Abstract
The inositol-depletion hypothesis proposes that lithium attenuates phosphatidylinositol signaling. Knockout (KO) mice of two genes (IMPA1 or Slc5a3), each encoding for a protein related to inositol metabolism, were studied in comparison with lithium-treated mice. Since we previously demonstrated that these KO mice exhibit a lithium-like neurochemical and behavioral phenotype, here we searched for pathways that may mediate lithium's/the KO effects. We performed a DNA-microarray study searching for pathways affected both by chronic lithium treatment and by the KO of each of the genes. The data were analyzed using three different bioinformatics approaches. We found upregulation of mitochondria-related genes in frontal cortex of lithium-treated, IMPA1 and Slc5a3 KO mice. Three out of seven genes differentially expressed in all three models, Cox5a, Ndufs7, and Ndufab, all members of the mitochondrial electron transfer chain, have previously been associated with bipolar disorder and/or lithium treatment. Upregulation of the expression of these genes was verified by real-time PCR. To further support the link between mitochondrial function and lithium's effect on behavior, we determined the capacity of chronic low-dose rotenone, a mitochondrial respiratory chain complex I inhibitor, to alter lithium-induced behavior as measured by the forced-swim and the amphetamine-induced hyperlocomotion paradigms. Rontenone treatment counteracted lithium's effect on behavior, supporting the proposition suggested by the bioinformatics analysis for a mitochondrial function involvement in behavioral effects of lithium mediated by inositol metabolism alterations.The results provide support for the notion that mitochondrial dysfunction is linked to bipolar disorder and can be ameliorated by lithium. The phenotypic similarities between lithium-treated wild-type mice and the two KO models suggest that lithium may affect behavior by altering inositol metabolism.
Keywords: lithium, bipolar disorder, inositol, DNA microarrays, knockout mice, mitochondria
INTRODUCTION
Lithium salts (Li) are among the main drugs used in the treatment of bipolar disorder, however, the mechanism by which Li exerts its mood-stabilizing effect is not yet unraveled. Li has been reported to have numerous effects on gene expression (Toker et al, 2012). The evolvement of large-scale gene expression analysis methodologies provided a promising tool to understand the cellular events underlying the mood-stabilizing effect of the drug. Several microarray studies of therapeutically relevant doses of Li have previously been performed in an attempt to find new potential drug targets, however, while the list of candidate genes increased, they were not replicated among the studies, suggesting that functional rather than single gene analysis might be more appropriate for analyzing large-scale gene expression data sets in this field (Konradi et al, 2012).
The inositol-depletion hypothesis proposes that Li acts by depletion of brain myo-inositol (Berridge et al, 1989). The inositol-depletion hypothesis is supported by the observation that decrease in inositol is common to two other widely used mood stabilizers, valproic acid (VPA) and carbamazepine (CBZ) (O'Donnell et al, 2000; Shaltiel et al, 2004). Several previous microarray studies of Li-induced changes in gene expression levels implied the involvement of inositol signaling-related genes. Brandish et al (2005) studied acute Li-treated rat cortical slices either with or without co-treatment with the muscarinic agonist carbachol. ADCYAP1, encoding for the pituitary adenylate cyclase activating polypeptide (PACAP), was among the genes affected by lithium and carbachol co-treatment. The effect could be prevented by inositol supplementation. Interestingly, ADCYAP1 knockout (KO) mice were shown to exhibit several bipolar disorder-related phenotypes (Hashimoto et al, 2009; Kawaguchi et al, 2003). In a more recent microarray study by McQuillin et al (2007) over 10 genes related to the phosphatidylinositol (PI) signaling system were shown to be affected by chronic lithium treatment in mouse brain. Changes in PI cycle-related genes were also reported in two additional microarray studies assessing Li's action (Bosetti et al, 2002; Seelan et al, 2008).
In an attempt to provide evidence for the inositol-depletion hypothesis, we have studied two strains of KO mice with deletion of genes involved in PI signal transduction, IMPA1 and Slc5a3 (Agam et al, 2009). Both of these genes encode for proteins known to be affected by chronic Li treatment (inositol monophosphatase1 designated as IMPase1 and sodium-myo-inositol transporter designated as SMIT1) (Hallcher and Sherman, 1980; Lubrich and van Calker, 1999). These KO mice share several mood stabilization-related behavioral and neurochemical characteristics with lithium treatment, favoring the notion that inositol metabolism is involved in the mechanism of Li-induced behavioral changes (Agam et al, 2009). Thus, they seemed to provide an opportunity to examine which of the many reported effects of lithium on gene expression could potentially be involved in Li-induced behavioral effects, specifically those mediated by the drug's effect on inositol metabolism.
Here, we present a first functional bioinformatics analysis of microarray data from frontal cortex of untreated wild-type (WT) mice, Li-treated WT mice, IMPA1 homozygote KO mice (IMPA1 KO), and Slc5a3 homozygote KO mice (SMIT1 KO). We further provide verification by q-PCR analysis of the expression of some single genes and present a pharmacological intervention to substantiate the bioinformatics results.
MATERIALS AND METHODS
Supplementary Figure S1 presents a schematic layout of the study.
Animals
Male, 8-week-old mice were used in all the experiments. Animals were maintained on a 12-h/12-h light/dark cycle (lights on between 0800 and 1200 h) with ad libitum access to food and water. All tests were performed during the light phase of the cycle between 0900 and 1900 h. SMIT1 and IMPA1 KO mice were originally generated as previously described (Berry et al, 2003; Cryns et al, 2008). Each KO strain is maintained in a separate colony in our animal-care facility by breeding heterozygote males and females. Myo-inositol supplementation in drinking water (3.5% w/v) to pregnant and lactating dams is used to rescue the KO mice. Genotyping was carried out as previously described (Bersudsky et al, 2008; Cryns et al, 2008). ICR mice (Harlan, Israel) were allowed to acclimatize to the new environment for 1 week before treatment initiation. All experiments were approved by the Ben-Gurion University animal experimentation ethics committee.
RNA Extraction
Total RNA was extracted from frontal cortex specimens using the TRI reagent (Sigma-Aldrich, St Louis, MO, USA) followed by purification using the RNeasy kit (Qiagen, Germantown, MD, USA). RNA concentration was determined spectrophotometrically (Nano Drop 2000, Thermo Fisher Scientific, Waltham, MA, USA).
DNA-Microarray Gene Expression
Since IMPA1 KO and SMIT1 KO mice were generated on different backgrounds (C57BL/129x1/Svj for SMIT1 and C57BL6/129Sv/EvBrd for IMPA1), the microarray analysis was carried out separately for each colony. In each colony, three groups with four animals per group were examined (except for WT-untreated mice of the SMIT1 colony of which three animals were studied): WT-untreated, WT-Li treated, and KO (IMPA1 or SMIT1). Extracted RNA of each mouse was hybridized to one Affymetrix Mouse Gene 1.0 ST chip. Hybridization was carried out at the DNA microarrays and DNA sequencing Laboratory of the National Institute for Biotechnology in the Negev, Beer-Sheva, Israel, affiliated to our university. Microarray chips and reagents used in the experiment of a given colony were from the same Affymetrix batch.
Statistical Analysis of the DNA Microarrays
Initial data analysis was supervised by VC-C from the Bioinformatics Core Facility, National Institute for Biotechnology in the Negev, using the Partek and SpotFire software. Briefly, signals were log2 transformed and preprocessed using the robust multi-chip average (RMA) method (Irizarry et al, 2003). Principal component analysis (PCA) was performed for each colony separately. It revealed clustering among the 12 samples (designated as ‘batch effect'). In both colonies, a batch effect was found which was balanced with regard to the biological groups. Probe sets with log2-transformed signals below a value of five in all samples were considered as ‘noise' and excluded from further analysis. Initially, three-way mixed model ANOVA was used to analyze the difference between the colonies regarding the effect of Li. The following effects were included in the analysis: WT-Li vs WT-untreated as the biological effect, colony (IMPA1 or SMIT1) and batch as random effects (batch effect relates to the overall differences between the experiment of the IMPA1 colony vs the SMIT1 colony in terms of the different colony background, the Affymetrix batches used for each of the colonies, the experimental dates, etc.). Subsequently, two-way mixed model ANOVA was carried out separately in each colony to assess the effect of Li treatment and of the KO of each of the genes. The following effects were used in the analysis: type (WT-untreated, WT Li-treated, and KO) as the biological effect and batch as the random effect. Differentially expressed genes were defined as those having P<0.05 compared with WT-untreated mice.
For clustering analysis, signals were batch-corrected using the Partek software. Genes showing differential expression in either Li-treated animals or KO animals were included in the analysis. The Hierarchical Clustering tool of the SpotFire software was used to create heatmap and dendrogram of Z-score transformed signals.
Network and Pathway Analyses
The Functional Annotation Tool in DAVID 6.7 (the Database for Annotation, Visualization and Integrated Discovery) bioinformatics software was used according to the publisher's recommendations (Huang da et al, 2009) for enrichment analysis of the genes differentially expressed in the same direction in the KO mice and Li-treated WT mice compared with WT-untreated mice in each colony. To gain further information about the direction of the enrichment change, two additional enrichment analyses were performed separately for the upregulated and the downregulated genes.
The raw data of the microarray results were uploaded onto the Gene Set Enrichment Analysis (GSEA) V2.5 software (Subramanian et al, 2005), a computational method that determines whether an a priori defined set of genes (‘gene sets') shows statistically significant concordant differences between two biological states. GSEA calculates an enrichment score (ES) for a given gene set using a ranked list of genes. The rank represents the correlation between the expression of the gene and the phenotype. ES is calculated by walking down the ranked list of genes, increasing a running-sum statistic when a gene is in the gene set and decreasing it when it is not. Further, the software infers the statistical significance of each ES against background ES distribution calculated by randomly shuffling the labels on the samples (phenotype permutation) or by creating random gene sets matching the actual size of a particular gene set (gene_set permutation). Given the relatively small sample size in our study, we used the set-up of 1000 gene-set permutations for analysis and a false discovery rate (FDR) cutoff of q-value<0.05 to identify differentially expressed sets of genes, as advised by the software team (http://www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html), using an a priori defined sets of genes (GO gene-set collection ‘c5.all.v2.5.symbols').
Data were further analyzed through the use of IPA (Ingenuity Systems, www.ingenuity.com) to identify pathways and functions affected by IMPA1 KO, Slc5a3 KO, and Li treatment. To reach the recommended number of genes to be used in the analysis, the cutoffs for P-value and fold of change (f.c.) were set to P<0.05, |f.c.|>1.1 and P<0.035, |f.c|>1.15 for the IMPA1 and the SMIT1 colonies, respectively. The functional analysis identifies the biological functions and/or diseases that were most significant to the data set. Genes from the uploaded data set that met the cutoffs mentioned above and were associated with biological functions and/or diseases in the Ingenuity Knowledge Base were considered for the analysis. Right-tailed Fisher's exact test was used to calculate a P-value determining the probability that each biological function and/or disease assigned to that data set is due to chance only. Canonical pathway analysis was used to identify the pathways in the IPA library of canonical pathways that were most significant to the uploaded data set. Genes from the data set that met the above-mentioned cutoffs and were associated with a canonical pathway in the Ingenuity Knowledge Base were considered for the analysis. The significance of the association between the data set and the canonical pathway was measured in two ways: (1) A ratio of the number of genes from the data set that map to the pathway, divided by the total number of molecules that map to the canonical pathway. (2) Fisher's exact test was used to calculate the P-value determining the probability that the association between the genes in the data set and the canonical pathway is explained by chance only.
Real-Time PCR
RNA samples were extracted from frontal cortex of the KO, Li-treated WT mice, and WT-untreated mice from IMPA1 and SMITA colonies (IMPA1 colony, n=13, 8, and 10, respectively; SMITA colony, n=10, 4 and 8, respectively), as described under RNA extraction. RNA was reverse transcribed using the Verso cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA). MAPK6 was used as the reference gene since it has been recommended as a brain control gene (Cai et al, 2007). Using our microarray data, we confirmed that the expression of this gene is not significantly different among the groups. Real-time PCR was performed using an ABsolute Blue Sybr mix (ABgene, Lithuania) and an Eco qPCR system (Illumina, San Diego, CA, USA). The relative expression of each gene was calculated by the Pfaffl method (Pfaffl, 2001) implemented in the Eco qPCR system software. A pool of all the samples was used for normalization. The thermal cycling program was as follows: hold on 95°C for 15 min. followed by 34 cycles of: 10 s 95°C, 30 s 58°C, 25 s 72°C. Supplementary Table S1 contains the primers' sequences for the genes examined and the respective efficiencies of their reactions.
Drugs
Rotenone (Sigma-Aldrich, St Louis, MO) was dissolved (0.1 mg/ml) in saline supplemented with 0.5% DMSO and injected subcutaneously (s.c.), 0.5 mg/kg. D-Amphetamine (Sigma-Aldrich, Gillinfhuam, UK) was dissolved (0.15 mg/ml) in saline and injected intraperitoneally (i.p.), 1.5 mg/kg. The dose and schedule of rotenone and amphetamine were chosen based on (a) the rationale that the model should represent gradual mitochondrial dysfunction preceding Li treatment, (b) doses previously reported in the literature, and (c) our preliminary results. We used a rotenone dose lower than previously reported to induce a Parkinson's-like phenotype (Greenamyre et al, 2010) to avoid motor dysfunction but high enough to obtain behavioral effects. The amphetamine dose was chosen so that it would be high enough to induce noticeable hyperactivity but lower than that resulting in a ceiling effect/stereotypic behavior.
Chronic Lithium Treatment
Chronic lithium treatment was carried out as previously described (O'Brien et al, 2004). Animals were weighed every 2 days during the treatment period. The measured serum Li levels following this treatment regime were 0.89±0.43 mmol/L (SD)
Rotenone Treatment
ICR mice were injected once daily s.c. with rotenone or vehicle for 4 weeks. After 2 weeks of injections, each of the groups was divided into two subgroups of Li treatment and regular food as described above for lithium treatment. All animals were daily weighed and their general well-being was assessed by examination of their fur and general appearance. After 4 weeks of treatment, all animals were evaluated for motor function by the hanging wire and rotarod tests a day before the mood-related behavioral tests. Animals with aberrant performance in either of the motor tests (exhibiting scores exceeding the mean±2SD of their group) were excluded from further experiments. Overall, five mice were excluded from the experiment based on the altered motor ability; three from the regular food+rotenone group and two from the regular food+vehicle group. The FST and amphetamine-induced hyperlocomotion tests were performed as described elsewhere (Bersudsky et al, 2007; Creson et al, 2011) on consecutive days.
Hanging Wire Test
Each animal was placed on a metal grid that was gently shaken to cause a grabbing reflex and slowly turned upside down. Latency to fall was counted during a session of 1 min, as previously described (Crawley, 1999).
Rotarod Test
The protocol for the test is a modified version of Monville et al (2006). Animals were subjected to a series of 1-min sessions separated by 10 min on the rotarod apparatus (ROTA-ROD/RS, Letica Scientific Instruments, Panlab, Barcelona, Spain) using the following protocol: 0 r.p.m., 4 r.p.m., three sessions of accelerating 4–40 r.p.m. (two training sessions followed by the test session). Latency to fall was counted for each session.
Statistical Analyses
Two-way ANOVA was used for the analyses of FST, hanging wire, and rotarod tests. Amphetamine-induced hyperlocomotion results were analyzed by two-way repeated measures ANOVA. Significant ANOVA results were followed by Fisher's LSD post hoc test.
In IMPA1 colony, statistical analysis of real-time PCR results was carried out by one-way ANOVA performed separately for each gene followed by post hoc Fisher's LSD test. ANOVA was not applicable in the SMIT1 colony due to heterogeneity of variances and the unbalanced sample size among the groups studied. Thus, the results were analyzed by Welch's t-test.
RESULTS
Gene Expression Analysis
Since IMPA1 KO and SMIT1 KO mice were generated on different backgrounds, the microarray analysis was carried out separately for each colony (further referred to as the SMIT1 colony or the IMPA1 colony). Three-way mixed model ANOVA of WT-untreated and WT Li-treated mice from both colonies revealed that the major source of variation between the groups was the colony (Supplementary Figure S2), supporting our a priori approach to perform the expression analysis separately for each colony.
IMPA1 colony: in the frontal cortex of both IMPA1 KO mice and Li-treated WT mice as compared with WT-untreated mice, 193 genes were similarly differentially expressed (either upregulated or downregulated) at a statistical significance level of P<0.05. SMIT1 colony: in the frontal cortex of both SMIT1 KO mice and Li-treated WT mice as compared with WT-untreated mice, 778 genes were similarly differentially expressed at a level of P<0.05. Hierarchical clustering of 4643 genes differentially expressed by Li treatment or the KO in either of the colonies showed two major clusters of the samples corresponding to SMIT1 and IMPA1 colonies (Supplementary Figure S3). Only one gene cluster consisting of 419 annotated genes (Supplementary Excel file) showed a similar expression pattern between the two colonies. Functional annotation of this gene cluster using the DAVID software revealed that almost all functional annotations with FDR<0.05 were related to mitochondria, specifically to respiratory function (Supplementary Figure S4). In addition, a small number of the functional annotations (6/36) were related to ribosomal function.
Analysis of the Differentially Expressed Genes Using the DAVID Software
For each colony, genes with similar differential expression (P<0.05) in the frontal cortex of the KO mice and the Li-treated WT mice as compared with WT-untreated mice were uploaded onto the software. Out of the uploaded genes, 159 and 654 genes from the IMPA1 and SMIT1 colonies, respectively, were identified by the software and used for the analysis. In both colonies, significantly enriched terms and significant KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways (Bonferroni corrected P<0.05, FDR<0.05) were related to mitochondria (Table 1; Supplementary Figure S5A and B). KEGG pathways significantly enriched in both colonies were Parkinson's disease, Alzheimer's disease, Huntington's disease, and oxidative phosphorylation. Hence, enrichment in oxidative phosphorylation-related genes is identified by the software as enrichment in the pathways related to these three diseases. Indeed, almost all of the genes included in those pathways were part of the mitochondrial electron transport chain. Interestingly, within the oxidative phosphorylation complexes, the differentially expressed genes belong to all complexes of the electron transfer chain except for complex II, consistent with the previously reported differential effect of lithium on the mitochondrial complexes (Maurer et al, 2009) (Supplementary Figure S6A and B).
Table 1. DAVID Functional Annotation Tool.
| Category | Term | P-value | FDR |
|---|---|---|---|
| IMPA1 colony | |||
| KEGG_pathway | Oxidative phosphorylation | 1.2 × 10−7 | 2.1 × 10−6 |
| KEGG_pathway | Parkinson's disease | 1.9 × 10−6 | 3.4 × 10−5 |
| KEGG_pathway | Huntington's disease | 5.7 × 10−6 | 1.0 × 10−4 |
| KEGG_pathway | Alzheimer's disease | 4.8 × 10−5 | 8.5 × 10−4 |
| Cellular compartment | Organelle envelope | 4.8 × 10−4 | 3 × 10−3 |
| Cellular compartment | Envelope | 5 × 10−4 | 3.1 × 10−3 |
| Cellular compartment | Mitochondrial envelope | 9.5 × 10−4 | 5.9 × 10−3 |
| Cellular compartment | Mitochondrial inner membrane | 1.4 × 10−3 | 8.5 × 10−3 |
| Cellular compartment | Organelle inner membrane | 2.3 × 10−3 | 1.5 × 10−2 |
| Cellular compartment | Mitochondrial membrane | 2.5 × 10−3 | 1.5 × 10−2 |
| Cellular compartment | Respiration chain | 3.4 × 10−3 | 2.1 × 10−2 |
| Cellular compartment | Mitochondrial part | 6.1 × 10−3 | 3.8 × 10−2 |
| SMIT1 colony | |||
| Cellular compartment | Mitochondrion | 4.1 × 10−9 | 1.6 × 10−8 |
| Cellular compartment | Mitochondrial part | 4.2 × 10−7 | 1.6 × 10−6 |
| Cellular compartment | Organelle membrane | 6.4 × 10−7 | 3.3 × 10−6 |
| KEGG_pathway | Parkinson's disease | 1.1 × 10−6 | 1.3 × 10−5 |
| Cellular compartment | Mitochondrial membrane | 3.6 × 10−6 | 1.4 × 10−5 |
| KEGG_pathway | Huntington's disease | 5.7 × 10−6 | 5 × 10−5 |
| Cellular compartment | Mitochondrial envelope | 1.7 × 10−5 | 6.6 × 10−5 |
| Cellular compartment | Mitochondrial inner membrane | 2.3 × 10−5 | 8.9 × 10−5 |
| KEGG_pathway | Oxidative phosphorylation | 1.1 × 10−5 | 9.2 × 10−5 |
| Cellular compartment | Organelle inner membrane | 7.6 × 10−5 | 3.0 × 10−4 |
| Biological process | Electron transport chain | 1.9 × 10−3 | 1.8 × 10−3 |
| Cellular compartment | Respiration chain | 7.9 × 10−4 | 3.1 × 10−3 |
| Cellular compartment | Organelle envelope | 2.8 × 10−3 | 1.1 × 10−2 |
| Cellular compartment | Envelope | 3 × 10−3 | 1.2 × 10−2 |
| Biological process | Generation of precursor metabolites and energy | 1.5 × 10−2 | 1.4 × 10−2 |
Abbreviation: FDR, false discovery rate.
The table summarizes the main findings obtained from DAVID Functional Annotation Tool analysis. For the snapshot of the original output, please refer to Supplementary Figure S5.
GO terms and KEGG pathways significantly enriched among genes with common differential expression in KO mice and WT Li-treated mice compared with WT-untreated mice.
Separate analyses of the upregulated and the downregulated genes showed that enrichment in mitochondria-related terms appears only among the upregulated genes (Supplementary Figure S7A and B). No significantly enriched terms (FDR<0.05) were found among the downregulated genes.
Analysis using the GSEA Software
For both KOs, the most significant gene set found using an FDR cutoff of q-value<0.05 was ‘structural_constituent_of ribosome' followed by various mitochondria-related gene sets (Table 2), all exhibiting upregulation as compared with WT-untreated mice. No significantly differentially expressed gene sets were obtained in the Li-treatment groups, a result possibly related to the observation that Li treatment in humans affects diseased individuals much more than healthy subjects, corresponding to greater behavioral effects of the drug in challenged animals than animals under baseline conditions (Marchese et al, 2013; Saul et al, 2012).
Table 2. GSEA Analysis.
| Name | Size | FDR q-value | P-value | NES |
|---|---|---|---|---|
| IMPA1 KO mice vs WT-untreated mice of the IMPA1 colony | ||||
| Structural constituent of ribosome | 77 | 2.14E−03 | <1.00E−04 | 2.15 |
| Mitochondrial membrane part | 50 | 1.02E−02 | <1.00E−04 | 2.01 |
| Mitochondrion | 300 | 8.56E−03 | <1.00E−04 | 1.99 |
| Mitochondrial part | 132 | 3.86E−02 | <1.00E−04 | 1.90 |
| Synapse organization and biogenesis | 18 | 8.90E−04 | <1.00E−04 | −2.30 |
| Synaptogenesis | 15 | 4.45E−03 | <1.00E−04 | −2.19 |
| Extracellular structure organization and biogenesis | 22 | 1.02E−02 | <1.00E−04 | −2.08 |
| SMIT1 KO mice vs WT-untreated mice of the SMIT1 colony | ||||
| Structural constituent of ribosome | 77 | <1.00E−04 | <1.00E−04 | 2.69 |
| Ribonucleoprotein complex | 121 | <1.00E−04 | <1.00E−04 | 2.35 |
| Ribosome | 35 | <1.00E−04 | <1.00E−04 | 2.34 |
| Organellar ribosome | 21 | 1.93E−04 | <1.00E−04 | 2.23 |
| Mitochondrial ribosome | 21 | 3.18E−04 | <1.00E−04 | 2.20 |
| Mitochondrial matrix | 42 | 2.11E−03 | <1.00E−04 | 2.12 |
| Mitochondrial lumen | 42 | 2.16E−03 | <1.00E−04 | 2.10 |
| Ribosomal subunit | 19 | 2.10E−03 | <1.00E−04 | 2.09 |
| Mitochondrial part | 130 | 5.68E−03 | <1.00E−04 | 2.02 |
| Mitochondrial membrane part | 50 | 6.55E−03 | <1.00E−04 | 2.00 |
| Active transmembrane transporter activity | 78 | 2.32E−03 | <1.00E−04 | −2.30 |
| Transmembrane receptor protein kinase activity | 42 | 1.72E−03 | <1.00E−04 | −2.24 |
| Receptor activity | 321 | 1.92E−03 | <1.00E−04 | −2.19 |
| Protein tyrosine kinase activity | 46 | 1.44E−03 | <1.00E−04 | −2.17 |
| Metallopeptidase activity | 29 | 2.32E−03 | <1.00E−04 | −2.14 |
| Transmembrane receptor protein tyrosine kinase activity | 34 | 1.94E−03 | <1.00E−04 | −2.14 |
| Secondary active transmembrane transporter activity | 27 | 3.17E−03 | <1.00E−04 | −2.10 |
| Substrate-specific transporter activity | 246 | 3.20E−03 | <1.00E−04 | −2.10 |
| Transmembrane transporter activity | 232 | 4.42E−03 | <1.00E−04 | −2.07 |
| Transmembrane receptor activity | 225 | 5.50E−03 | <1.00E−04 | −2.05 |
| Substrate-specific transmembrane transporter activity | 216 | 5.21E−03 | <1.00E−04 | −2.04 |
| Ion transmembrane transporter activity | 167 | 7.01E−03 | <1.00E−04 | −2.01 |
| Symporter activity | 16 | 7.54E−03 | 3.63E−03 | −2.00 |
| Cation transmembrane transporter activity | 138 | 7.43E−03 | <1.00E−04 | −2.00 |
| Cell projection biogenesis | 20 | 1.24E−02 | 1.94E−03 | −1.96 |
| Anion transmembrane transporter activity | 26 | 1.90E−02 | <1.00E−04 | −1.92 |
| Membrane fraction | 224 | 2.51E−02 | <1.00E−04 | −1.89 |
Gene_Sets showing a significant difference between KO mice and WT-untreated mice.
Size=number of genes included in a particular gene set; FDR q-value=false discovery rate, adjusted to the gene-set size and multiple hypotheses testing; NES=normalized enrichment score (ES) calculated by dividing the actual ES by the mean ES against all permutations performed. A positive score indicates upregulation of the gene set in KO mice as compared with WT-untreated mice. A negative score indicates downregulation of the gene set in KO mice as compared with WT-untreated mice.
Analysis Using the IPA Software
As seen in Supplementary Figure S8A, using the cutoffs of P<0.05, f.c.>1.1 and P<0.035, f.c>1.15 for the IMPA1 and SMIT1 colonies, respectively (the choice of these cutoffs is justified under Materials and methods, and Network and Pathway Analyses sections), multiple biological functions were significantly affected both by Li treatment in the two colonies and by IMPA1 or Slc5a3 KO. However, the common significantly affected canonical pathways in the four groups (WT Li-treated mice from the SMIT1 colony, WT Li-treated mice from the IMPA1 colony, SMIT1 KO, and IMPA1 KO) were oxidative phosphorylation, mitochondrial dysfunction, and ubiquinone biosynthesis, all related to mitochondria (Table 3; Supplementary Figure S8B). Supplementary Figure S9 represents the Venn diagram of the significantly upregulated (A) and downregulated (B) genes.
Table 3. Canonical Pathways Found by the IPA Software to be Significantly Affected in the Frontal Cortex of Li-Treated WT Mice, IMPA1 KO Mice, and SMIT1 KO Mice.
| Canonical pathway | Ratioa IMPA1 KO | P-value IMPA1 KO | Ratio IMPA1_Li | P-value IMPA_Li | Ratio SMIT1 KO | P-value SMIT1 KO | Ratio SMIT_Li | P-value SMIT_Li |
|---|---|---|---|---|---|---|---|---|
| Oxidative phosphorylation | 17/166 | 6.77E−06 | 13/166 | 2.43E−04 | 10/166 | 5.14E−03 | 19/166 | 3.86E−06 |
| Mitochondrial dysfunction | 14/125 | 8.17E−05 | 8/125 | 2.25E−02 | 12/125 | 1.26E−04 | 17/125 | 3.89E−06 |
| Ubiquinone biosynthesis | 10/119 | 2.33E−04 | 8/119 | 1.48E−03 | 7/119 | 1.41E−02 | 9/119 | 2.12E−03 |
Ratio indicates the number of genes from the data set that map to the pathway divided by the total number of genes that map to the canonical pathway.
Gene expression data of the IMPA1 KO mice and Li-treated WT mice from the IMPA1 colony (IMPA1_Li), SMIT1 KO mice and Li-treated WT mice from the SMIT1 colony (SMIT_Li) were subjected to the IPA canonical pathway analysis. Genes from the uploaded data sets that met the cutoffs (P<0.05, f.c.>1.1 and P<0.035, f.c.>1.15 for the IMPA1 and the SMIT1 colonies, respectively, as justified under Materials and methods) and that were associated with a canonical pathway in the Ingenuity Knowledge Base were considered for the analysis. Fisher's exact test was used to calculate the P-value determining the probability that the association between the genes in the data set and the canonical pathway is explained by chance only.
Single Gene Expression
The expression of seven annotated genes was significantly changed (P<0.05) in the same direction (upregulation or downregulation) in IMPA1 and in SMIT1 KO mice and in Li-treated WT mice of both colonies as compared with their respective WT-untreated littermates (Table 4). Four of the genes, Cox5a, Ndufab1, Ndufs7, and Uqcrfs1, participate in the oxidative phosphorylation pathway and were all upregulated compared with WT-untreated mice. Three of these genes, Cox5a, Ndufs7, and Ndufab1, have previously been indicated by at least two independent studies to be upregulated by lithium treatment, downregulated in bipolar patients or genetically associated with bipolar disorder (Andreazza et al, 2010; Konradi et al, 2004; Sun et al, 2006; Washizuka et al, 2005; WTCCC, 2007).
Table 4. Genes with Common Expression Change in IMPA1 KO, SMIT1 KO, and Li-Treated Mice.
| Gene symbol | Entrez gene name | Fold of change SMIT KO | P-value SMIT KO | Fold of change SMIT_Li | P-value SMIT_Li | Fold of change IMPA KO | P-value IMPA KO | Fold of change IMPA_Li | P-value IMPA_Li |
|---|---|---|---|---|---|---|---|---|---|
| Cox5aa | Cytochrome C oxidase, subunit Va | 1.27 | 0.004 | 1.22 | 0.007 | 1.16 | 0.02 | 1.18 | 0.016 |
| Gaa | Glucosidase, alpha, acid | −1.32 | 0.03 | −1.4 | 0.01 | −1.2 | 0.015 | −1.16 | 0.036 |
| Gemin4 | gem (nuclear organelle) associated protein 4 | −1.18 | 0.016 | −1.25 | 0.003 | −1.12 | 0.013 | −1.15 | 0.006 |
| Ndufab1a | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, | 1.16 | 0.019 | 1.2 | 0.005 | 1.2 | 0.01 | 1.14 | 0.03 |
| Ndufs7a | NADH dehydrogenase (ubiquinone) Fe-S protein 7 | 1.2 | 0.042 | 1.22 | 0.02 | 1.15 | 0.014 | 1.13 | 0.02 |
| Rc3h1 | RING CCCH (C3H) domains 1 | −1.11 | 0.02 | −1.22 | 0.0005 | −1.08 | 0.04 | −1.1 | 0.02 |
| Uqcrfs1 | Ubiquinol-cytochrome C reductase, Rieske iron-sulfur polypeptide 1 | 1.15 | 0.03 | 1.3 | 0.0008 | 1.2 | 0.009 | 1.14 | 0.04 |
Genes previously reported to be upregulated by lithium treatment or downregulated in/genetically associated with bipolar disorder in at least two independent studies. All of them are related to mitochondrial function.The designations SMIT_Li and IMPA_Li as in Table 3.
We performed quantitative real-time PCR analysis of Cox5a, Ndufs7, and Ndufab1 in the frontal cortex of IMPA1 KO mice, SMIT KO mice and their WT Li-treated and untreated littermates (Supplementary Figure S10A and B). In general, the results were consistent with those obtained in the microarrays analysis: the expression of the three genes was modestly higher both in IMPA1 KO and in SMIT1 KO and in WT Li-treated mice compared with their WT-untreated littermates. Yet, possibly due to the relatively small expression changes and the greater variability in the results obtained using a real-time PCR, these differences reached statistical significance only for some of the groups and some of the genes studied.
The non-mitochondrial genes with a similar significant expression change in all four groups, Gaa, Gemin4, and Rc3h1, have not previously been reported to be related to lithium treatment and/or to bipolar disorder, and did not cluster to any particular function. Thus, their relevance to lithium's therapeutic effect remains to be investigated.
Supplementary Table S2 presents genes differentially expressed in some, but not in all four groups of mice (IMPA1 and SMIT1 KO mice and Li-treated WT mice of both colonies as compared with their respective WT-untreated littermates). These genes were shown to be affected by lithium treatment in previous microarray studies.
Rotenone-Induced Behavioral Effects
On the basis of our microarray results and previous reports of reduced mitochondrial function in bipolar patients (Andreazza et al, 2010; Stork and Renshaw, 2005), we hypothesized that chronic mild inhibition of oxidative phosphorylation would induce a bipolar disorder-like behavioral phenotype that would be ameliorated by chronic lithium treatment. Chronic treatment with rotenone, an oxidative phosphorylation complex I inhibitor, is used to model Parkinson's disease. However, the doses shown to elicit Parkinson's-like effects are higher than those used in our study.
Mice were divided into groups according to two main factors: (1) rotenone injections (yes or no) and (2) lithium-supplemented food (yes or no), creating four groups: regular food+vehicle, chronic Li treatment+vehicle, regular food+chronic rotenone treatment, and chronic Li treatment+chronic rotenone treatment.
No difference was found between rotenone-treated and untreated mice in weight, well-being (monitored by the examination of their fur and general appearance) or general motor function (Supplementary Table S3), confirming that rotenone concentration used did not affect the general condition of the animals. Sporadic mice exhibiting altered motor ability in either the hanging wire or the rotarod tests were excluded from further experiments.
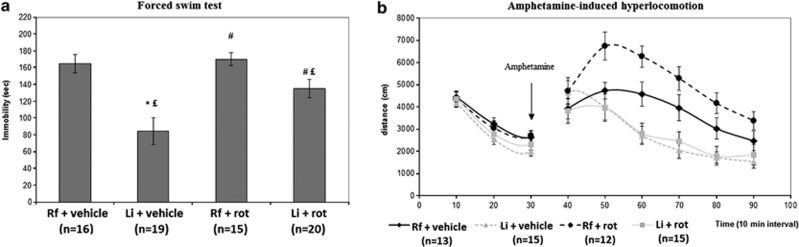
In the FST, as expected (Bersudsky et al, 2007; O'Brien et al, 2004), lithium treatment exerted an antidepressant-like effect, reducing the immobility time as compared with both regular food groups, either rotenone pretreated or vehicle pretreated (Figure 1a). Two-way ANOVA revealed a significant treatment effect of lithium (F1,68=29.04, P<0.000001), a significant effect of rotenone (F1,68=6.92, P<0.011), and a significant lithium × rotenone interaction (F1,68=4.5, P<0.038). Lithium reduced the immobility time of vehicle-pretreated mice by 48% (regular food+vehicle, 165±39 sec; Li+vehicle, 85±49 s, Fisher's LSD post hoc test, P<10−5), an effect twice stronger than in rotenone-pretreated animals (regular food+rotenone, 170±39 s; Li+rotenone, 135±49 s. P<0.03, 21% reduction). Chronic treatment with rotenone (regular food+rotenone) did not have a significant effect. However, rotenone treatment significantly prevented the effect of lithium (P<0.0008) (Figure 1a).
Figure 1.
Behavioral effects of chronic rotenone treatment. Chronic rotenone treatment abolished antidepressant-like effect of lithium in the FST (a) and augmented amphetamine-induced hyperlocomotion response (b) in mice. Rf, regular food; Li, lithium-supplemented regular food; vehicle, 0.5% DMSO in saline; rot, rotenone. Results are means±SEM. (a) Rotenone pretreatment prevents the antidepressant-like effect of lithium. Two-way ANOVA revealed a significant treatment effect of lithium (F1,68=29.04, P<0.000001), a significant effect of rotenone (F1,68=6.92, P<0.011), and a significant lithium × rotenone interaction (F1,68=4.5, P<0.038). Fisher's LSD post hoc test: *P<0.05 compared with rf+vehicle, #P<0.05 compared with Li+vehicle, £P<0.05 compared with rf+rot. (b) Lithium treatment abolishes rotenone-augmented amphetamine-induced hyperlocomotion. No difference was found among the groups before amphetamine injection. Repeated measures ANOVA of the results following amphetamine treatment revealed a significant treatment effect (F3,51=7.78,P<0.0003) and time × treatment interaction (F15,255=3.33, P<0.00004). Rotenone significantly augmented the effect of amphetamine (Fisher's LSD post hoc test, P<0.026). Li treatment prevented the effect of rotenone on amphetamine-induced hyperactivity (Fisher's LSD post hoc test P<0.0002).
In the amphetamine-induced hyperlocomotion test, there was no difference among the groups before amphetamine administration. Rotenone augmented amphetamine-induced hyperlocomotion and Li treatment abolished this effect (Figure 1b). Two-way repeated measures ANOVA of the results following amphetamine administration indicated a significant treatment effect (F3,51=7.78, P<0.0003) and time × treatment interaction (F15,255=3.33, P<0.00004). Post hoc analysis indicated that rotenone's augmentation of the effect of amphetamine was statistically significant (Fisher's LSD post hoc test, P<0.026) and so was Li's effect to abolish rotenone's effect on amphetamine-induced hyperlocomotion (Fisher's LSD post hoc test, P<0.0002) (Figure 1b). Two-way repeated measures ANOVA of the total distance during the baseline phase and the total distance during the first 30 min after amphetamine administration revealed a significant effect of amphetamine (F1,53=23.627, P<0.00001) and a significant amphetamine × lithium × rotenone interaction (F1,53=4.26, P<0.04).
DISCUSSION
The first part of the present study was a gene expression microarray analysis looking for inositol-modulated pathways related to the mood-stabilizing effect of Li in mouse frontal cortex. We were able to evaluate inositol-mediated gene expression changes because of our simultaneous analysis of expression changes in Li-treated mice and in mice harboring KO of either IMPA1 or Slc5a3, both encoding proteins related to the PI cycle and shown to be affected by therapeutic levels of the drug. Our group has previously reported Li-like behavioral and neurochemical phenotypes exhibited by these two strains of KO mice (Agam et al, 2009).
At the single gene level, out of seven genes with similar differential expression in IMPA1 and SMIT1 KO mice and in Li-treated WT mice from the two colonies, four are part of the oxidative phosphorylation pathway. Three of them, Cox5a, Ndufab1, and Ndufs7, all found to be upregulated in the present study both in the microarray data and by real-time PCR, were previously reported to be associated with Li treatment (McQuillin et al, 2007) and bipolar disorder (Andreazza et al, 2010; Konradi et al, 2004; Sun et al, 2006; Washizuka et al, 2005; WTCCC, 2007). However, these results need to be considered with caution. Measurements of respective protein levels encoded by these mitochondria-related genes and measurements of their function are needed to verify functional significance of the expression changes of these particular genes.
Surprisingly, our microarray data did not show a significant upregulation of Adcyap1 as observed by Brandish et al (2005) after 24 h of Li-induced inositol depletion. However, a trend for significant upregulation of this gene was observed in SMIT1 KO and Li-treated WT mice from the SMIT1 colony. Thus, it is possible that changes in Adcyap1 expression are proximal to the cascade of events modulated by inositol depletion. In general, out of 29 inositol-regulated genes reported by Brandish et al, 15 genes did not pass the expression level threshold in our study, partly explaining the low replicability between the two studies.
Some of our findings at the level of single genes corroborate findings of other microarray studies on the effect of Li, as summarized in Supplementary Table S4. For example: (1) Our finding of significantly upregulated Cox5a in IMPA1 KO, SMIT1 KO, and Li-treated mice from both colonies corroborates the report of McQuillin et al (2007) in mouse brain following chronic lithium treatment. (2) The downregulation of Bai3 reported by McQuillin et al (2007) is replicated in IMPA1 KO mice and in Li-treated mice from both colonies. (3) Three previous microarray studies (Brandish et al, 2005; McQuillin et al, 2007; Seelan et al, 2008) reported differential expression of the Ptprk gene. Similarly, we found differential expression of Ptprk in SMIT1 KO mice and in their littermate WT Li-treated mice, but not in IMPA1 KO mice or in their littermate WT Li-treated mice. (4) Additional examples are summarized in Supplementary Table S2.
It could have been expected that commonly differentially expressed genes in our study would be enriched in genes related to PI signaling. Unexpectedly, only mitochondrial function was shown by all three bioinformatics approaches to be enriched among these genes. This finding corroborates previous reports of downregulation of mitochondria-related genes (Konradi et al, 2004; Sun et al, 2006), mitochondrial dysfunction (Stork and Renshaw 2005), and mitochondrial structural abnormalities (Cataldo et al, 2010) in bipolar patients. The results are also consistent with studies showing beneficial effects of mood stabilizers on mitochondrial function (Maurer et al, 2009; Valvassori et al, 2010) and oxidative stress (Lai et al, 2006).
Our results in the two Li-treatment groups did not replicate previously reported gene expression changes, neither in BDNF nor in Bcl-2 (Machado-Vieira et al, 2009). It is noticeable that while upregulation of these two genes following chronic Li treatment is seemingly well established (Machado-Vieira et al, 2009), it is not robustly reflected in microarray studies (Chetcuti et al, 2008; Fatemi et al, 2009; McQuillin et al, 2007), possibly due to the methodological differences between microarrays and real-time PCR analyses. As recently discussed by Konradi et al (2012), the strength of transcriptome studies, such as microarrays, is the examination of the expression pattern of interrelated groups of genes. The statistical chance for a concordant expression change in a group of genes with a similar function or location is much smaller than the likelihood to reach a significant change for an individual gene. The use of stringent statistical tests such as correction for multiple testing and of relatively high cutoff values for the analysis of microarray data decreases the number of ‘false positive‘ differentially expressed genes at the cost of an increased number of ‘false negatives', excluding genes with small but true fold of change and relatively high variance. Moreover, an organism is a complex network with multiple regulation targets. The activity of an enzyme or the level of a metabolite may be affected at the level of genetics, epigenetics, change in the levels of transcription factors, alternative splicing, existence of RNAi, translation or degradation rate, activity of isoenzymes, etc. Since any of the above can be altered, we suggest that each individual might have his own ‘network setup' which defines his personal response to environmental factors intervening with the network. Thus, particularly in multifactorial disorders, the expression level of a single gene might be a poor indicator of upstream causative factors or of the downstream consequences responsible for a final outcome. Several microarray studies were carried out to unravel the pathophysiology of bipolar disorder using different species, tissues, and mood-stabilizer treatment regimens (Chetcuti et al, 2008; Konradi et al, 2004; McQuillin et al, 2007; Sun et al, 2006). Among the reported candidate genes almost none replicated between the studies. However, if one considers functional gene groups, quite a few of the studies indicated downregulation of mitochondria-related genes in bipolar patients, with components of the electron transport chain in particular (Konradi et al, 2004; Sun et al, 2006) and upregulation of these biological components following chronic Li treatment (Valvassori et al, 2010).
In the second part of the present study, we aimed to model the reported mitochondrial dysfunction in bipolar patients (Andreazza et al, 2010; Konradi et al, 2004; Stork and Renshaw, 2005) using chronic low doses of rotenone. This reflects the concept that the disorder involves a gradual decrease in mitochondrial function, with symptoms beginning only when a threshold is reached or when an event occurs which requires fully functional cells, making the subject more vulnerable to environmental factors targeting mitochondrial function. Our results showing counteracting effects of lithium and rotenone in the FST model of depression and in the amphetamine-induced hyperlocomotion model of mania are in favor of an interrelationship between the mood-stabilizing effect of Li and mitochondrial function.
This is the first report of a whole-genome expression study carried out concomitantly in Li-treated mice and in mice exhibiting Li-like behavior induced by KO of either of two genes affecting inositol metabolism. Our results indicate upregulation of genes related to mitochondrial function in Li-treated mice, with similar effects in the two KO mice strains exhibiting a common phenotype to lithium-treated WT mice. The results raise the possibility that the effect of lithium on mitochondrial function may be due to altered inositol metabolism.
FUNDING AND DISCLOSURE
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Agam G, Bersudsky Y, Berry GT, Moechars D, Lavi-Avnon Y, Belmaker RH. Knockout mice in understanding the mechanism of action of lithium. Biochem Soc Trans. 2009;37 (Pt 5:1121–1125. doi: 10.1042/BST0371121. [DOI] [PubMed] [Google Scholar]
- Andreazza AC, Shao L, Wang JF, Young LT. Mitochondrial complex I activity and oxidative damage to mitochondrial proteins in the prefrontal cortex of patients with bipolar disorder. Arch Gen Psychiatry. 2010;67:360–368. doi: 10.1001/archgenpsychiatry.2010.22. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Downes CP, Hanley MR. Neural and developmental actions of lithium: a unifying hypothesis. Cell. 1989;59:411–419. doi: 10.1016/0092-8674(89)90026-3. [DOI] [PubMed] [Google Scholar]
- Berry GT, Wu S, Buccafusca R, Ren J, Gonzales LW, Ballard PL, et al. Loss of murine Na+/myo-inositol cotransporter leads to brain myo-inositol depletion and central apnea. J Biol Chem. 2003;278:18297–18302. doi: 10.1074/jbc.M213176200. [DOI] [PubMed] [Google Scholar]
- Bersudsky Y, Shaldubina A, Agam G, Berry GT, Belmaker RH. Homozygote inositol transporter knockout mice show a lithium-like phenotype. Bipolar Disord. 2008;10:453–459. doi: 10.1111/j.1399-5618.2007.00546.x. [DOI] [PubMed] [Google Scholar]
- Bersudsky Y, Shaldubina A, Belmaker RH. Lithium's effect in forced-swim test is blood level dependent but not dependent on weight loss. Behav Pharmacol. 2007;18:77–80. doi: 10.1097/FBP.0b013e32801416ed. [DOI] [PubMed] [Google Scholar]
- Bosetti F, Seemann R, Bell JM, Zahorchak R, Friedman E, Rapoport SI, et al. Analysis of gene expression with cDNA microarrays in rat brain after 7 and 42 days of oral lithium administration. Brain Res Bull. 2002;57:205–209. doi: 10.1016/s0361-9230(01)00744-4. [DOI] [PubMed] [Google Scholar]
- Brandish PE, Su M, Holder DJ, Hodor P, Szumiloski J, Kleinhanz RR, et al. Regulation of gene expression by lithium and depletion of inositol in slices of adult rat cortex. Neuron. 2005;45:861–872. doi: 10.1016/j.neuron.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Cai JH, Deng S, Kumpf SW, Lee PA, Zagouras P, Ryan A, et al. Validation of rat reference genes for improved quantitative gene expression analysis using low density arrays. Biotechniques. 2007;42:503–512. doi: 10.2144/000112400. [DOI] [PubMed] [Google Scholar]
- Cataldo AM, McPhie DL, Lange NT, Punzell S, Elmiligy S, Ye NZ, et al. Abnormalities in mitochondrial structure in cells from patients with bipolar disorder. Am J Pathol. 2010;177:575–585. doi: 10.2353/ajpath.2010.081068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetcuti A, Adams LJ, Mitchell PB, Schofield PR. Microarray gene expression profiling of mouse brain mRNA in a model of lithium treatment. Psychiatr Genet. 2008;18:64–72. doi: 10.1097/YPG.0b013e3282fb0051. [DOI] [PubMed] [Google Scholar]
- Crawley JN. Behavioral phenotyping of transgenic and knockout mice: experimental design and evaluation of general health, sensory functions, motor abilities, and specific behavioral tests. Brain Res. 1999;835:18–26. doi: 10.1016/s0006-8993(98)01258-x. [DOI] [PubMed] [Google Scholar]
- Creson TK, Austin DR, Shaltiel G, McCammon J, Wess J, Manji HK, et al. Lithium treatment attenuates muscarinic M(1) receptor dysfunction. Bipolar Disord. 2011;13:238–249. doi: 10.1111/j.1399-5618.2011.00915.x. [DOI] [PubMed] [Google Scholar]
- Cryns K, Shamir A, Van Acker N, Levi I, Daneels G, Goris I, et al. IMPA1 is essential for embryonic development and lithium-like pilocarpine sensitivity. Neuropsychopharmacology. 2008;33:674–684. doi: 10.1038/sj.npp.1301431. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Reutiman TJ, Folsom TD. The role of lithium in modulation of brain genes: relevance for aetiology and treatment of bipolar disorder. Biochem Soc Trans. 2009;37 (Pt 5:1090–1095. doi: 10.1042/BST0371090. [DOI] [PubMed] [Google Scholar]
- Greenamyre JT, Cannon JR, Drolet R, Mastroberardino PG.2010Lessons from the rotenone model of Parkinson's disease Trends Pharmacol Sci 31141–142.author reply 142-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallcher LM, Sherman WR. The effects of lithium ion and other agents on the activity of myo-inositol-1-phosphatase from bovine brain. J Biol Chem. 1980;255:10896–10901. [PubMed] [Google Scholar]
- Hashimoto H, Hashimoto R, Shintani N, Tanaka K, Yamamoto A, Hatanaka M, et al. Depression-like behavior in the forced swimming test in PACAP-deficient mice: amelioration by the atypical antipsychotic risperidone. J Neurochem. 2009;110:595–602. doi: 10.1111/j.1471-4159.2009.06168.x. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kawaguchi C, Tanaka K, Isojima Y, Shintani N, Hashimoto H, Baba A, et al. Changes in light-induced phase shift of circadian rhythm in mice lacking PACAP. Biochem Biophys Res Commun. 2003;310:169–175. doi: 10.1016/j.bbrc.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- Konradi C, Sillivan SE, Clay HB. Mitochondria, oligodendrocytes and inflammation in bipolar disorder: Evidence from transcriptome studies points to intriguing parallels with multiple sclerosis. Neurobiol Dis. 2012;45:37–47. doi: 10.1016/j.nbd.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JS, Zhao C, Warsh JJ, Li PP. Cytoprotection by lithium and valproate varies between cell types and cellular stresses. Eur J Pharmacol. 2006;539:18–26. doi: 10.1016/j.ejphar.2006.03.076. [DOI] [PubMed] [Google Scholar]
- Lubrich B, van Calker D. Inhibition of the high affinity myo-inositol transport system: a common mechanism of action of antibipolar drugs. Neuropsychopharmacology. 1999;21:519–529. doi: 10.1016/S0893-133X(99)00037-8. [DOI] [PubMed] [Google Scholar]
- Machado-Vieira R, Manji HK, Zarate CA., Jr The role of lithium in the treatment of bipolar disorder: convergent evidence for neurotrophic effects as a unifying hypothesis. Bipolar Disord. 2009;11 (Suppl 2:92–109. doi: 10.1111/j.1399-5618.2009.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese G, Scheggi S, Secci ME, De Montis MG, Gambarana C. Anti-anhedonic activity of long-term lithium treatment in rats exposed to repeated unavoidable stress. Int J Neuropsychopharmacol. 2013;16:1611–1621. doi: 10.1017/S1461145712001654. [DOI] [PubMed] [Google Scholar]
- Maurer IC, Schippel P, Volz HP. Lithium-induced enhancement of mitochondrial oxidative phosphorylation in human brain tissue. Bipolar Disord. 2009;11:515–522. doi: 10.1111/j.1399-5618.2009.00729.x. [DOI] [PubMed] [Google Scholar]
- McQuillin A, Rizig M, Gurling HM. A microarray gene expression study of the molecular pharmacology of lithium carbonate on mouse brain mRNA to understand the neurobiology of mood stabilization and treatment of bipolar affective disorder. Pharmacogenet Genomics. 2007;17:605–617. doi: 10.1097/FPC.0b013e328011b5b2. [DOI] [PubMed] [Google Scholar]
- Monville C, Torres EM, Dunnett SB. Comparison of incremental and accelerating protocols of the rotarod test for the assessment of motor deficits in the 6-OHDA model. J Neurosci Methods. 2006;158:219–223. doi: 10.1016/j.jneumeth.2006.06.001. [DOI] [PubMed] [Google Scholar]
- O'Brien WT, Harper AD, Jove F, Woodgett JR, Maretto S, Piccolo S, et al. Glycogen synthase kinase-3beta haploinsufficiency mimics the behavioral and molecular effects of lithium. J Neurosci. 2004;24:6791–6798. doi: 10.1523/JNEUROSCI.4753-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell T, Rotzinger S, Nakashima TT, Hanstock CC, Ulrich M, Silverstone PH. Chronic lithium and sodium valproate both decrease the concentration of myo-inositol and increase the concentration of inositol monophosphates in rat brain. Brain Res. 2000;880:84–91. doi: 10.1016/s0006-8993(00)02797-9. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul MC, Gessay GM, Gammie SC. A new mouse model for mania shares genetic correlates with human bipolar disorder. PLoS ONE. 2012;7:e38128. doi: 10.1371/journal.pone.0038128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelan RS, Khalyfa A, Lakshmanan J, Casanova MF, Parthasarathy RN. Deciphering the lithium transcriptome: microarray profiling of lithium-modulated gene expression in human neuronal cells. Neuroscience. 2008;151:1184–1197. doi: 10.1016/j.neuroscience.2007.10.045. [DOI] [PubMed] [Google Scholar]
- Shaltiel G, Shamir A, Shapiro J, Ding D, Dalton E, Bialer M, et al. Valproate decreases inositol biosynthesis. Biol Psychiatry. 2004;56:868–874. doi: 10.1016/j.biopsych.2004.08.027. [DOI] [PubMed] [Google Scholar]
- Stork C, Renshaw PF. Mitochondrial dysfunction in bipolar disorder: evidence from magnetic resonance spectroscopy research. Mol Psychiatry. 2005;10:900–919. doi: 10.1038/sj.mp.4001711. [DOI] [PubMed] [Google Scholar]
- Su B, Wang X, Zheng L, Perry G, Smith MA, Zhu X. Abnormal mitochondrial dynamics and neurodegenerative diseases. Biochim Biophys Acta. 2010;1802:135–142. doi: 10.1016/j.bbadis.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wang JF, Tseng M, Young LT. Downregulation in components of the mitochondrial electron transport chain in the postmortem frontal cortex of subjects with bipolar disorder. J Psychiatry Neurosci. 2006;31:189–196. [PMC free article] [PubMed] [Google Scholar]
- Toker L, Belmaker RH, Agam G. Gene-expression studies in understanding the mechanism of action of lithium. Expert Rev Neurother. 2012;12:93–97. doi: 10.1586/ern.11.184. [DOI] [PubMed] [Google Scholar]
- Valvassori SS, Rezin GT, Ferreira CL, Moretti M, Goncalves CL, Cardoso MR, et al. Effects of mood stabilizers on mitochondrial respiratory chain activity in brain of rats treated with d-amphetamine. J Psychiatr Res. 2010;44:903–909. doi: 10.1016/j.jpsychires.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Washizuka S, Kakiuchi C, Mori K, Tajima O, Akiyama T, Kato T. Expression of mitochondria-related genes in lymphoblastoid cells from patients with bipolar disorder. Bipolar Disord. 2005;7:146–152. doi: 10.1111/j.1399-5618.2005.00184.x. [DOI] [PubMed] [Google Scholar]
- WTCCC Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.