Abstract
Background
Glioma is rarely curable, and factors that influence the prognosis of glioma patients are not fully understood. Loss of heterozygosity (LOH) of 1p/19q has long been known to be a typical molecular signature of oligodendroglial neoplasms. However, whether LOH of 1p/19q is associated with survival in gliomas remains controversial. Here our goal was to evaluate the association between LOH of 1p/19q and progression-free survival (PFS) and overall survival (OS) by conducting a meta-analysis among glioma cases.
Methods
The PubMed and Embase databases were searched from the earliest records to May 2013 to identify studies that met prestated inclusion criteria. Reference lists of retrieved articles were also reviewed. Three authors independently extracted information needed for further analysis. Either a fixed- or a random-effects model was used to calculate the overall combined hazard ratio (HR) estimates.
Results
Twenty-eight eligible studies involving 3 408 cases were included in the meta-analysis. Compared with the chromosomal intact group, codeletion of 1p and 19q was associated with a better PFS (HR = 0.63; 95% CI, 0.52–0.76) and OS (HR = 0.43; 95% CI, 0.35–0.53). Subgroup analyses showed this association to be independent of detection methods and the grades and subtypes of gliomas. Furthermore, isodeletion of chromosome 1p predicted a similar favorable disease outcome (PFS: HR = 0.68; 95% CI, 0.47–0.97) (OS: HR = 0.51; 95% CI, 0.35–0.75), especially in low-grade gliomas, whereas isodeletion of 19q only indicated longer PFS (HR = 0.70; 95% CI, 0.56–0.87).
Conclusion
Codeletion of 1p and 19q is associated with better survival rates in glioma. Isodeletion of 1p predicts similar outcomes but to a lesser extent, whereas the effects of isodeletion of 19q remained only marginal.
Keywords: 1p, 19q, glioma, meta-analysis, survival
As the most frequent primary brain tumor, gliomas account for approximately 30% of all brain and central nervous system tumors and 80% of all malignant brain tumors.1 Based on their histological appearance, gliomas can be divided into 2 major subtypes according to the 2007 WHO classification:2 One is astrocytic tumors including pilocytic astrocytomas (PA), astrocytomas, and glioblastomas (GBM); the other is oligodendroglial tumors including pure oligodendrogliomas and mixed oligoastrocytic tumors. Tumors can be further divided into grades I (PA), II (low grade), III (anaplastic), and IV (GBM) depending on the presence of anaplastic features.2
Factors that influence the prognosis for glioma patients are not completely understood. The combination of surgery, radiotherapy, and chemotherapy remains the standard treatment in most cases; however, not all patients derive clinical benefit from it. It has been recognized that acquired molecular abnormalities have been associated with histological subtype and grade.1,3 Specific molecular abnormalities and/or genetic mutation patterns are believed to be associated with the prognosis of gliomas and can differentiate not only among histological subtypes but also between low-grade (grades I and II) and high-grade (grades III and IV) tumors. Combined loss of genetic materials on chromosomal arms 1p and 19q from an unbalanced translocation can lead to the loss of 1 hybrid chromosome and thereby loss of heterozygosity (LOH).4 LOH of 1p/19q has long been known to be a typical molecular signature of oligodendroglial neoplasms;5 now, accumulating data suggest that LOH of 1p/19q is present in about 80% of low-grade oligodendrogliomas, 60% of anaplastic oligodendrogliomas (AO), 30%–50% of oligoastrocytomas, 30% of anaplastic oligoastrocytomas, and 10% of diffuse astrocytic gliomas including GBM.6 In 1998, it was first reported that LOH of 1p/19q could predict better response to chemotherapy and longer survival in AO patients.7 Soon thereafter, studies including prospective randomized phase III trials demonstrated that a LOH of 1p/19q status was associated with good outcome and might consequently play an important role in the treatment.8,9 However, the magnitudes of the association varied between studies: some studies10,11 even failed to show such a positive association.
An improved understanding of this issue could have important public health and clinical implications considering that gliomas are still rarely curable. With recently accumulating evidence, our goal therefore, was to evaluate the association between LOH of 1p/19q and progression-free survival (PFS) and overall survival (OS) by conducting a meta-analysis among patients with different grades and subtypes of gliomas. Additionally, the relationships between isodeletion of chromosomal arms 1p or 19q and clinic outcomes were examined.
Method
Search Strategy and Selection of Studies
The current meta-analysis was carried out in accordance with the PRISMA statement for reporting systematic reviews and meta-analyses.12 PubMed and Embase were searched for studies evaluating the deletions involving chromosomal arms 1p/19q and survival in brain tumors from the earliest records to May 2013. The following search terms were used: (i) glioma, brain tumor, oligodendroglioma, oligodendroglial tumor, glioblastoma, astrocytoma, astrocytic tumor, oligoastrocytoma, oligodendroglial neoplasm; and (ii) chromosomal arms deletion, codeletion, loss of heterozygosity, 1p, 19q. Only one restriction was imposed, that for clinical studies. In addition, the reference lists of retrieved papers were reviewed.
The search results were first screened for titles and/or abstracts. A second screening was based on full-text review. Studies were considered eligible if they met the following criteria: (i) the exposures of interest were glioma and chromosomal arms 1p and 19q; (ii) the outcomes of interests were progression-free survival and overall survival; (iii) hazard ratio (HR) and the corresponding 95% confidence interval (CI) (or data sufficient to calculate them) were reported; and (iv) exclusion of letters to the editor, reviews, and articles published in non-English language books or papers.
Data Collection and Extraction
Three authors (J. Zhao, W. Ma and H. Zhao) independently extracted information using predefined data abstraction forms. The following details were extracted: first author's full name, year of publication, country of origin, tumor subtype, tumor grade, median age at the time of diagnosis, duration of follow-up, method to detect 1p/19q, total number of cases, number of LOH of 1p/19q cases and controls, subtype of 1p/19q experiment vs control, number of 1p loss cases and controls, number of 19q loss cases and controls, and assessments of outcomes (HR and the corresponding 95% CI of PFS and/or OS. When the statistical variables were not given explicitly in an article, they were estimated from available data using methods reported by Tierney et al.13 If one study reported both the results of univariate and multivariate analysis, the latter was selected.
The key exposure variable was the codeletion of 1p/19q at baseline, but patients with 1p/19q isodeletion and codeletion served as the experimental group in 7 studies.,14–20 In addition, patients with intact 1p/19q were treated as reference groups in most studies, while in 2 studies,21,22 those with isodeletion and intact 1p/19q served as controls. The rest of the papers assessed only the outcomes of both codeletion and intact 1p/19q. The studies selected for specific meta-analysis were indicated in the context. We required a minimum of 3 studies to carry out pooled analyses.
Statistical Analysis
Homogeneity of HRs across the studies was tested by Q statistic (significance level at P < .05). The I2 statistic, a quantitative measure of inconsistency across studies,23 was also calculated. The combined risk estimates were computed by fixed-effects models if the I2 statistic was <50%; otherwise, the random-effects models were used.24
Because characteristics of populations, ascertainment of different glioma subtypes, and adjustments for confounding factors were not consistent between studies, we further conducted a sensitivity analysis to explore possible explanations for heterogeneity and to examine the influence of various exclusion criteria on the overall risk estimate. We also investigated the influence of a single study on the outcome estimate by omitting one study in each turn.
Potential publication bias was assessed by visual inspection of the Begg's funnel plots. We also performed the Begg rank correlation test at the P < .10 level of significance.25 All analyses were performed using STATA version 12.0 (StataCorp LP). A P < .05 was considered statistically significant, except where otherwise specified.
Result
Literature Search
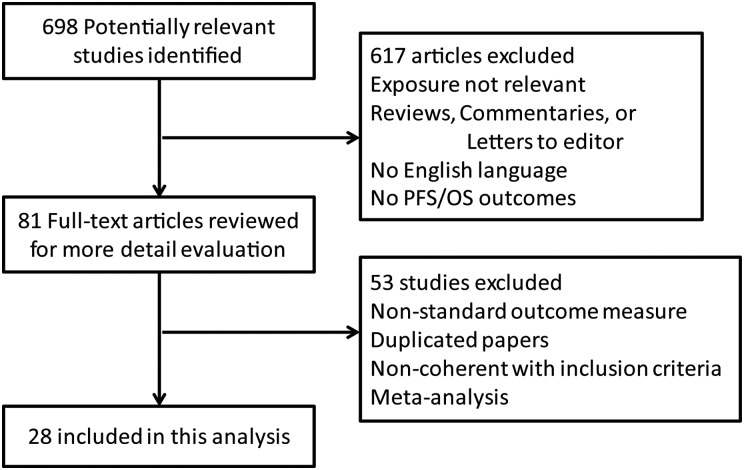
Of 698 titles initially retrieved, 617 were excluded because they did not meet the inclusion criteria. Of the remaining 81 studies, all were found in full text and were evaluated by 3 independent investigators; 53 papers were excluded because of duplication, nonstandard outcome measure, and noncoherence with inclusion criteria. A total of 28 studies8–11,14–22,26–40were ultimately included in this meta-analysis. A flow chart showing the study selection is presented in Fig. 1.
Fig. 1.
Flow chart of study selection. Flow chart shows literature search for studies of loss of 1p/19q heterozygosity in relation to survivals in glioma.
Study Characteristics
The characteristics of the selected 28 studies are presented in Table 1. These papers were published between 2004 and 2013. Six studies were conducted in the US and Canada, 16 in Europe, 3 in Asia, 2 in Australia, and 1 in multiple countries. The mean length of follow-up ranged from 14 to 107 months. A total of 3 408 cases were included in the current meta-analysis, of which 898 were low-grade glioma, 1 725 were high-grade glioma, and the remaining 785 were not specified. When these 3 408 cases were classified by their histological feature, 791 were astrocytic tumors, 1 451 were oligodendroglial tumors, and the tumor subtypes of the remaining 1166 cases were not indicated. Of the total 28 studies, 12 studies detected the chromosomal arm 1p/19q status by PCR or RT-PCR, 11 studies used fluorescent in situ hybridization (FISH), and the remaining 5 studies detected 1p/19q by other methods such as denaturing high-performance liquid chromatography, array comparative genomic hybridization, and multiplex ligation–dependent probe amplification. As a result, 1 333 cases showed LOH of 1p/19q, of which 765 demonstrated codeletion of 1p and 19q.
Table 1.
Main characteristics of all studies included in current meta-analysis
| Author | Year | Country | Tumor Subtypea | Tumor Grade | Median Age (range) | Median Follow-up (months) | Method to Detect 1p/19q | No. of Patients (LOH 1p/19q: control) | 1p/19q Experimental vs Control Subtypeb |
|---|---|---|---|---|---|---|---|---|---|
| Fallon | 2004 | US and Canada | 2 | High | 39.2 (20–71) | 106.8 | FISH | 76 (54:22) | 2 |
| Walker | 2005 | UK | 2 | Mixed | 40 (21–73) | 39.6 | PCR | 100 (46:54) | 1 |
| Kujas | 2005 | France | 3 | Low | 39 (17–66) | 63.3 | FISH | 127 (49:68) | 1 |
| Dehais | 2006 | France | 1 | High | 45.5 (20–83) | 57.3 | PCR | 143 (27:116) | 1 |
| Brandes | 2006 | Italy | 2 | High | 47.8 (18–70) | NR | FISH | 67(32:35) | 2 |
| Miller | 2006 | US | 3 | High | 52.4 (21–90) | 16.8 | FISH | 389(212:177) | 1 |
| Homma | 2006 | Switzerland | 1 | Mixed | 55.4 | NR | PCR | 209(6:203) | 2 |
| Mariani | 2006 | Switzerland | 3 | Low | 38.3 (20–76) | 71 | PCR | 66 (17:49) | 1 |
| Kaloshi | 2007 | France | 1 | Low | 44 (24–72) | 30.4 | PCR | 86 (36:50) | 3 |
| Iwamoto | 2008 | US | 3 | Low; high | 38.9 | 56.1 | PCR | 111 (39:72) | 1 |
| Idbaih | 2008 | France | 2 | High | 50.3 (23–84) | NR | PCR | 52 (15:37) | 1 |
| Wick | 2009 | German | 3 | High | 43.0 (20–77) | 54(max) | Other | 181 (74:107) | 1 |
| Weller | 2009 | German | 1 | Mixed | 61.5 (19–87) | 29.4 | PCR | 281 (24:257) | 1 |
| Mikkelsen | 2009 | US | 2 | High | 45.5 (18–81) | 32 | Other | 48 (36:12) | 2 |
| Kesari | 2009 | US | 2 | Low | 43 (20–68) | 39.4 | FISH | 28 (18:10) | 1 |
| Kuo | 2009 | Taiwan | 2 | Mixed | 38 (4–82) | 83 | PCR | 49 (34:15) | 1 |
| Gravendeel | 2009 | Netherlands | 3 | Mixed | 50.2 (12–81) | NR | FISH | NR | 1 |
| Gan | 2010 | Australia | 2 | Low | 43 (17–71) | 34 | PCR | 37 (18:19) | 1 |
| Ji | 2010 | China | 2 | Low; high | 43.1 (10–76) | NR | PCR | 131 (100:31) | 2 |
| Kim | 2010 | Switzerland, German, Japan | 3 | Low | 39 ( > 20) | NR | PCR | 268 (112:156) | 2 |
| Scheie | 2011 | Norway | 2 | Mixed | 43 (19–72) | 50 | FISH | 95 (52:43) | 2 |
| Parkinson | 2011 | Australia | 2 | Mixed | 52.4 (25–80) | NR | FISH | 51 (26:25) | 1 |
| Grauer | 2011 | German | 3 | High | 40.5 (18–66) | 52.9 | Other | 24 (21:3) | 1 |
| Taal | 2011 | Netherlands | 1 | Low | 38 (25–59) | 14 | FISH | NR | 1 |
| Hartmann | 2011 | German | 1,2 | Low | 39 | 64.8 | FISH | 133 (51:82) | 1 |
| Li | 2012 | China | 2 | High | 43 (16–71) | 38.2 | Other | 77 (28:49) | 1 |
| Cairncross | 2013 | US and Canada | 2 | High | 43 (18–76) | NR | FISH | 263 (126:137) | 3 |
| Erdem-Eraslan | 2013 | Europe | 2 | High | 49.5 (19–69) | NR | Other | 316 (80:236) | 1 |
aTumor subtype. Type 1: astrocytic tumors including pilocytic astrocytomas, astrocytomas, and glioblastomas; type 2: oligodendroglial tumors including pure oligodendroglial tumors and mixed oligoastrocytic tumors; type 3: mixed subtype.
b1p/19q experimenal vs control subtype. Type 1: 1p/19q codeletion vs intact 1p/19q; type 2: 1p/19q isodeletion and codeletion vs intact 1p/19q; type 3: 1p/19q codeletion vs 1p/19q isodeletion and intact 1p/19q.
Abbrevations: FISH, fluorescent in situ hybridization; LOH, loss of heterozygosity; NR, not reported.
LOH of 1p/19q and PFS
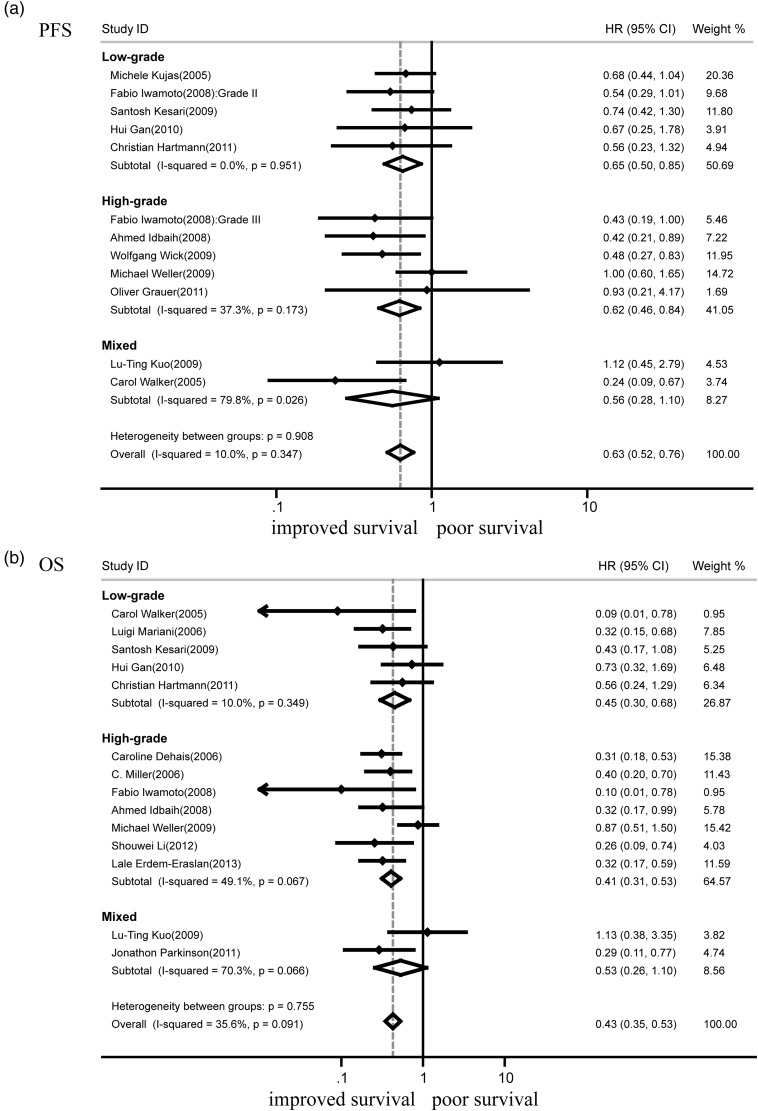
Since the key exposure variable was the codeletion of chromosomal arms 1p and 19q, 11 studies that assessed PFS for both codeletion and intact 1p/19q were strictly analyzed first (Fig. 2a). The absence of both 1p and 19q was significantly associated with better PFS (HR = 0.63; 95% CI, 0.52–0.76; n = 11; P < .001). Further subgroup analysis showed that codeletion of 1p/19q indicated better PFS in both low-grade tumors (HR = 0.65; 95% CI, 0.50–0.85; n = 5; P < .001) and high-grade gliomas (HR = 0.62; 95% CI, 0.46–0.84; n = 4; P < .001). In current meta-analysis, heterogeneity, the variation in study outcomes between studies was measured by P value and I2, the percentage of variation across studies that is due to heterogeneity rather than random chance. No heterogeneities were observed between different subgroups (P = .91) and individual studies (I2 = 10.0%; P = .35). Similar analysis suggested that codeletion of 1p/19q indicated good PFS in oligodendroglial tumors (HR = 0.65; 95% CI, 0.48–0.87; n = 6, P < .01) (Table 2). Patients with 1p/19q isodeletion and codeletion served as the experimental group in 4 other studies. Analysis on all these 15 papers showed that LOH of 1p/19q was associated with better PFS (HR = 0.56; 95% CI, 0.47–0.67; P < .001) (Table 2); there was no substantial heterogeneity for the outcome (I2 = 27.2%, P = .15).
Fig. 2.
Forest plots showing hazard ratios of codeletion of 1p/19q for progression-free survival (a) and overall survival (b). Each study is shown by the point estimate of the hazard ratio and 95% confidence intervals (extending lines). The diamonds represent the estimated pooled effect (labeled total).
Table 2.
Hazard ratios of different human chromosome 1p/19q heterozygosity vs intact 1p/19q status for progression-free survival and overall survival
| Studies | HR (95% CI) |
|---|---|
| PFS | |
| Codeletion of 1p/19q in oligodendroglial tumors (16, 22, 25, 26, 28, 32) | 0.65 (0.48–0.87) |
| LOH of 1p/19q in glioma (5–7, 9, 16, 17, 21–26, 28, 30, 32) | 0.56 (0.47–0.67) |
| PCR to detect 1p/19q (16, 21, 22, 24, 26, 28) | 0.63 (0.47–0.83) |
| FISH to detect 1p/19q (17, 25, 32) | 0.68 (0.49–0.94) |
| OS | |
| Codeletion of 1p/19q in astrocytic tumors (18, 24, 32) | 0.52 (0.36–0.75) |
| Codeletion of 1p/19q in oligodendroglial tumors (16, 22, 25, 26, 28, 29, 32–34) | 0.41 (0.30–0.56) |
| LOH of 1p/19q in gliomas (4–7, 10, 16, 18–22, 24–26, 28, 29, 32, 34) | 0.41 (0.34–0.48) |
| 1p deletion in astrocytic tumors (8, 18, 24) | 0.65 (0.38–1.12) |
| 19q deletion in astrocytic tumors (8, 18, 24, 31) | 0.64 (0.43–0.95) |
| 19q deletion in oligodendroglial tumors (4, 25, 26) | 1.12 (0.10–12.09) |
| PCR to detect 1p/19q (16, 18, 20–22, 24, 26, 28, 33) | 0.46 (0.35–0.60) |
| FISH to detect 1p/19q (19, 25, 29, 32) | 0.39 (0.25–0.60) |
Abbreviations: FISH, fluorescent in situ hybridization; LOH, loss of heterozygosity
LOH of 1p/19q and OS
A total of 14 studies that assessed OS of both codeletion and intact 1p/19q were analyzed (Fig. 2b). The deletion of both 1p and 19q was clearly associated with good OS (HR = 0.43; 95% CI, 0.35–0.53; n = 14, P < .001). Subsequent subgroup analysis showed that codeletion of 1p/19q also indicated favorable outcomes in both low-grade tumors (HR = 0.45; 95% CI, 0.30–0.68; n = 5, P < .001) and high-grade gliomas (HR = 0.41; 95% CI, 0.31–0.53; n = 6, P < .001). No heterogeneities were observed between subgroups (P = .76) and all 14 studies (I2 = 35.6%, P = .09). Similar analysis suggested that codeletion of 1p/19q indicated good OS in astrocytic tumors (HR = 0.52; 95% CI, 0.36–0.75; n = 3; P < .001) and oligodendroglial tumors (HR = 0.41; 95% CI, 0.30–0.56; n = 9, P < .001) (Table 2). Patients with 1p/19q isodeletion and codeletion served as the experimental group in 5 other studies. Analysis on these 19 papers showed that LOH of 1p/19q was associated with better OS (HR = 0.41; 95% CI, 0.34–0.48; P < .001) (Table 2), and there was no substantial heterogeneity for the outcome (I2 = 36.3%; P = .06).
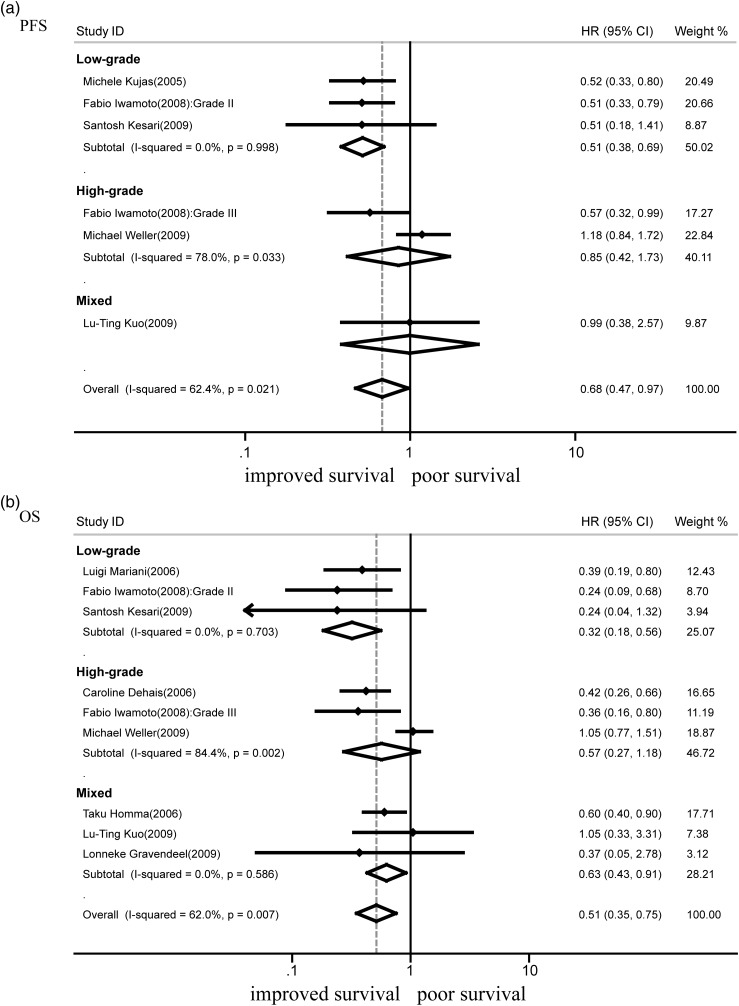
1p Deletion and PFS
Nine studies assessed the outcomes of chromosomal arm 1p deletion. A total of 1 282 cases were included, of which 442 showed 1p loss; the other 840 cases had intact 1p. The absence of 1p was associated with better PFS (HR = 0.68; 95% CI, 0.47–0.97; n = 5: P < .05) (Fig. 3a). There was heterogeneity between studies in this analysis (I2 = 62.4%; P = .02). Sensitivity analyses were immediately conducted to explore potential sources of heterogeneity in the association between 1p deletion and PFS and to examine the influence of various exclusion criteria on the overall risk estimate. GBM was the most deadly of malignant primary brain tumors. It was reported that median survival with standard-of-care radiation and chemotherapy with temozolomide was about 15 months, and median survival without treatment was 4.5 months.41 Exclusion of the study that only focused on GBM10 yielded better results (HR = 0.55; 95% CI, 0.43–0.71; P < .001) with no evidence of heterogeneity observed among the remaining studies (I2 = 0%, P = .79). Further subgroup analysis showed that deletion of 1p indicated good PFS in low-grade tumors (HR = 0.51; 95% CI, 0.38–0.69; n = 3; P < .001) (Fig. 3a). After excluding the GBM study,10 there were not enough data to conduct meta-analysis in high-grade gliomas, oligodendroglial tumors, and astrocytic tumors.
Fig. 3.
Forest plots showing hazard ratios of isodeletion of 1p for progression-free survival (a) and overall survival (b).
1p Deletion and OS
A total of 8 studies that assessed OS for deletion and intact 1p were analyzed (Fig. 3b). The absence of 1p was associated with good OS (HR = 0.51; 95% CI, 0.35–0.75; n = 8; P < .001). Heterogeneity was observed for the outcome (I2 = 62.0%; P < .01). The exclusion of one study focused on GBM10 hardly changed the overall hazard ratio (HR = 0.47; 95% CI, 0.36–0.60; n = 7; P < .001) with no substantial heterogeneity (I2 = 0%; P = .49). Subgroup analysis showed that deletion of 1p indicated good OS in low-grade tumors (HR = 0.32; 95% CI,.018–0.56; n = 3; P < .001). Based on the available data, the associations between 1p loss and OS were inconclusive in high-grade gliomas (HR = 0.57; 95% CI, 0.27–1.18; n = 3; P > .05) (Fig. 3b) and astrocytic tumors (HR = 0.65; 95% CI, 0.38–1.12; n = 3, P > .05) (Table 2).
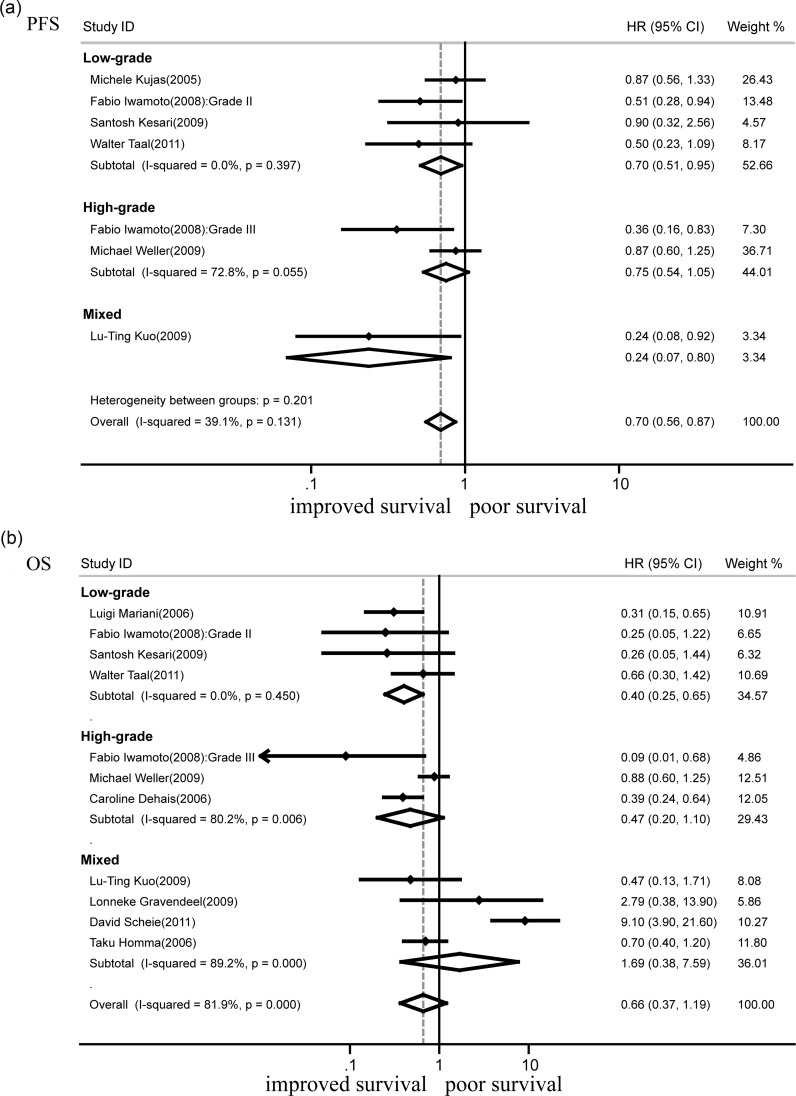
19q deletion and PFS
Eleven studies assessed the outcomes of chromosomal arm 19q deletion. A total of 1 311 glioma cases were included, of which 387 showed 19q loss, while 924 had intact 19q. The absence of 19q was associated with better PFS (HR = 0.70; 95% CI, 0.56–0.87; n = 6, P < .01) (Fig. 4a). There was no substantial heterogeneity between studies (I2 = 39.1%; P = .13). Subgroup analysis showed that deletion of 19q indicated good PFS in low-grade tumors (HR = 0.70; 95% CI, 0.51–0.95; n = 4; P < .01) (Fig. 4a).
Fig. 4.
Forest plots showing hazard ratios of isodeletion of 19q for progression-free survival (a) and overall survival (b).
19q deletion and OS
A total of 10 studies that assessed OS for deleted and intact 19q were analyzed (Fig. 4b). Based on the available data, no association of 19q loss and OS was evident (HR = 0.66; 95% CI, 0.37–1.19; n = 10, P > .05). Substantial heterogeneity was observed for the outcome (I2 = 81.9%; P < .001). Sensitivity analyses were conducted to explore potential sources; it turned out that exclusion of any 1 or 2 studies did not change the heterogeneity. Exclusion of all 7 investigations that contained high-grade gliomas showed a better OS (HR = 0.40; 95% CI, 0.25–0.65; n = 4; P < .001) with no substantial heterogeneity (I2 = 0%; P = .45). Loss of 19q was associated with OS in astrocytic tumors (HR = 0.64; 95% CI, 0.43–0.95; n = 4; P < .05) but not in oligodendroglial tumors (HR = 1.12; 95% CI, 0.10–12.09; n = 3; P > .05) (Table 2).
Methods of 1p/19q Detection and Outcome
There was no difference in the hazard ratio for PFS or OS (Table 2) between studies utilizing PCR and those using FISH to assess the status of chromosomal arms 1p and 19q (P values for subgroup differences were.71 and .53, respectively).
Publication Bias
Potential publication bias was assessed by Begg's funnel plots and Begg's rank correlation. Visual inspection of the Begg's funnel plot did not identify substantial asymmetry. The Begg's rank correlation test also indicated no evidence of publication bias among studies of gliomas and LOH of 1p/19q (P = .92).
Discussion
In this study, we meta-analyzed the published data about chromosomal arms 1p/19q status in gliomas and their association with survival. Results show that codeletion of 1p and 19q was associated with a more favorable disease outcome and that this association was independent of detection methods, grades, and subtypes of gliomas. Isodeletion of chromosomal arm 1p also predicted longer PFS and OS, especially in low-grade gliomas. The effects of isodeletion of 19q remained only marginal.
LOH of 1p/19q and Survival
Loss of 1p and 19q due to an unbalanced centromeric translocation t (1;19) (q10; p10) were first associated with sensitivity to alkylating agent chemotherapy7 and later to sensitivity to radiotherapy as well.42 Here, our results confirmed that codeletion of 1p/19q was associated with favorable clinic outcomes. Further subgroup meta-analysis suggested that codeletion of 1p/19q was an independent predictive biomarker.
Although usually both chromosomes 1p and 19q are lost, isodeletion of 1p or 19q is still very common in gliomas.43 Unfortunately, most research merely focused on codeletion of 1p/19q rather than single loss of 1p or 19q. Our data demonstrated that single loss of 1p could predict longer survival in the low-grade tumors. High-grade glioma patients, especially GBM patients, could not derive clinical benefit from isodeletion of 1p. There are several reasons to explain the inconsistency. First, it was reported there were 2 types of 1p loss with opposite significance in gliomas.44 Type II 1p loss, mainly observed in GBM and not associated with 19q loss, had an unfavorable prognostic value. Second, the presence of other prognostically unfavorable genetic alterations (ie, chromosomal arm 9p or 10q loss) could compromise the prognostic utility.45,46 Third, only GBM-O, one subtype of GBM, contained 1p and/or 19q deletion,2 but the reported LOH of 1p/19q frequencies varied from 5%47,48 to about 25%.49,50 The rare cases of 1p/19q deletion in GBM made it difficult to collect sufficiently powered studies.
Sources of Heterogeneity
No substantial heterogeneity was observed among studies of 1p/19q codeletion status and clinical outcome, even when there were significant differences in characteristics of populations, ascertainment of tumor subtype, and adjustment for confounding factors. As to 19q loss analysis, substantial heterogeneity was observed and no single major source could be detected through the sensitivity analysis, only excluded all the high-grade tumors could change the heterogeneity. Both statistical and clinical heterogeneity could contribute to this result. Clinical heterogeneity may derive from the different patients (with different ages, tumor sizes, ethnicity, physical condition, etc.), diverse types of treatments, various treatment protocols, different doses and types of drugs, and so on. Further multicenter research, using standardized methods, is encouraged.
Assessment of chromosomal arm 1p/19q status is most frequently accomplished by FISH, PCR-based LOH analysis, and multiplex ligation-dependent probe amplification. Of these methods, FISH has proven to be robust, easy to implement, and cost effective.51 In the current study, we found that there was no observed difference between studies using PCR and those using FISH when assessing the clinical outcome of glioma cases with LOH of 1p/19q. This suggested that chromosomal arm detection methods could not be the source of heterogeneity and that the 1p/19q codeletion pattern would be a powerful prognostic and predictive biomarker in neuro-oncology practice.
Study Strengths and Limitations
Our meta-analyses generated several important implications. First, analyzing LOH for 1p and/or 19q and PFS and OS in gliomas made our results more extensive and valid. Second, the association of 1p/19q codeletion with survival persists and remains statistically significant based on various classification criteria. Third, with the accumulating evidence and enlarged sample size, we have enhanced statistical power to provide more precise and reliable hazard ratio estimates. Fourth, all of the analyses were conducted by random-effects modelsand fixed-effects models (data not shown). Both models showed similar results, which indicated that the statistical results were robust.
Despite its advantages, our meta-analysis had some limitations. First, publication bias was a major concern that may cause bias. In this research, Begg's funnel plot did not show any evidence of publication bias. Nevertheless, we still need to consider the fact that studies with positive data are more easily accepted. First, the languages of the published studies included in this meta-analysis were restricted to English, and other potentially eligible studies that met our inclusion criteria could not be included. Second, the number of studies concerning some histological subtypes (such as pilocytic astrocytomas and glioblastomas) were too small to perform a pooled analysis. Third, most of the glioma patients were treated with different combination therapies. Some patients were treated with neurosurgery alone. Fourth, evaluation standards and cut-off values for LOH chromosomal arm 1p/19q may also be different in various studies.
Conflict of interest statement. None declared.
Funding
None declared.
Acknowledgment
The authors thank Dr. Bin Zhao for assistance in data collection.
References
- 1.Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205(12):613–621. doi: 10.1016/j.cancergen.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenkins RB, Blair H, Ballman KV, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66(20):9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 5.Reifenberger J, Reifenberger G, Liu L, James CD, Wechsler W, Collins VP. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol. 1994;145(5):1175–1190. [PMC free article] [PubMed] [Google Scholar]
- 6.Riemenschneider MJ, Jeuken JW, Wesseling P, Reifenberger G. Molecular diagnostics of gliomas: state of the art. Acta Neuropathol. 2010;120(5):567–584. doi: 10.1007/s00401-010-0736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90(19):1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 8.Wick W, Hartmann C, Engel C, et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009;27(35):5874–5880. doi: 10.1200/JCO.2009.23.6497. [DOI] [PubMed] [Google Scholar]
- 9.Erdem-Eraslan L, Gravendeel LA, de Rooi J, et al. Intrinsic molecular subtypes of glioma are prognostic and predict benefit from adjuvant procarbazine, lomustine, and vincristine chemotherapy in combination with other prognostic factors in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. J Clin Oncol. 2013;31(3):328–336. doi: 10.1200/JCO.2012.44.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27(34):5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 11.Kuo LT, Kuo KT, Lee MJ, et al. Correlation among pathology, genetic and epigenetic profiles, and clinical outcome in oligodendroglial tumors. Int J Cancer. 2009;124(12):2872–2879. doi: 10.1002/ijc.24303. [DOI] [PubMed] [Google Scholar]
- 12.Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6(7):e1000100. doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheie D, Meling TR, Cvancarova M, et al. Prognostic variables in oligodendroglial tumors: a single-institution study of 95 cases. Neuro Oncol. 2011;13(11):1225–1233. doi: 10.1093/neuonc/nor114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandes AA, Tosoni A, Cavallo G, et al. Correlations between O6-methylguanine DNA methyltransferase promoter methylation status, 1p and 19q deletions, and response to temozolomide in anaplastic and recurrent oligodendroglioma: a prospective GICNO study. J Clin Oncol. 2006;24(29):4746–4753. doi: 10.1200/JCO.2006.06.3891. [DOI] [PubMed] [Google Scholar]
- 16.Fallon KB, Palmer CA, Roth KA, et al. Prognostic value of 1p, 19q, 9p, 10q, and EGFR-FISH analyses in recurrent oligodendrogliomas. J Neuropathol Exp Neurol. 2004;63(4):314–322. doi: 10.1093/jnen/63.4.314. [DOI] [PubMed] [Google Scholar]
- 17.Mikkelsen T, Doyle T, Anderson J, et al. Temozolomide single-agent chemotherapy for newly diagnosed anaplastic oligodendroglioma. J Neurooncol. 2009;92(1):57–63. doi: 10.1007/s11060-008-9735-x. [DOI] [PubMed] [Google Scholar]
- 18.Homma T, Fukushima T, Vaccarella S, et al. Correlation among pathology, genotype, and patient outcomes in glioblastoma. J Neuropathol Exp Neurol. 2006;65(9):846–854. doi: 10.1097/01.jnen.0000235118.75182.94. [DOI] [PubMed] [Google Scholar]
- 19.Xiong J, Liu Y, Wang Y, Ke RH, Mao Y, Ye ZR. Chromosome 1p/19q status combined with expression of p53 protein improves the diagnostic and prognostic evaluation of oligodendrogliomas. Chin Med J (Engl) 2010;123(24):3566–3573. [PubMed] [Google Scholar]
- 20.Kim YH, Nobusawa S, Mittelbronn M, et al. Molecular classification of low-grade diffuse gliomas. Am J Pathol. 2010;177(6):2708–2714. doi: 10.2353/ajpath.2010.100680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaloshi G, Benouaich-Amiel A, Diakite F, et al. Temozolomide for low-grade gliomas: predictive impact of 1p/19q loss on response and outcome. Neurology. 2007;68(21):1831–1836. doi: 10.1212/01.wnl.0000262034.26310.a2. [DOI] [PubMed] [Google Scholar]
- 22.Cairncross G, Wang M, Shaw E, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31(3):337–343. doi: 10.1200/JCO.2012.43.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 25.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 26.Walker C, du Plessis DG, Joyce KA, et al. Molecular pathology and clinical characteristics of oligodendroglial neoplasms. Ann Neurol. 2005;57(6):855–865. doi: 10.1002/ana.20496. [DOI] [PubMed] [Google Scholar]
- 27.Kujas M, Lejeune J, Benouaich-Amiel A, et al. Chromosome 1p loss: a favorable prognostic factor in low-grade gliomas. Ann Neurol. 2005;58(2):322–326. doi: 10.1002/ana.20543. [DOI] [PubMed] [Google Scholar]
- 28.Dehais C, Laigle-Donadey F, Marie Y, et al. Prognostic stratification of patients with anaplastic gliomas according to genetic profile. Cancer. 2006;107(8):1891–1897. doi: 10.1002/cncr.22211. [DOI] [PubMed] [Google Scholar]
- 29.Miller CR, Dunham CP, Scheithauer BW, Perry A. Significance of necrosis in grading of oligodendroglial neoplasms: a clinicopathologic and genetic study of newly diagnosed high-grade gliomas. J Clin Oncol. 2006;24(34):5419–5426. doi: 10.1200/JCO.2006.08.1497. [DOI] [PubMed] [Google Scholar]
- 30.Mariani L, Deiana G, Vassella E, et al. Loss of heterozygosity 1p36 and 19q13 is a prognostic factor for overall survival in patients with diffuse WHO grade 2 gliomas treated without chemotherapy. J Clin Oncol. 2006;24(29):4758–4763. doi: 10.1200/JCO.2006.05.9238. [DOI] [PubMed] [Google Scholar]
- 31.Iwamoto FM, Nicolardi L, Demopoulos A, et al. Clinical relevance of 1p and 19q deletion for patients with WHO grade 2 and 3 gliomas. J Neurooncol. 2008;88(3):293–298. doi: 10.1007/s11060-008-9563-z. [DOI] [PubMed] [Google Scholar]
- 32.Idbaih A, Criniere E, Marie Y, et al. Gene amplification is a poor prognostic factor in anaplastic oligodendrogliomas. Neuro Oncol. 2008;10(4):540–547. doi: 10.1215/15228517-2008-022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kesari S, Schiff D, Drappatz J, et al. Phase II study of protracted daily temozolomide for low-grade gliomas in adults. Clin Cancer Res. 2009;15(1):330–337. doi: 10.1158/1078-0432.CCR-08-0888. [DOI] [PubMed] [Google Scholar]
- 34.Gravendeel LA, Kouwenhoven MC, Gevaert O, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 35.Gan HK, Rosenthal MA, Dowling A, et al. A phase II trial of primary temozolomide in patients with grade III oligodendroglial brain tumors. Neuro Oncol. 2010;12(5):500–507. doi: 10.1093/neuonc/nop065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkinson JF, Afaghi V, Payne CA, et al. The impact of molecular and clinical factors on patient outcome in oligodendroglioma from 20 years’ experience at a single centre. J Clin Neurosci. 2011;18(3):329–333. doi: 10.1016/j.jocn.2010.07.101. [DOI] [PubMed] [Google Scholar]
- 37.Grauer O, Pascher C, Hartmann C, et al. Temozolomide and 13-cis retinoic acid in patients with anaplastic gliomas: a prospective single-arm monocentric phase-II study (RNOP-05) J Neurooncol. 2011;104(3):801–809. doi: 10.1007/s11060-011-0548-y. [DOI] [PubMed] [Google Scholar]
- 38.Taal W, Dubbink HJ, Zonnenberg CB, et al. First-line temozolomide chemotherapy in progressive low-grade astrocytomas after radiotherapy: molecular characteristics in relation to response. Neuro Oncol. 2011;13(2):235–241. doi: 10.1093/neuonc/noq177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartmann C, Hentschel B, Tatagiba M, et al. Molecular markers in low-grade gliomas: predictive or prognostic? Clin Cancer Res. 2011;17(13):4588–4599. doi: 10.1158/1078-0432.CCR-10-3194. [DOI] [PubMed] [Google Scholar]
- 40.Li S, Yan C, Huang L, Qiu X, Wang Z, Jiang T. Molecular prognostic factors of anaplastic oligodendroglial tumors and its relationship: a single institutional review of 77 patients from China. Neuro Oncol. 2012;14(1):109–116. doi: 10.1093/neuonc/nor185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson DR, O'Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107(2):359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- 42.Bauman GS, Ino Y, Ueki K, et al. Allelic loss of chromosome 1p and radiotherapy plus chemotherapy in patients with oligodendrogliomas. Int J Radiat Oncol Biol Phys. 2000;48(3):825–830. doi: 10.1016/s0360-3016(00)00703-3. [DOI] [PubMed] [Google Scholar]
- 43.Gupta K, Salunke P. Molecular markers of glioma: an update on recent progress and perspectives. J Cancer Res Clin Oncol. 2012;138(12):1971–1981. doi: 10.1007/s00432-012-1323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Idbaih A, Marie Y, Pierron G, et al. Two types of chromosome 1p losses with opposite significance in gliomas. Ann Neurol. 2005;58(3):483–487. doi: 10.1002/ana.20607. [DOI] [PubMed] [Google Scholar]
- 45.Trost D, Ehrler M, Fimmers R, et al. Identification of genomic aberrations associated with shorter overall survival in patients with oligodendroglial tumors. Int J Cancer. 2007;120(11):2368–2376. doi: 10.1002/ijc.22574. [DOI] [PubMed] [Google Scholar]
- 46.Houillier C, Mokhtari K, Carpentier C, et al. Chromosome 9p and 10q losses predict unfavorable outcome in low-grade gliomas. Neuro Oncol. 2010;12(1):2–6. doi: 10.1093/neuonc/nop002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinto LW, Araujo MB, Vettore AL, et al. Glioblastomas: correlation between oligodendroglial components, genetic abnormalities, and prognosis. Virchows Arch. 2008;452(5):481–490. doi: 10.1007/s00428-007-0562-9. [DOI] [PubMed] [Google Scholar]
- 48.Kraus JA, Lamszus K, Glesmann N, et al. Molecular genetic alterations in glioblastomas with oligodendroglial component. Acta Neuropathol. 2001;101(4):311–320. doi: 10.1007/s004010000258. [DOI] [PubMed] [Google Scholar]
- 49.Bigner SH, Matthews MR, Rasheed BK, et al. Molecular genetic aspects of oligodendrogliomas including analysis by comparative genomic hybridization. Am J Pathol. 1999;155(2):375–386. doi: 10.1016/S0002-9440(10)65134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He J, Mokhtari K, Sanson M, et al. Glioblastomas with an oligodendroglial component: a pathological and molecular study. J Neuropathol Exp Neurol. 2001;60(9):863–871. doi: 10.1093/jnen/60.9.863. [DOI] [PubMed] [Google Scholar]
- 51.Horbinski C, Miller CR, Perry A. Gone FISHing: clinical lessons learned in brain tumor molecular diagnostics over the last decade. Brain Pathol. 2011;21(1):57–73. doi: 10.1111/j.1750-3639.2010.00453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]