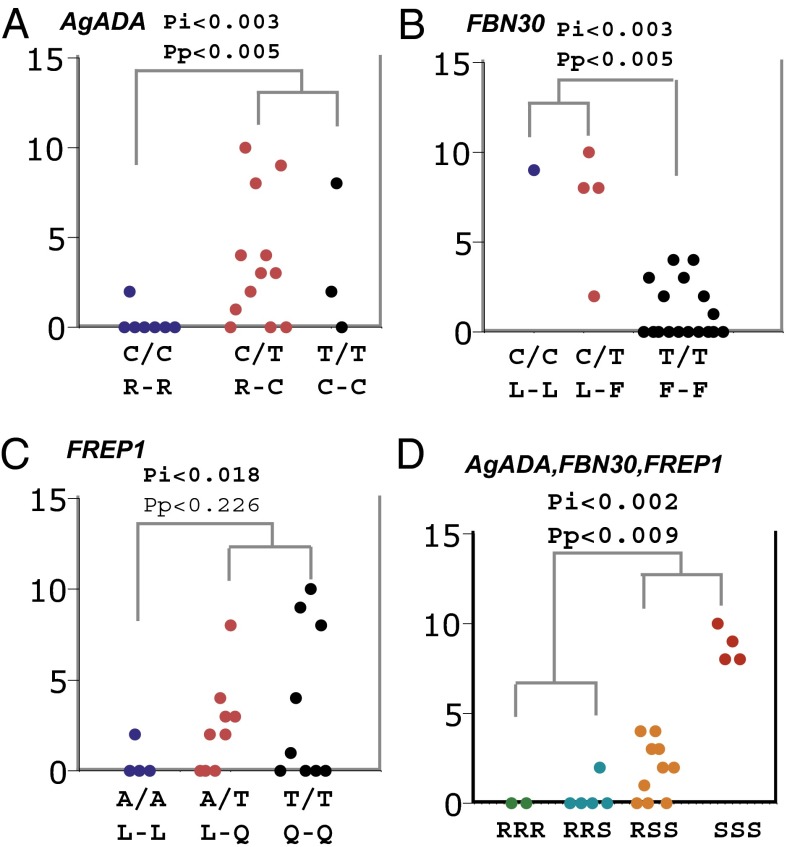
We cloned the five candidate genes from 22 individual wild mosquitoes infected with P. falciparum and genotyped them (phenotypes and genotypes are shown in Table S5). The data support the conclusion that three of five candidate SNPs in three genes are associated significantly with P. falciparum infection (Table 3; Fig. 3). The SNPs, C427T in An. gambiae adenosine deaminase (AgADA) (AGAP006906) and T28C in fibrinogen-related protein 30 (FBN30) (AGAP006914), are associated significantly with the prevalence and mean intensity of infection. The T1325A SNP in fibrinogen-related protein 1 (FREP1) (AGAP007031) is associated significantly with the mean intensity of infection. These mutations were designated mosquitoes against P. falciparum parasites (maplap): maplap1, maplap2, and maplap3, respectively. maplap1 changes amino acid 143 from arginine to cysteine (R143C), maplap2 changes amino acid 10 from phenylalanine to leucine (F10L), and maplap3 changes amino acid 442 from glutamine to leucine (Q442L). As anticipated, the infection intensity in mosquitoes increased when mosquitoes have more susceptible alleles (Fig. 3D).
Fig. 3.
Direct association analysis of P. falciparum infection and genetic variation within candidate genes in wild-derived An. gambiae. (A) AgADA gene SNP 427 (2L, 39,852,810 bp). (B) FBN30 gene SNP 28 (2L, 39,966,795 bp). (C) FREP1 gene SNP 1325 (2L, 41,165,983 bp). (D) Combined effect of the SNPs in all three genes has a stronger effect on parasite resistance than individual genes. RRR, all three genes are resistant genotypes in mosquitoes; RRS, two of three genes are resistant genotypes; RSS, one of three genes is resistant genotype; SSS, all three genes are susceptible genotypes. The y-axis gives the number of oocysts in each mosquito (dots), and the x-axis specifies the genotypes (top) and amino acids (bottom) for each mosquito. Mosquitoes (n = 22) were grouped according to their genotypes and phenotypes; closely related phenotypes joined first. Pi, mean intensity of infection-associated P value calculated with a t test. Pp, prevalence-associated P value calculated with the Fisher exact test. Bold letters indicate statistically significant associations.
On the basis of the genotypes from 22 mosquitoes (Table S5), the correlation coefficients between maplap1 and maplap2, maplap1 and maplap3, and maplap2 and maplap3 are 0.009, 0.005, and 0.02, respectively, according to Haploview analysis (28), supporting the conclusion that the effects of the three genes are independent of one another and do not result from linkage effects. This result of the small haplotype block size in wild-derived An. gambiae populations is consistent with previous reports (29) and also supports the proposal that a direct-association approach may be the best way to search candidate genes for traits of interest in mosquito populations.
Effect of FREP1, FBN30, and AgADA dsRNA-Mediated Knockdown on Plasmodium Parasite Infection.
There is a report of immune-related genes whose products exhibit different effects on P. falciparum and Plasmodium berghei (30). The three genes in this study are associated significantly with P. falciparum infection, and we used the rodent parasite in RNAi experiments to test whether they function across species (Fig. 4). FREP1 mRNA was reduced ∼80% in treated mosquitoes after injection of gene-specific dsRNAs, and surprisingly, this resulted in a reduction in infection prevalence from 70% and 83% to 30% and 31%, respectively, in two replicates of control and experimental mosquitoes. In contrast, ablation of FBN30 transcripts to undetectable levels resulted in a twofold increase in the oocyst mean intensities of infection (P < 0.004). The prevalence was similar, at 82% versus 95% and 95% versus 96%, in two replicates for control and experimental samples, respectively. Knock-down of AgADA transcripts slightly reduced P. berghei mean intensity of infection when comparing control and experimental groups. Prevalence also was lower (82% and 85% to 76% and 70%, respectively) in AgADA dsRNA-treated mosquitoes.
Fig. 4.
Verification of the phenotypes of dsRNA-mediated knock-downs of FREP1, FBN30, and AgADA mRNA on Plasmodium infection. (A, E, and I) Quantitative reverse-transcription PCR detection of FREP1, FBN30, and AgADA mRNAs in dsRNA-treated experimental and control mosquitoes. Ag S7 mRNA was used as a loading control. (B, F, and J) Fluorescent microscopic detection of GFP-P. berghei oocysts (green spots) in mosquitoes treated with the GFP dsRNA. (C, G, and K) Fluorescent microscopic detection of GFP-P. berghei oocysts in mosquitoes treated with FREP1, FBN30, or AgADA dsRNA. (D, H, and L) Statistical analyses of the number of oocysts in mosquitoes treated with the dsRNA of FREP1, FBN30, or AgADA, respectively, and GFP. The experiments were repeated twice and showed similar results. The black lines indicate the means of oocyst number under different treatment groups.
Discussion
Identification of genetic variation underlying vector competence is anticipated to be useful for the development of parasite-control strategies. Several QTL for Plasmodium parasite infection of An. gambiae have been identified. However, the high genetic variation and small available sample size make the identification of natural resistance genes a major challenge. Microarray-based differential expression often is used to detect candidate genes (4). However, the genes underlying QTL may not be regulated by a parasite infection. We integrated multiple independent expression profiles to find immune signature patterns using a set of training genes, and thereby unlinked the basal parasite-resistance phenotype from that elicited in response to infection. Microarray data at the gene level also may be imprecise because of the coexistence of genuine biological variation. The method of combining related genes as modules was used previously to find robust signatures (18, 31). We used syntenic chromosomal domains as higher-level modules to detect the immunity signature expression patterns against malaria parasites in An. gambiae and also narrowed the candidate genomic regions.
We found three SNPs (maplap1, maplap2, maplap3) in AgADA, FBN30, and FREP1, respectively, related to parasite infection in An. gambiae. maplap1 in AgADA is associated significantly with P. falciparum mean intensity of infection and prevalence in wild-derived mosquitoes from Kenya. It is interesting to note that mutations in adenosine deaminase also cause severe immunodeficiency in humans (32), supporting the conclusion that this gene is conserved in metazoan immunity. However, knock-down of AgADA did not greatly alter mosquito susceptibility to P. berghei, as anticipated. This may result from differences between the local P. falciparum isolates and rodent malaria P. berghei (33) or from physiological differences between the naturally occurring mutation and the experimental transient down-regulation of AgADA expression.
The fibrinogen-related gene family has been found to defend against pathogens in invertebrates as pattern recognition proteins, and there are 59 putative fibrinogen-related genes (FREPs or FBNs) in An. gambiae (21). Fibrinogen-related genes were screened previously for roles in immunity against parasites by ablating several genes simultaneously, and it was proposed that FBN9 encoded the most-potent anti-Plasmodium FREP protein (34). Our one-by-one analysis of the candidate genes supports the conclusion that FBN30 and FREP1 also play important roles during parasite infection.
Although FBN30 and FREP1 proteins are members of FBNs, and both have signal peptides and fibrinogen-like domains at their N and C termini, respectively, they differ in length and expression profiles and have contrasting effects against parasites. FREP1 and FBN30 have 738 and 280 amino acids, respectively. FBN30 is abundant in mosquito fat body tissues and inhibits parasite development (35), supporting the hypothesis that it may be involved in humoral innate immunity. In contrast, FREP1 is abundant in midguts (36) and facilitates parasite infection, supporting the hypothesis that it may act as a receptor of Plasmodium parasite to facilitate ookinete invasion. The detailed molecular mechanisms of these proteins against Plasmodium require further elucidation. In conclusion, we analyzed the PRI in An. gambiae populations from malaria-endemic areas and identified and confirmed that the AgADA, FBN30, and FREP1 genes play roles in the Plasmodium parasite infections in An. gambiae. FREP1 was determined to be essential for Plasmodium invasion of An. gambiae.
Materials and Methods
Mosquito Reference Genome Sequences, Oligonucleotide Array Data, and Analytical Software.
The An. gambiae reference genome sequences (assembly version AgamP3, PEST strain) and C. quinquefasciatus supercontigs (version CpipJ1, Johannesburg JHB strain) were downloaded from VectorBase (www.vectorbase.org). Affymetrix oligo array (GeneChip Plasmodium/Anopheles Genome Array) data were obtained from www.angaged.bio.uci.edu (35). The R (version 2.6.1; www.r-project.org/) and Bioconductor (version 2.8, www.bioconductor.org/) packages were used for statistical analyses. The MUMmer software (version 3.0) was downloaded from sourceforge (mummer.sourceforge.net/). Programs implemented for this project are available for download (http://omics.ou.edu/download/.ExpressionPattern/scripts.zip).
Detecting Synteny Between An. gambiae and C. quinquefasciatus.
Genome sequence comparisons between An. gambiae and C. quinquefasciatus were used to identify conserved chromosomal domains. The “promer” function (with parameters –mum–coords -c 41 -g 1000 -l 8) in the MUMmer package (37) was used to detect the conserved genomic regions on the basis of derived protein sequences. After removal of isolated and small (<150 bp) conserved regions, neighboring conserved regions were joined recursively to form a single, larger region if they were on the same An. gambiae chromosome and included in the same C. quinquefasciatus contigs. Syntenic blocks were verified manually, using the graphic presentation generated by the “mummerplot” in the MUMmer package. Genomic regions between syntenic blocks in An. gambiae are not conserved and were included as internal controls in the signature expression profile detection.
Detecting Signature Expression Profiles of Genomic Blocks Containing Immunity-Related Genes.
Probes on the Affymetrix array were mapped to the An. gambiae reference genome, using blast-like alignment tool (38). Cross-hybridizing probes mapping to multiple genomic regions were removed from further analysis. No infections are required because we used a set of known genes related to infection as a training set to identify characteristic expression patterns. However, a minimum number of independent genome-wide expression profiles (samples) are needed to obtain high resolution of genomic block expression patterns. Because we identified a total of 2,494 An. gambiae genomic blocks, and the expression of each block in the samples can be higher, lower, or unchanged, more than eight sample conditions are needed for this study (38 = 6,561). We used array data from 14 samples of different developmental stages and tissue origins (35). The probe ID, sequence, and signal data were assembled into one table (http://omics.ou.edu/download/.ExpressionPattern/OligoSignalAndPosition.tab). The array data then were normalized with the quantile normalization procedure (39) and mapped to the syntenic and nonconserved regions on the basis of their positions in the An. gambiae genome. The expression values of probes were condensed to syntenic and nonconserved blocks, using the Robust Multichip Average algorithm (40). The hierarchical clustering algorithm (41) was used to construct an expression tree of genomic blocks.
Seventy genes known or proposed to have roles in mosquito immune defense (positive or negative) were used to detect signature expression profiles in the constructed expression tree (Table S2). The genes were used to designate genomic blocks as “immunity” blocks. The immunity block enrichment P values based on the binomial distribution for a leaf were calculated at different levels of branches, and the smallest P value was used to represent the “immunity”-associated value (p) for that leaf. The enrichment scores were generated using the equation score = −ln(p). If a genomic block in a branch contained more than one immune gene, it was counted only once in statistical analysis. The enrichment-score profile was smoothed using the Ramer-Douglas-Peucker line smooth algorithm, with a window size of 40 leaves (42, 43).
Direct Association Between Genetic Variation Within Candidate Genomic Blocks and P. falciparum Infection in Wild-Derived An. gambiae.
An. gambiae larvae were collected from diverse habitats in a malaria-hyperendemic region in highland areas near Kisumu, western Kenya. Fewer than four larvae per habitat were collected, and only one female from each habitat was used to avoid the effects of siblings on the association studies. The distance between any two habitats was ≥10 m. Morphologically identified An. gambiae larvae were brought to the laboratory at the International Centre for Insect Physiology and Ecology and reared to adults. The resulting 3–5-d postemergence female mosquitoes were challenged with P. falciparum-infected blood drawn from patients with >100 gametocytes per microliter blood [approved institutional review board protocols No. 163 from Kenya Medical Research Institute (Kenya) and 0906M68726 from the University of Minnesota)]. All mosquitoes for the direct-association study were fed simultaneously on the same source of infected human blood. The blood serum was replaced with the same amount of nonimmunized human serum (AB-type; Sigma-Aldrich) before mosquito challenges. Fully engorged mosquitoes were maintained in the laboratory with cotton soaked in an 8% (wt/vol) glucose solution, midguts were dissected 7 d after blood feeding and stained with 0.1% mercurochrome (Sigma-Aldrich), and the number of oocysts was counted with light microscopy. Mosquito carcasses were preserved in 75% (vol/vol) ethanol for subsequent DNA analysis, including molecular species identification (44), whole-genome sequencing by an Illumina paired-end approach, and DNA amplification followed by Sanger sequencing. The high-throughput sequencing 100-bp reads from individual mosquitoes were mapped to the An. gambiae reference genome, using the short oligonucleotide analysis package program (v2.21) (45), and the “soapsnp” program (v1.02) was used to detect nucleotide variations at each position. The scores for base-calling (phred score) were greater than 30 to avoid SNPs caused by sequencing errors. At least two reads from one mosquito at a SNP site were required. SNPs within the candidate genomic regions were verified visually, using GBrowse (v2.26) (27). The genotypes of candidate SNPs were analyzed further by PCR cloning and Sanger sequencing. The highly conserved regions (no variations in nine wild mosquitoes based on Illumina sequencing) adjacent to candidate SNPs were selected manually to design oligonucleotide primers (Table S6) to clone the genomic regions by DNA amplification. The amplification products were used to genotype the candidate SNPs in individual mosquitoes, using Sanger sequencing. Finally, the Fisher exact test was used to assess the significance of association of SNPs with the prevalence of P. falciparum infection. The t and Wilcox tests were used to assess the significance of association between SNPs and the P. falciparum mean intensities of infection.
Validation of Candidate Genes by RNAi.
Challenge experiments with P. berghei and RNAi-mediated ablation of candidate gene mRNA accumulation were performed as described previously (22). Briefly, the An. gambiae G3 strain was maintained at 27 °C, 80% humidity with a 12-h alternating day–night cycle and fed on mice for egg production. The coding regions of AgADA, FBN30, and FREP1 were amplified from DNA derived from an adult cDNA library, using gene-specific primers and a nested iterative strategy and were used as templates for production in vitro of dsRNA (Table S6). No off-target sequences for the designed dsRNA were found in the An. gambiae genome. dsRNA was injected into the hemocoel of ∼100 1-d-old adult G3 mosquitoes, and specific transcript levels were assayed quantitatively 72 h later in a sample of five of them. The An. gambiae ribosomal S7 gene (AgS7) was used as an internal control for quantitative expression analyses. The remaining mosquitoes were fed the same day with a blood meal containing the GFPcon P. berghei strain (MRA-865). Midguts were dissected 7 d later and the number of oocysts was counted under fluorescence microscopy.
Supplementary Material
Acknowledgments
We thank Dr. Kenneth Clinkenbeard (Oklahoma State University) for comments on the manuscript and Dr. Baolin Wu (University of Minnesota) for consulting in biostatistical analyses. This work was supported by the University of Oklahoma and the National Institutes of Health (NIH) (1R56AI081829-01A1, to J.L.). A.A.J. was supported in part by a grant from the NIH (AI29746).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321024110/-/DCSupplemental.
References
- 1.Cibulskis RE, Aregawi M, Williams R, Otten M, Dye C. Worldwide incidence of malaria in 2009: Estimates, time trends, and a critique of methods. PLoS Med. 2011;8(12):e1001142. doi: 10.1371/journal.pmed.1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt RA, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298(5591):129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 3.Richman AM, Dimopoulos G, Seeley D, Kafatos FC. Plasmodium activates the innate immune response of Anopheles gambiae mosquitoes. EMBO J. 1997;16(20):6114–6119. doi: 10.1093/emboj/16.20.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riehle MM, et al. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science. 2006;312(5773):577–579. doi: 10.1126/science.1124153. [DOI] [PubMed] [Google Scholar]
- 5.Blandin S, et al. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004;116(5):661–670. doi: 10.1016/s0092-8674(04)00173-4. [DOI] [PubMed] [Google Scholar]
- 6.Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science. 2009;324(5924):258–261. doi: 10.1126/science.1171400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science. 2004;303(5666):2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- 8.Ito J, Ghosh A, Moreira LA, Wimmer EA, Jacobs-Lorena M. Transgenic anopheline mosquitoes impaired in transmission of a malaria parasite. Nature. 2002;417(6887):452–455. doi: 10.1038/417452a. [DOI] [PubMed] [Google Scholar]
- 9.Menge DM, et al. Quantitative trait loci controlling refractoriness to Plasmodium falciparum in natural Anopheles gambiae mosquitoes from a malaria-endemic region in western Kenya. Genetics. 2006;173(1):235–241. doi: 10.1534/genetics.105.055129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA. 1997;94(26):14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beutler B, Ulevitch RJ. Genetic Analysis of Host Responses in Sepsis. Curr Infect Dis Rep. 2001;3(5):419–426. [PubMed] [Google Scholar]
- 12.Williams EJ, Bowles DJ. Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 2004;14(6):1060–1067. doi: 10.1101/gr.2131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Q, Halfon MS. Complex organizational structure of the genome revealed by genome-wide analysis of single and alternative promoters in Drosophila melanogaster. BMC Genomics. 2009;10:9. doi: 10.1186/1471-2164-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quijano C, et al. Selective maintenance of Drosophila tandemly arranged duplicated genes during evolution. Genome Biol. 2008;9(12):R176. doi: 10.1186/gb-2008-9-12-r176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Besansky NJ, Fahey GT. Utility of the white gene in estimating phylogenetic relationships among mosquitoes (Diptera: Culicidae) Mol Biol Evol. 1997;14(4):442–454. doi: 10.1093/oxfordjournals.molbev.a025780. [DOI] [PubMed] [Google Scholar]
- 16.Arensburger P, et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330(6000):86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buffa FM, Harris AL, West CM, Miller CJ. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br J Cancer. 2010;102(2):428–435. doi: 10.1038/sj.bjc.6605450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302(5643):249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 19.Sharakhov IV, et al. Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion (2La) in the Anopheles gambiae complex. Proc Natl Acad Sci USA. 2006;103(16):6258–6262. doi: 10.1073/pnas.0509683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang MH, et al. Genome-wide patterns of gene expression during aging in the African malaria vector Anopheles gambiae. PLoS ONE. 2010;5(10):e13359. doi: 10.1371/journal.pone.0013359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christophides GK, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298(5591):159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 22.Michel K, Budd A, Pinto S, Gibson TJ, Kafatos FC. Anopheles gambiae SRPN2 facilitates midgut invasion by the malaria parasite Plasmodium berghei. EMBO Rep. 2005;6(9):891–897. doi: 10.1038/sj.embor.7400478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volz J, Müller HM, Zdanowicz A, Kafatos FC, Osta MA. A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol. 2006;8(9):1392–1405. doi: 10.1111/j.1462-5822.2006.00718.x. [DOI] [PubMed] [Google Scholar]
- 24.Zieler H, Dvorak JA. Invasion in vitro of mosquito midgut cells by the malaria parasite proceeds by a conserved mechanism and results in death of the invaded midgut cells. Proc Natl Acad Sci USA. 2000;97(21):11516–11521. doi: 10.1073/pnas.97.21.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valanne S, Wang JH, Rämet M. The Drosophila Toll signaling pathway. J Immunol. 2011;186(2):649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- 26.Fullaondo A, et al. Spn1 regulates the GNBP3-dependent Toll signaling pathway in Drosophila melanogaster. Mol Cell Biol. 2011;31(14):2960–2972. doi: 10.1128/MCB.01397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein LD. Using GBrowse 2.0 to visualize and share next-generation sequence data. Brief Bioinform. 2013;14(2):162–171. doi: 10.1093/bib/bbt001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Weetman D, et al. Association mapping of insecticide resistance in wild Anopheles gambiae populations: Major variants identified in a low-linkage disequilbrium genome. PLoS ONE. 2010;5(10):e13140. doi: 10.1371/journal.pone.0013140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitri C, et al. Fine pathogen discrimination within the APL1 gene family protects Anopheles gambiae against human and rodent malaria species. PLoS Pathog. 2009;5(9):e1000576. doi: 10.1371/journal.ppat.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segal E, Friedman N, Kaminski N, Regev A, Koller D. From signatures to models: Understanding cancer using microarrays. Nat Genet. 2005;37(Suppl):S38–S45. doi: 10.1038/ng1561. [DOI] [PubMed] [Google Scholar]
- 32.Hirschhorn R, Vawter GF, Kirkpatrick JA, Jr, Rosen FS. Adenosine deaminase deficiency: Frequency and comparative pathology in autosomally recessive severe combined immunodeficiency. Clin Immunol Immunopathol. 1979;14(1):107–120. doi: 10.1016/0090-1229(79)90131-4. [DOI] [PubMed] [Google Scholar]
- 33.Molina-Cruz A, et al. Some strains of Plasmodium falciparum, a human malaria parasite, evade the complement-like system of Anopheles gambiae mosquitoes. Proc Natl Acad Sci USA. 2012;109(28):E1957–E1962. doi: 10.1073/pnas.1121183109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong Y, Dimopoulos G. Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J Biol Chem. 2009;284(15):9835–9844. doi: 10.1074/jbc.M807084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dissanayake SN, Marinotti O, Ribeiro JM, James AA. angaGEDUCI: Anopheles gambiae gene expression database with integrated comparative algorithms for identifying conserved DNA motifs in promoter sequences. BMC Genomics. 2006;7:116. doi: 10.1186/1471-2164-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker DA, et al. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics. 2011;12:296. doi: 10.1186/1471-2164-12-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 40.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.ERamer U. An iterative procedure for the polygonal approximation of plane curves. Computer Graphics and Image Processing. 1972;1(3):244–256. [Google Scholar]
- 43.Graber JH, Cantor CR, Mohr SC, Smith TF. Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res. 1999;27(3):888–894. doi: 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49(4):520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 45.Li R, et al. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.