Abstract
Ubiquitination is crucial for cellular processes, such as protein degradation, apoptosis, autophagy, and cell cycle progression. Dysregulation of the ubiquitination network accounts for the development of numerous diseases, including cancer. Thus, targeting ubiquitination is a promising strategy in cancer therapy. Both apoptosis and autophagy are involved in tumorigenesis and response to cancer therapy. Although both are categorized as types of cell death, autophagy is generally considered to have protective functions, including protecting cells from apoptosis under certain cellular stress conditions. This review highlights recent advances in understanding the regulation of apoptosis and autophagy by ubiquitination.
Keywords: Apoptosis, autophagy, BRUCE, caspase, NF-κB, p53, ubiquitin, ubiquitination
Failure in apoptotic cell death is one of the major causes of tumorigenesis. A primary strategy for cancer therapy is to specifically induce apoptosis in cancer cells, whereas the resistance of certain cancer cells to therapy can be at least partially due to the cytoprotective role of autophagy against apoptosis[1]. Autophagy not only inhibits the initiation of tumorigenesis by limiting cytoplasmic damage, genomic instability, and inflammation, but also promotes the survival of certain cancer cells by enabling adaptation to stressful metabolic environments. Ubiquitination is a post-translational modification that impacts almost all cellular activities, including protein degradation, cell cycle progression, apoptosis, and autophagy. This review highlights recent researches on the regulation of apoptosis and autophagy by ubiquitination, with particular emphasis on how this regulation affects tumorigenesis.
Targeting Ubiquitination and Related Pathways in Cancer Therapy
Ubiquitination is a process in which one or multiple ubiquitin moieties are covalently attached to a substrate through an enzymatic cascade involving ubiquitin-activating enzyme (E1), ubiquitin-carrier protein (E2), and ubiquitin-protein ligase (E3). Formation of a ubiquitin Lys48 chain on the ε-NH2 group of a substrate's internal Lys residue (polyubiquitination) can target the substrate for degradation by the 26S proteasome. Ubiquitin can also be attached to the free α-NH2 group in a substrate's N-terminus to promote proteasomal degradation[2]. The ubiquitin-proteasome pathway degrades most cellular proteins in eukaryotic cells. However, ubiquitination may not always target proteins for degradation. For example, polyubiquitination at Lys63 is involved in inhibitor of NF-κB (IκB) kinase (IKK) activation[3]. In addition, a linear polyubiquitin chain can be achieved by conjugating the C-terminal glycine of ubiquitin and the a-NH2 group of the N-terminal methionine of its neighbor ubiquitin[4]. Substrates can also undergo monoubiquitination or multi-monoubiquitination—adding one ubiquitin to one or multiple Lys residues, respectively. Recent evidence suggests that ubiquitin can be linked to Cys, Ser, or Thr residues in a substrate through thio- or oxy-ester bonds (i.e., esterification), though the physiological relevance of these modifications remains to be defined[5]–[7]. Ubiquitin moieties can be released from a substrate by deubiquitinating enzymes.
For an organism to function properly, proteins must be degraded after they undergo specific functions. Moreover, proteins that are misfolded or damaged during translation, folding, or translocation must be degraded and eliminated in time. Many regulatory proteins related to tumorigenesis are proteosomal substrates. Either blocked degradation of oncogenic proteins/growth-enhancing factors or accelerated degradation of growth-suppressing proteins may disrupt the pathways controlling cell cycle progression, cell death, or survival, leading to cancer development[8],[9] (Table 1). For example, the tumor suppressor CYLD is mutated in several cancers, including cylindromatosis. The deubiquitinating activity of CYLD for IKKγ is critical for its cylindromatosis-suppressive function[10]. The ubiquitin ligase Itch promotes the polyubiquitination and degradation of large tumor suppressor 1 (LTSA1), which is closely related to enhanced cell growth and epithelial-to-mesenchymal transition.
Table 1. Deregulated ubiquitination of key substrates in different cancer types.
| Deregulatedprotein | Substrate | Modification | Tumors | Reference(s) | |
| MDM2 (HDM2) | ↑ | p53 | Polyubiquitination | Non-small cell lung cancer, breast cancer, soft tissue carcinoma, colorectal cancer | [71],[72] |
| HAUSP | ↓ | p53, MDM2 | De-ubiquitination | Non-small cell lung cancer, lymphoma | [73] |
| APC | ↓ | Cyclin B, securin | Polyubiquitination | Colorectal cancer | [8] |
| FANCL | ↓ | FANCD2 | Monoubiquitination | Fanconi anaemia related cancers | [74] |
| CYLD | ↓ | IKKγ | De-ubiquitination | Cylindromatosis | [10] |
| IAP2 | ↓ | BCL10 | Polyubiquitination | MALT lymphomas | [75] |
| CBL | ↓ | RTKs | Multiple monoubiquitination | Lymphoma, AML, gastric carcinoma | [76] |
| pVHL | ↓ | HIF | Polyubiquitination | von Hippel-Lindau disease | [77],[78] |
| E6-AP | p53 | Polyubiquitination | Human papillomavirus-positive cancer | [79] | |
| SCFβ−TRCP | ↑ | IκB | Polyubiquitination | Colon cancer, prostate cancer, melanoma | [80] |
| KLHL20 | ↑ | PML | Polyubiquitination | Human prostate cancer | [81] |
| USP9X | ↑ | MCL1 | De-ubiquitination | Diffuse large B-cell lymphomas, human follicular lymphomas | [82] |
| FBW7 | ↓ | KLF5 | Polyubiquitination | Breast cancer | [83] |
| ITCH | ↑ | LATS1 | Polyubiquitination | Cancer cell lines (HeLa, MCF10A and MCF7) | [84],[85] |
| SIAH2 | ↑ | C/EBPδ | Polyubiquitination | Breast cancer | [86] |
| ASB2α | ↑ | Filamin | Polyubiquitination | Myeloid leukemia | [87] |
| FBXO11 (mutation) | BCL6 | Polyubiquitination | Diffuse large B-cell lymphoma | [88] | |
| Ubiquilin-1 | ↑ | BCL2L10/BCLb | Monoubiquitination | Lung adencarcinomas | [32] |
↑ stands for up-regulation, and ↓ for down-regulation. MALT, mucosa-associated lymphoid tissue; AML, acute myeloid leukemia.
Due to the critical roles of ubiquitination and the ubiquitin-mediated proteolysis in tumorigenesis and cell growth, targeting the components involved in these processes is a powerful approach for cancer therapy. Bortezomib is the first proteasome inhibitor for clinical use in human cancers[11]. It is a dipeptide boronate that specifically and reversibly blocks chymotrypsin-like activity of the proteasome in a variety of cancer cells[12]. Although bortezomib inhibits NF-κB activation and results in autophagy[13], this lethal effect of proteasome inhibition is probably due to loss of amino acid homeostasis[14]. Notably, bortezomib has been used successfully as an anticancer drug for multiple myeloma and mantle cell lymphoma in the clinic[12],[15]–[17].
Because ubiquitination is generally substrate-specific, the components of the ubiquitination pathway might be more specific drug targets for cancer therapy than the proteasome. Cullin-RING ubiquitin ligases (CRLs) are involved in cellular processes such as cell cycle progression, cell death signaling, DNA damage, and stress responses[18]. NEDD8 is a ubiquitin-like protein that modifies Cullin and is required for the activity of CRLs[19]. Because NEDD8-activating enzyme (NAE) catalyzes the first step in the NEDD8 pathway, targeting CRLs via inhibition of NAE may be a promising anticancer strategy. Indeed, MLN4924, a selective inhibitor of NAE, has potent tumor-suppressing activity in a wide range of tumors, including acute myeloid leukemia and diffuse large B cell lymphomas[20],[21].
Regulation of Apoptosis by Ubiquitination
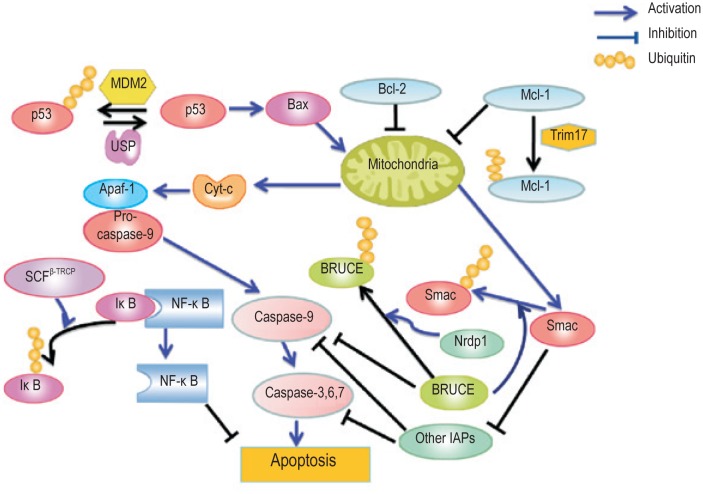
Apoptosis (i.e., programmed cell death) is a cellular suicide process that is important for embryonic development and maintaining the size of cell populations. There are two primary apoptotic pathways: extrinsic and intrinsic. The extrinsic pathway involves members of the tumor necrosis factor (TNF) receptor gene superfamily, which bind extracellular ligands and transduce intracellular signals during cell destruction. This pathway involves several caspases, cysteine proteases with specific cellular targets[22]. The intrinsic pathway does not involve receptor-mediated intracellular signaling, but induces signaling in mitochondria. In mammals, the intrinsic pathway is regulated by the Bcl-2 family of proteins, the adaptor protein apoptotic protease-activating factor-1 (Apaf-1), and the caspases[23].
Bcl-2 family members include both anti-apoptotic (Bcl-2, Bcl-xL, Bcl-w, and Mcl-1) and pro-apoptotic proteins (Bax, Bak, Bad, Bid, and Bim). Caspases are crucial intracellular executioners of apoptosis. The release of cytochrome C from mitochondria causes the formation of the apoptosome (Apaf-1/caspase-9 complex), activates the downstream effector caspases, and finally results in cleavage of crucial substrates[24]. Degradation of anti-apoptotic members is necessary for apoptotic progression[25]–[27], whereas degradation of pro-apoptotic members is required for the suppression of apoptosis[28],[29]. The levels of anti- and pro-apoptotic molecules can be regulated by ubiquitination and proteasomal degradation. Bcl-2 family proteins can be polyubiquitinated and degraded by the 26S proteasome. For example, Trim17-mediated ubiquitination and subsequent degradation of Mcl-1, an anti-apoptotic Bcl-2 family member, triggers neuronal apoptosis[30]. The pro-apoptotic Bcl-2 member Bax can be regulated by ubiquitination indirectly; the ubiquitin ligase Trim39 inhibits APC/C Cdh1-mediated ubiquitination and degradation of the Bax activator MOAP-1, thus enhancing Bax activation and apoptosis[31]. Moreover, the levels of Bcl2L10/Bclb, an anti-apoptotic Bcl2-like protein, are inversely correlated with survival in patients with several cancer types, including lung adenocarcinomas. Bcl2L10/Bclb can be specifically monoubiquitinated and stabilized by ubiquilin-1 (UBQLN1)[32].
The inhibitors of apoptosis proteins (IAPs) have one to three baculovirus IAP repeat (BIR) domains and can block apoptosis by directly binding and inhibiting caspases[33],[34]. Furthermore, almost all IAPs have ubiquitin ligase activity, which is required for the ubiquitination of certain substrates involved in apoptosis[35]. X-linked inhibitor of apoptosis protein (XIAP) catalyzes the ubiquitination and degradation of caspase-3[36],[37]. cIAP1 promotes autoubiquitination and self-degradation[38]. Apoptosis inducing factor (AIF) is also a substrate of XIAP, and ubiquitination at K255 of AIF shows a non-degradable role of ubiquitination in caspase-independent cell death[39]. On the other hand, IAPs can be regulated by deubiquitinating enzymes. For example, ubiquitin-specific protease 19 (USP19) is responsible for the inhibition of TNF-α-induced caspase activation and apoptosis in a cIAP-dependent manner[40].
The activity of IAPs can be suppressed by pro-apoptotic factors, such as second mitochondria-derived activator of caspase (Smac)[41]. Conversely, some IAPs promote Smac ubiquitination and degradation[42]. BRUCE/Apollon is a large (528 kDa), membrane-associated, essential IAP in mammals. A decrease in BRUCE levels promotes apoptosis[43]. BRUCE inhibits the Smac-induced apoptosis by promoting Smac ubiquitination and degradation[44],[45]. Furthermore, BRUCE/Apollon can be degraded in a ubiquitin-dependent manner by the ubiquitin ligase Nrdp1 during apoptosis.
The tumor suppressor p53 maintains the integrity of the genome and regulates cell cycle, DNA repair, and apoptosis. p53 promotes the activation of the pro-apoptotic Bcl-2 family proteins and the release of cytochrome C. Dysregulation of p53 is reported in numerous types of cancer. Several ubiquitin ligases, including MDM2, have been reported to promote ubiquitination and degradation of p53, while p53 is deubiquitinated and stabilized by ubiquitin-specific proteases (USPs). Evasion of apoptosis is a primary cause of tumorigenesis. Thus, inhibiting the activity of p53 ubiquitin ligases or activating p53 USPs can be a strategy for cancer therapy. Otub1 and nucleolin play direct roles in suppressing MDM2-mediated ubiquitination of p53[46],[47]. HAUSP regulates the activities of MDM2 and p53 by deubiquitination, while vif1 and vif2 antagonize HAUSP and promote p53-dependent apoptosis[48]. Translationally controlled tumor protein (TCTP), which is down-regulated in tumor progression, inhibits MDM2 autoubiquitination and promotes MDM2-mediated ubiquitination and degradation of p53[49]. In addition, Fanconi anemia complementation group F (FANCF) monoubiquitinates FANCD2, which is involved in the FA/BRCA DNA damage response pathway. Silencing FANCF elevates p53 activation in mitoxantrone-treated breast cancer cells[50].
As a transcription factor involved in the extrinsic apoptosis pathway, NF-κB activates the expression of genes that contribute to cell proliferation, metastasis, and suppression of apoptosis. SHARPIN, a ubiquitin-binding and ubiquitin-like-domain-containing protein, promotes linear ubiquitination of NEMO/IKBKG, an adaptor of IKKs, and subsequent activation of NF-κB signaling[51]. IκB, which inactivates NF-κB under normal physiological conditions, can be phosphorylated by activated IKKβ, ubiquitinated by SCFβ−TRCP, and finally degraded by the proteasome in response to DNA damage[52],[53]. Nrdp1 promotes ubiquitination and degradation of the epidermal growth factor receptor family member ErbB3, which is upstream of NF-κB activation[54],[55]. In a word, ubiquitination plays an important role in the regulation of apoptosis, and the components involved in the ubiquitination of key substrates can be potential targets for cancer therapy (Figure 1).
Figure 1. Regulation of apoptosis by ubiquitination.
Apoptosis is controlled by both pro-apoptotic and anti-apoptotic factors. Ubiquitination regulates almost all of these factors and promotes their proteasomal degradation. IAPs, inhibitors of apoptosis proteins.
Regulation of Autophagy by Ubiquitination
Autophagy, once categorized as programmed cell death type II, is a cellular process by which intracellular proteins, lipids, and organelles are degraded in the lysosomal compartment after delivery from other cellular compartments[56]. Autophagy can both suppress cancer initiation and promote the growth of established cancers[57]. There are three types of autophagy: macroautophagy, microautophagy, and chaperone-mediated autophagy. Although autophagy is generally thought to be non-selective, certain ubiquitinated proteins (e.g., catalase), organelles (e.g., peroxisomes and mitochondria), and invading bacteria have been shown to be selectively targeted for autophagic degradation[58].
Macroautophagy is mediated by a unique organelle—the autophagosome. To date, 18 autophagy-related proteins (Atgs) in yeast, namely Atg1–10, Atg12–14, Atg16–18, Atg29, and Atg31, have been found to play a role in autophagosome formation. Atg8, called LC3 in mammals, is a ubiquitin-like protein present on autophagic membranes as a phosphatidylethanolamine (PE)–conjugate. Ubiquitination plays important roles in selective autophagy. p62/SQSTM1 or NBR1 binds both ubiquitin and LC3, probably providing a selective link between ubiquitinated substrates and autophagy[59]. Nuclear dot protein 52 (NDP52), an autophagy receptor, targets intracellular ubiquitinated bacterial proteins for autophagic degradation[60].
Misfolded polypeptides are usually recognized by molecular chaperones and degraded by the proteasome following polyubiquitination by ubiquitin ligases, such as CHIP and Parkin. However, when misfolded proteins cannot be sufficiently removed by chaperone-mediated proteasomal degradation, protein aggregation occurs and may in turn inactivate the proteasome, resulting in cytotoxicity. Thus, p62/NBR1-mediated autophagic degradation may serve as an important compensatory mechanism for degradation of these ubiquitinated protein aggregates[59].
Role of Ubiquitination in Mutual Regulation of Apoptosis and Autophagy
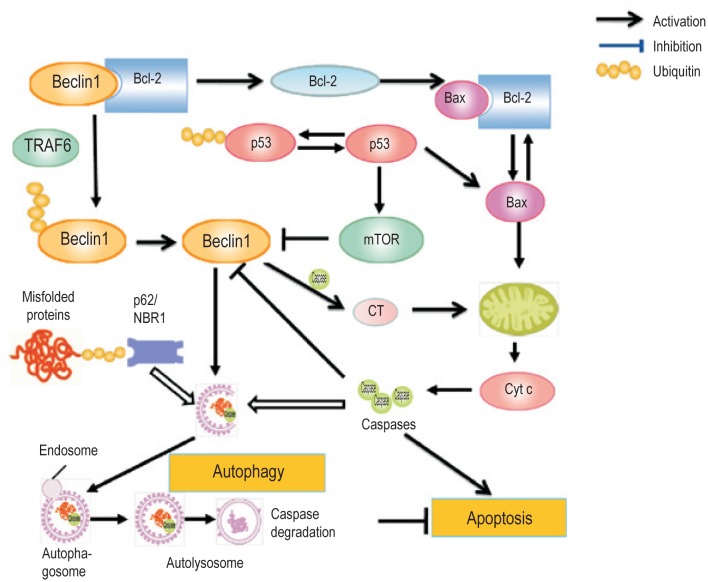
The crosstalk between autophagy and apoptosis is necessary for controlling the balance between cell survival and death. These two processes share common stimuli and signaling pathways (Figure 2). Beclin 1, a mammalian Atg6 ortholog, is a subunit of the class III PI3-kinase complex. Beclin 1 interacts with Bcl-2 via the BH3 domain in Beclin 1 but can be released in starvation conditions to activate autophagy. This interaction can be terminated through c-Jun N-terminal kinase (JNK)–mediated phosphorylation of Bcl-2 and TNF receptor-associated factor 6 (TRAF6)–mediated ubiquitination of Beclin 1[61],[62]. Phosphorylated Bcl-2 binds the pro-apoptotic protein Bax to inhibit apoptosis. Under extreme conditions that cannot be rescued by autophagy, JNK promotes hyperphosphorylation of Bcl-2, resulting in the release of Bax to execute apoptosis[63]. Caspase-mediated cleavage of Beclin 1 inhibits Beclin 1–induced autophagy, and the cleavage product, the C-terminal region (CT), enhances apoptosis by promoting the release of pro-apoptotic factors from mitochondria[64]. Beclin 1 can also indirectly affect the crosstalk between apoptosis and autophagy by controlling the levels of p53, a tumor suppressor that promotes apoptosis under genotoxic stress[12],[65]. p53 induces the synthesis of mTOR and DRAM[66]. Inhibition of mTOR induces autophagy, whereas knockout of DRAM reduces autophagy[67],[68]. Furthermore, p53 can down-regulate LC3 levels in starved cells, preventing the “autophagy burst” that may be dangerous for cells[69]. Under normal conditions, p53 is kept at low levels by the ubiquitin ligase MDM2[70]. However, p53 levels can also be controlled by Beclin 1 via regulating the deubiquitinating activity of USP10 and USP13[65].
Figure 2. Model for a role of ubiquitination in mutual regulation of apoptosis and autophagy.
Autophagy can target ubiquitinated misfolded proteins, caspases, and other cargo (such as damaged mitochondria and invading bacteria) for degradation, probably through p62 and NBR1. Apoptosis and autophagy are counter-regulated in multiple steps, such as at p53, the Beclin 1/Bcl-2 interaction, the cleavage of Beclin 1 into the C-terminal region by caspases, and the autophagic degradation of caspases. Ubiquitination can promote degradation of both p53 and Beclin 1 and thus, controls the mutual regulation of apoptosis and autophagy.
Concluding Remarks
Dysregulation of ubiquitination can lead to the development of several types of cancer. Targeting ubiquitination is therefore a promising strategy for cancer therapy. Ubiquitination can occur on not only the ε-NH2 group of an internal Lys residue, but also the α-NH2 group of the N-terminal residue of a substrate. Moreover, recent evidence suggests that ubiquitin can be attached to Cys, Ser, or Thr residue on a substrate by esterification. These non-Lys ubiquitinations might provide another layer of the regulation of protein functions, and further studies should focus on the identification of relevant substrates and physiological roles of these modifications.
Future studies should also further explore how the ubiquitination of the critical proteins is involved in tumorigenesis and cancer therapy. Of course, these studies will require better understanding of tumorigenesis mechanisms. Recent research efforts on cancer stem cells and personalized cancer genome sequencing are expected to help in this regard. The mechanisms governing the selectivity in autophagy remain to be further explored. Because the cytoprotection of autophagy and the evasion of apoptosis contribute to resistance to cancer therapy, it is important to unravel how these two pathways are mutually regulated. The investigation on this issue has just begun and deserves more attention, especially with regard to how ubiquitination is involved in the counter-regulation of these critical processes.
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China (No. 2012CB910300), the National Natural Science Foundation of China (No. 30525033), and the Fundamental Research Funds for the Central Universities of China to X.-B. Q.
References
- 1.Giampietri C, Petrungaro S, Padula F, et al. Autophagy modulators sensitize prostate epithelial cancer cell lines to TNF-alpha-dependent apoptosis. Apoptosis. 2012:1–13. doi: 10.1007/s10495-012-0752-z. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Deng L, Wang C, Spencer E, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 4.Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-κB activation. EMBO reports. 2009;10:706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wenzel DM, Lissounov A, Brzovic PS, et al. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Herr RA, Chua WJ, et al. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J Cell Biol. 2007;177:613–624. doi: 10.1083/jcb.200611063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tokarev AA, Munguia J, Guatelli JC. Serine-threonine ubiquitination mediates downregulation of BST-2/tetherin and relief of restricted virion release by HIV-1 Vpu. J Virol. 2011;85:51–63. doi: 10.1128/JVI.01795-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wäsch R, Engelbert D. Anaphase-promoting complex-dependent proteolysis of cell cycle regulators and genomic instability of cancer cells. Oncogene. 2005;24:1–10. doi: 10.1038/sj.onc.1208017. [DOI] [PubMed] [Google Scholar]
- 9.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 10.Evans PC, Ovaa H, Hamon M, et al. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem J. 2004;378:727. doi: 10.1042/BJ20031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer cell. 2004;5:417–421. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 12.Ostrowska H. The ubiquitin-proteasome system: a novel target for anticancer and anti-inflammatory drug research. Cell Mol Biol Lett. 2008;13:353–365. doi: 10.2478/s11658-008-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia L, Gopinathan G, Sukumar JT, et al. Blocking autophagy prevents bortezomib-induced NF-kappaB activation by reducing I-kappaBalpha degradation in lymphoma cells. PloS one. 2012;7:e32584. doi: 10.1371/journal.pone.0032584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suraweera A, Munch C, Hanssum A, et al. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell. 2012;48:242–253. doi: 10.1016/j.molcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams J, Kauffman M. Development of the proteasome inhibitor Velcade™(Bortezomib) Cancer Invest. 2004;22:304–311. doi: 10.1081/cnv-120030218. [DOI] [PubMed] [Google Scholar]
- 16.Richardson PG, Barlogie B, Berenson J, et al. A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med. 2003;348:2609–2617. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 17.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 18.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Revi Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 19.Sakata E, Yamaguchi Y, Miyauchi Y, et al. Direct interactions between NEDD8 and ubiquitin E2 conjugating enzymes upregulate cullin-based E3 ligase activity. Nat Struct Mol Biol. 2007;14:167–168. doi: 10.1038/nsmb1191. [DOI] [PubMed] [Google Scholar]
- 20.Soucy TA, Smith PG, Milhollen MA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 21.Swords RT, Kelly KR, Smith PG, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 22.Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer. 2002;2:420–430. doi: 10.1038/nrc821. [DOI] [PubMed] [Google Scholar]
- 23.Yuan J, Yankner BA. Apoptosis in the nervous system. Nature. 2000;407:802–809. doi: 10.1038/35037739. [DOI] [PubMed] [Google Scholar]
- 24.Zou H, Henzel WJ, Liu X, et al. Apaf-1, a human protein homologous to C. Elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–414. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 25.Dimmeler S, Breitschopf K, Haendeler J, et al. Dephosphorylation targets Bcl-2 for ubiquitin-dependent degradation: a link between the apoptosome and the proteasome pathway. J Exp Med. 1999;189:1815–1822. doi: 10.1084/jem.189.11.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu S, Takehara T, Hikita H, et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52:698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 27.Nakano H, Miyazawa T, Kinoshita K, et al. Functional screening identifies a microRNA, miR-491 that induces apoptosis by targeting Bcl-X(L) in colorectal cancer cells. Int J Cancer. 2010;127:1072–1080. doi: 10.1002/ijc.25143. [DOI] [PubMed] [Google Scholar]
- 28.Li B, Dou QP. Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proc Natl Acad Sci U S A. 2000;97:3850–3855. doi: 10.1073/pnas.070047997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh JM, Kim SH, Cho EA, et al. Human papillomavirus type 16 E5 protein inhibits hydrogen peroxide-induced apoptosis by stimulating ubiquitin-proteasome-mediated degradation of Bax in human cervical cancer cells. Carcinogenesis. 2010;31:402–410. doi: 10.1093/carcin/bgp318. [DOI] [PubMed] [Google Scholar]
- 30.Magiera M, Mora S, Mojsa B, et al. Trim17-mediated ubiquitination and degradation of Mcl-1 initiate apoptosis in neurons. Cell Death Differ. 2013;20:281–292. doi: 10.1038/cdd.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang NJ, Zhang L, Tang W, et al. The Trim39 ubiquitin ligase inhibits APC/CCdh1-mediated degradation of the Bax activator MOAP-1. J Cell Biol. 2012;197:361–367. doi: 10.1083/jcb.201111141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beverly LJ, Lockwood WW, Shah PP, et al. Ubiquitination, localization, and stability of an anti-apoptotic BCL2-like protein, BCL2L10/BCLb, are regulated by Ubiquilin1. Proc Natl Acad Sci U S A. 2012;109:E119–E126. doi: 10.1073/pnas.1119167109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson R, Goyal L, Ditzel M, et al. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- 34.Hawkins CJ, Wang SL, Hay BA. A cloning method to identify caspases and their regulators in yeast: identification of Drosophila IAP1 as an inhibitor of the Drosophila caspase DCP-1. Proc Natl Acad Sci U S A. 1999;96:2885. doi: 10.1073/pnas.96.6.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamm I, Kornblau SM, Segall H, et al. Expression and prognostic significance of IAP-family genes in human cancers and myeloid leukemias. Clin Cancer Res. 2000;6:1796–1803. [PubMed] [Google Scholar]
- 36.Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc Natl Acad Sci U S A. 2001;98:8662. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schile AJ, García-Fernández M, Steller H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev. 2008;22:2256–2266. doi: 10.1101/gad.1663108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi Y, Xia P. Cellular inhibitor of apoptosis protein-1 (cIAP1) plays a critical role in β-cell survival under endoplasmic reticulum stress: promoting ubiquitination and degradation of C/EBP homologous protein (CHOP) J Biol Chem. 2012;287:32236–32245. doi: 10.1074/jbc.M112.362160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis EM, Wilkinson AS, Davis NY, et al. Nondegradative ubiquitination of apoptosis inducing factor (AIF) by X-linked inhibitor of apoptosis at a residue critical for AIF-mediated chromatin degradation. Biochemistry. 2011;50:11084–11096. doi: 10.1021/bi201483g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mei Y, Hahn AA, Hu S, et al. The USP19 deubiquitinase regulates the stability of c-IAP1 and c-IAP2. J Biol Chem. 2011;286:35380–35387. doi: 10.1074/jbc.M111.282020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du C, Fang M, Li Y, et al. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 42.Olson MR, Holley CL, Yoo SJ, et al. Reaper is regulated by IAP-mediated ubiquitination. J Biol Chem. 2003;278:4028–4034. doi: 10.1074/jbc.M209734200. [DOI] [PubMed] [Google Scholar]
- 43.Qiu XB, Markant SL, Yuan J, et al. Nrdp1-mediated degradation of the gigantic IAP, BRUCE, is a novel pathway for triggering apoptosis. EMBO J. 2004;23:800–810. doi: 10.1038/sj.emboj.7600075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hao Y, Sekine K, Kawabata A, et al. Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat Cell Biol. 2004;6:849–860. doi: 10.1038/ncb1159. [DOI] [PubMed] [Google Scholar]
- 45.Qiu XB, Goldberg AL. The membrane-associated inhibitor of apoptosis protein, BRUCE/Apollon, antagonizes both the precursor and mature forms of Smac and caspase-9. J Biol Chem. 2005;280:174–182. doi: 10.1074/jbc.M411430200. [DOI] [PubMed] [Google Scholar]
- 46.Bhatt P, d'Avout C, Kane NS, et al. Specific domains of nucleolin interact with Hdm2 and antagonize Hdm2-mediated p53 ubiquitination. FEBS J. 2012;279:370–383. doi: 10.1111/j.1742-4658.2011.08430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun XX, Challagundla KB, Dai MS. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 2011;31:576–592. doi: 10.1038/emboj.2011.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee HR, Choi WC, Lee S, et al. Bilateral inhibition of HAUSP deubiquitinase by a viral interferon regulatory factor protein. Nat Struct Mol Biol. 2011;18:1336–1344. doi: 10.1038/nsmb.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amson R, Pece S, Lespagnol A, et al. Reciprocal repression between P53 and TCTP. Nat Med. 2011;18:91–99. doi: 10.1038/nm.2546. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Zhao L, Sun H, et al. Gene silencing of FANCF potentiates the sensitivity to mitoxantrone through activation of JNK and p38 signal pathways in breast cancer cells. PloS one. 2012;7:e44254. doi: 10.1371/journal.pone.0044254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ikeda F, Deribe YL, Skånland SS, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wertz IE, Dixit VM. Signaling to NF-κB: regulation by ubiquitination. Cold Spring Harb Perspect Biol. 2010;2:a003350. doi: 10.1101/cshperspect.a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen F, Demers LM, Shi X. Upstream signal transduction of NF-kappaB activation. Curr Drug Targets Inflamm Allergy. 2002;1:137–149. doi: 10.2174/1568010023344706. [DOI] [PubMed] [Google Scholar]
- 54.Chen L, Siddiqui S, Bose S, et al. Nrdp1-mediated regulation of ErbB3 expression by the androgen receptor in androgen-dependent but not castrate-resistant prostate cancer cells. Cancer Res. 2010;70:5994–6003. doi: 10.1158/0008-5472.CAN-09-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Limpert AS, Carter BD. Axonal neuregulin 1 type III activates NF-κB in schwann cells during myelin formation. J Biol Chem. 2010;285:16614–16622. doi: 10.1074/jbc.M109.098780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lockshin RA, Zakeri Z. Apoptosis, autophagy, and more. Int J Biochem Cell Biol. 2004;36:2405–2419. doi: 10.1016/j.biocel.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 57.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 59.Kirkin V, McEwan DG, Novak I, et al. A role for Ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 60.Thurston TLM, Ryzhakov G, Bloor S, et al. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 61.Wei Y, Pattingre S, Sinha S, et al. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. SciSignal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei Y, Sinha S, Levine B. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy. 2008;4:949–951. doi: 10.4161/auto.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Djavaheri-Mergny M, Maiuri M, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene. 2010;29:1717–1719. doi: 10.1038/onc.2009.519. [DOI] [PubMed] [Google Scholar]
- 65.Liu J, Xia H, Kim M, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crighton D, Wilkinson S, O'Prey J, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 67.Ravikumar B, Vacher C, Berger Z, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 68.Maiuri MC, Malik SA, Morselli E, et al. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8:1571–1576. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- 69.Scherz-Shouval R, Weidberg H, Gonen C, et al. p53-dependent regulation of autophagy protein LC3 supports cancer cell survival under prolonged starvation. Proc Natl Acad Sci U S A. 2010;107:18511–18516. doi: 10.1073/pnas.1006124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bond GL, Hu W, Levine AJ. MDM2 is a central node in the p53 pathway: 12 years and counting. Curr Cancer Drug Targets. 2005;5:3–8. doi: 10.2174/1568009053332627. [DOI] [PubMed] [Google Scholar]
- 71.Menin C, Scaini MC, De Salvo GL, et al. Association between MDM2-SNP309 and age at colorectal cancer diagnosis according to p53 mutation status. J Natl Cancer Inst. 2006;98:285–288. doi: 10.1093/jnci/djj054. [DOI] [PubMed] [Google Scholar]
- 72.Lind H, Zienolddiny S, Ekstrøm PO, et al. Association of a functional polymorphism in the promoter of the MDM2 gene with risk of nonsmall cell lung cancer. Int J Cancer. 2006;119:718–721. doi: 10.1002/ijc.21872. [DOI] [PubMed] [Google Scholar]
- 73.Masuya D, Huang C, Liu D, et al. The HAUSP gene plays an important role in non–small cell lung carcinogenesis through p53-dependent pathways. J Pathol. 2006;208:724–732. doi: 10.1002/path.1931. [DOI] [PubMed] [Google Scholar]
- 74.Meetei AR, de Winter JP, Medhurst AL, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 75.Hu S, Du M, Park S, et al. cIAP2 is a ubiquitin protein ligase for BCL10 and is dysregulated in mucosa-associated lymphoid tissue lymphomas. J Clin Invest. 2006;116:174–181. doi: 10.1172/JCI25641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Casas S, Nagy B, Elonen E, et al. Aberrant expression of HOXA9, DEK, CBL and CSF1R in acute myeloid leukemia. Leuk Lymphoma. 2003;44:1935–1941. doi: 10.1080/1042819031000119299. [DOI] [PubMed] [Google Scholar]
- 77.Hoffman MA, Ohh M, Yang H, et al. von Hippel-Lindau protein mutants linked to type 2C VHL disease preserve the ability to downregulate HIF. Hum Mol Genet. 2001;10:1019–1027. doi: 10.1093/hmg/10.10.1019. [DOI] [PubMed] [Google Scholar]
- 78.Kondo K, Klco J, Nakamura E, et al. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 79.Hengstermann A, D'silva MA, Kuballa P, et al. Growth suppression induced by downregulation of E6-AP expression in human papillomavirus-positive cancer cell lines depends on p53. J Virol. 2005;79:9296–9300. doi: 10.1128/JVI.79.14.9296-9300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Winston JT, Strack P, Beer-Romero P, et al. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yuan WC, Lee YR, Huang SF, et al. A Cullin3-KLHL20 Ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell. 2011;20:214–228. doi: 10.1016/j.ccr.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 82.Schwickart M, Huang XD, Lill JR, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2009;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 83.Zhao D, Zheng HQ, Zhou Z, et al. The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 2010;70:4728–4738. doi: 10.1158/0008-5472.CAN-10-0040. [DOI] [PubMed] [Google Scholar]
- 84.Ho KC, Zhou Z, She YM, et al. Itch E3 ubiquitin ligase regulates large tumor suppressor 1 tumor-suppressor stability. Proc Natl Acad Sci U S A. 2011;108:4870–4875. doi: 10.1073/pnas.1101273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Salah Z, Melino G, Aqeilan RI. Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Res. 2011;71:2010–2020. doi: 10.1158/0008-5472.CAN-10-3516. [DOI] [PubMed] [Google Scholar]
- 86.Sarkar TR, Sharan S, Wang J, et al. Identification of a Src tyrosine kinase/SIAH2 E3 ubiquitin ligase pathway that regulates C/EBPδ expression and contributes to transformation of breast tumor cells. Mol Cell Biol. 2012;32:320–332. doi: 10.1128/MCB.05790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Razinia Z, Baldassarre M, Bouaouina M, et al. The E3 ubiquitin ligase specificity subunit ASB2α targets filamins for proteasomal degradation by interacting with the filamin actin-binding domain. J Cell Sci. 2011;124:2631–2641. doi: 10.1242/jcs.084343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Duan S, Cermak L, Pagan JK, et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2011;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]