Abstract
Objectives
The objective of the study was to identify pancreatic islet-selective gene(s) that may play a functional role in islet biology and diabetes development.
Methods
Through bioinformatics, we identified and cloned a pancreas-enriched complementary DNA encoding transmembrane emp24 protein transport domain 6 (TMED6) and examined its mRNA and protein expression in tissues and islet cell lines by Northern analysis and immunofluorescence histochemistry. We also studied die role of TMED6 in insulin secretion using a knockdown approach and its gene expression changes during the development of diabetes in Goto-Kakizaki rats.
Results
TMED6 is selectively expressed in pancreatic islets and belongs to the EMP24_GP25L superfamily, which is known to be involved in protein trafficking and secretion. Northern analysis revealed that TMED6 mRNA is highly and selectively expressed in pancreas. Immunofluorescence histochemistry of mouse pancreas showed that TMED6 expression is restricted to pancreatic islets with higher levels in α cells than β cells. Knockdown of TMED6 gene expression in Min6 β cells decreased insulin secretion. Moreover, TMED6 gene expression was significantly lower in diabetic Goto-Kakizaki rats.
Conclusions
TMED6 may play a functional role in islet biology, particularly in hormone production or secretion, and its dysregulation may be implicated in the development of diabetes.
Keywords: TMED6, expression, islets, insulin, diabetes, Goto-Kakizaki rats
Pancreatic islets consist of specialized types of endocrine cells whose main function is to secrete hormones to maintain glucose homeostasis. Decades of intensive research have identified many islet-specific or cell type–selective genes, such as transcription factors Pdx-1 and neurogenin 3, and final hormone products insulin and glucagon, which confer unique functional or morphologic characteristics to islet cells.1–3 However, our current understanding of these genes is largely confined to either transcription factors (eg, Pdx-1 and neurogenin 3) or final hormone products (eg, insulin from β cells and glucagon from α cells). A remarkable functional aspect of β cells and α cells is that these cells can produce and store large amounts of insulin and glucagon, respectively, and rapidly release these hormones in response to immediate or long-term demands. Presumably, islet cells are equipped with a unique protein-processing machinery that is conducive to performing these functions, but our knowledge about this machinery and whether there is a cell-specific component is still limited.
During a bioinformatics search for pancreas-specific genes, we discovered that transmembrane emp24 protein transport domain 6 (TMED6), a member of protein family known to participate in transporting protein cargo forward from the endoplasmic reticulum (ER), is selectively expressed in pancreatic islets, and investigated its role in insulin secretion and diabetes development.
MATERIALS AND METHODS
Molecular Cloning and Bacterial Expression of Recombinant TMED6
Total RNA was extracted from C57/BL mouse pancreas using RNAqueous–4PCR (Ambion, Austin, Tex). First-strand complementary DNA (cDNA) was made by reverse transcription in a reaction of 20-μL mixture containing 1 μg of total RNA, poly(dT) primer, and Moloney murine leukemia virus reverse transcription using the Advantage kit (Clontech, Mountain View, Calif) and used it as a template for subsequent polymerase chain reaction (PCR) amplification. On the basis of TMED6 DNA sequence (NM_025458, GenBank), forward primer 5′-NdeI–linked ccatATGTTCCCTTTGCTCCTCGT-3′ and reverse XhoI-linked primer 5′-agtcctcgag GCATCTTGGCTTCTTTGTG-3′ were designed to amplify the full-length TMED6 coding sequence using the high-fidelity PCR system (Boehringer Mannheim, Indianapolis, Ind). The resulting sequence was digested with NdeI and XhoI and then subcloned into His(6)-tag bacterial expression vector PET28 (Novagen, Gibbstown, NJ) to create plasmid p6608. After verifying the sequence, the clone was transformed into Escherichia coli Turner (Novagen). The His-tagged protein was overexpressed by isopropyl betathiogalactoside induction4 and purified with Ni2+-NTA resin (Qiagen) under denaturing conditions (8 mol/L urea) to homogeneity for polyclonal antibody production in rabbits (AbboMax, San Jose, Calif).
Northern Analysis and Real-Time PCR
For Northern analysis, 3 male C57BL/6J mice (Jackson Laboratory) aged 7 to 9 weeks were euthanized with CO2. Tissue from pancreas, brain, white and brown fat, heart, kidney, intestine, liver, lung, and muscle were excised and snap-frozen in liquid nitrogen. Total RNA was extracted with Trizol (Invitrogen, Carlsbad Calif) according to manufacturers instructions. Fifteen micrograms of total RNA was loaded per lane for Northern analysis. The mouse TMED6 cDNA probe corresponding to full-length cDNA was randomly labeled (Stratagene, La Jolla, Calif) with 32P-dCTP, hybridized to the blot at 65°C in Rapid-hyb buffer (Amersham, Piscataway, NJ), washed twice with 0.5 × SSC/1% sodium dodecyl sulfate at 65°C (stringent wash), and then exposed to x-ray film.
Histoimmunofluorescence Analysis
Anti-TMED6 serum was raised against the recombinant mouse TMED6 protein. To obtain antigen-specific antibody, 2 mL of diluted antiserum was incubated with a polyvinylidene fluoride blot (Bio-Rad, Hercules, Calif) containing 1 mg of the recombinant TMED6 protein. The blot strip was washed, and bound antibody was eluted with 500 μL of Tris-HCl buffer (pH 5) into a tube containing neutralizing buffer (pH 9). The resulting antibody eluate was used for subsequent immunostaining. Mouse pancreas was cryosectioned (20 μm), fixed with 4% paraformaldehyde, and permeabilized with 0.5% cold Triton X-100. The tissue slides were incubated with primary anti-TMED6 (1:20 dilution), anti-insulin (MAB107; Chemicon, Temecula, Calif), or anti-glucagon (A0565, DAKO, Carpinteria, Calif) and then washed and reincubated with a second antibody conjugated with fluorescent chromophorc (goat anti–rabbit IgG, Alexa568, or Alexa488; Molecular Probes). For cell staining, Min6 β cells or TC1.6 α cells were plated on a chamber slide, fixed with 4% paraformaldehyde for 30 minutes, and stained with anti-TMED6/insulin or anti-TMED6/glucagon antibodies. The slides were counterstained with DAPI (4′-6-diamidino-2-phenylindole) to reveal nuclei. Confocal imaging was performed with a Zeiss LSM510 microscope (Carl Zeiss MicroImaging, Thornwood, NY).
MIN6 β-Cell and TC1.6 α-Cell Culture
MIN6 β cells5 and TC1.6 α cells2 were grown in Dulbecco’s modified Eagle medium containing 10% fetal calf serum, 25 mmol/L of glucose, penicillin, and streptomycin in a humidified atmosphere at 37°C with 5% CO2.
TMED6 Knockdown and Insulin Secretion in Min6 β Cells
To decrease TMED6 gene expression, an RNA interference (RNAi) approach was used. MIN6 β cells were plated onto a 24-well cell culture plate at 2 × 105 cells per well in Dulbecco’s modified Eagle medium (25 mmol/L glucose) with 10% fetal bovine serum for 2 days. At 90% cell confluence, 150 nmol/L of predesigned small interfering RNAs (siRNAs) targeting TMED6 (Tmed6_1: CAG ATTAACTTTGCTACACAA; Qiagen) or scrambled control siRNA (catalog no. 1022563; Qiagen) was transfected into the cells using Hyperfectamine transfection reagents (Qiagen) according to the manufacturer’s instructions. Culture medium was collected 48 hours after the transfection to measure insulin concentration by enzyme-linked immunosorbent assay (Mercodia, Uppsala, Sweden). Total RNAs were extracted, and reverse transcription was performed. Quantitative real-time PCR for TMED6 was conducted on a LightCycler 480 (Roche) using 1 μL of the cDNA and TaqMan Gene Expression Assay primer and probe set (assay ID: Mm00481395_ml; Applied Biosystems, Foster, Calif). Relative fold changes of TMED6 mRNA expression levels were calculated using 2−ΔΔCT method with β-actin as the reference gene.
Animal Studies
Animal studies were approved by the Institutional Animal Care and Use Committee of the University of Maryland School of Medicine. Goto-Kakizaki (GK) breeding pairs were a gift from Dr R. V Farese at the University of South Florida and expanded at the University of Maryland. Wistar rats (Charles River, Wilmington, Mass) were used as controls.6 Intraperitoneal glucose tolerance tests (IPGTTs; glucose, 2 g/kg) were performed on male rats at 4 or 8 weeks of ages after an overnight fast. Glucose levels were measured in tail vein blood using a glucometer (Accu-Check, Roche, Indianapolis, Ind). For quantitative PCR of rat TMED6, total rat pancreas RNAs were reverse transcribed for TaqMan assay by using a mouse TMED6 probe set (Mm00481395_m, match rat TMED6 sequence) and reference probe β-actin (Rn00667869_ml).
Statistical Analysis
Results are expressed as mean ± SE. Student t test was used to examine the differences between means (GraphPad Software, San Diego, Calif). Differences were considered to be significant P < 0.05.
RESULTS
Molecular Cloning and Bacterial Expression of TMED6
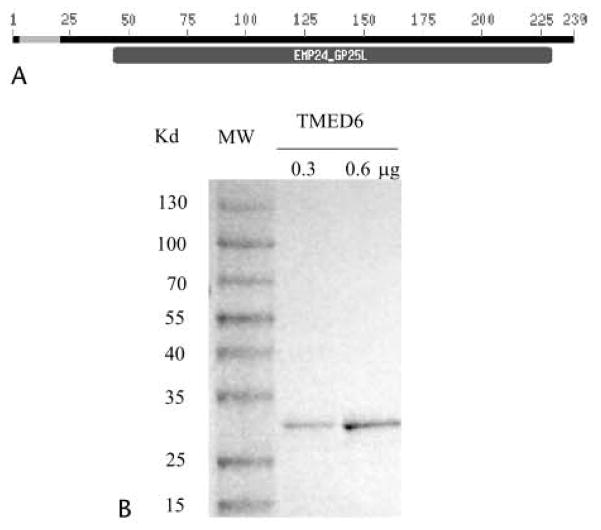
To discover new transcripts expressed predominantly in the pancreas and islet, we pooled and catalogued all expressed sequence tags (ESTs) from human (~200,000 ESTs) and mouse (~120,000 ESTs) in GenBank (build no. 34) and ranked the frequency of their expression in the pancreas compared with other tissues. As expected, known pancreas-specific genes such as insulin, glucagon, pancreatic lipase, and amylase were at the top of the list. In the next group of genes, the TMED6 gene stood out because of its relative abundance and the specificity of its expression in the pancreas and the absence of information on its role in the pancreas and islets. Sequence analysis revealed that the full-length TMED6 cDNA encodes a 239–amino acid protein. Analysis of the putative protein sequence indicated that a large portion of the protein contains a highly hydrophobic region that is typical of a secretory signal sequence, followed by a domain belonging to the EMP24_GP25L superfamily (Fig. 1A). The EMP24_GP25L domain has been implicated in transporting protein cargo forward from the ER and binding to coat proteins by their cytoplasmic domains.7 The mouse TMED6 protein shares 77% and 94% identity to its human and rat counterparts (data not shown), respectively.
FIGURE 1.
TMED6 protein structure and bacterial expression. A, TMED6 protein sequence contains a hydrophobic secretory leader sequence (amino acids 3–22, blue), followed by an EMP24_GP25L domain, which is known to participate in protein transportation. B, Purified recombinant His-TMED6 recombinant protein revealed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis with molecular ladder.
Next, we expressed the entire mouse TMED6 protein as a His-tag fusion recombinant protein in bacteria. Using nickel-affinity chromatography, we purified protein to more than 95% homogeneity on a denaturing gel at a size of approximately 28 kd (Fig. 1B), which is expected from the fusion protein.
Tissue and Cellular Expression Pattern of TMED6
Northern analysis revealed a restricted expression of TMED6 in mouse tissues, with the highest expression in whole pancreas, lower levels of expression in white adipose tissue and intestine, and no expression in the brain, liver, lung, kidney, and heart (Fig. 2).
FIGURE 2.
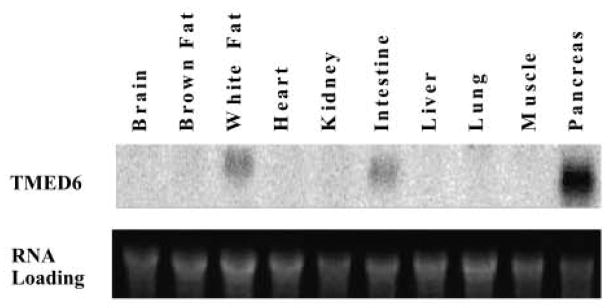
Tissue distribution of TMED mRNA. Total RNA 15 μg) from the indicated tissues was probed with P32-labeled TMED6 (upper panel). Lower panel indicates RNA loading.
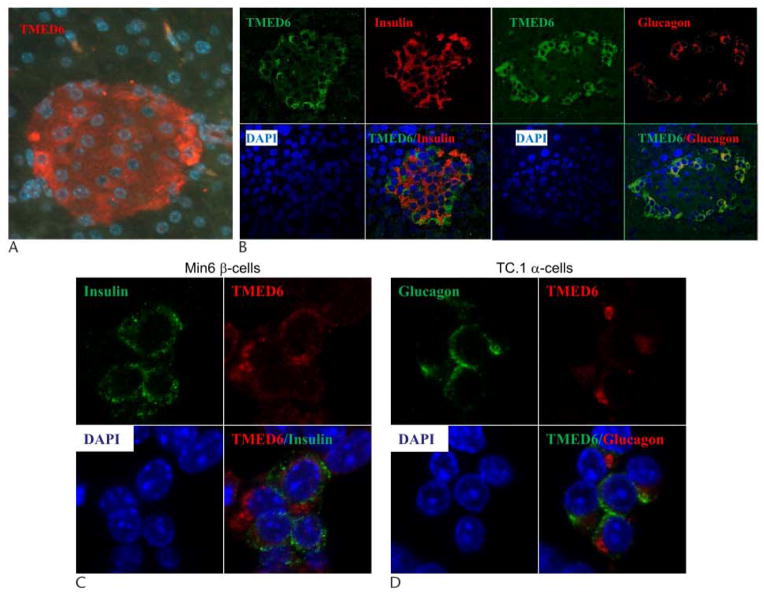
Using TMED6 antibody generated from the recombinant proteins, we conducted immunofluorescence studies to localize TMED6 expression within the pancreas. As shown in Figure 3A, TMED6 is expressed in islets and may be expressed at very low levels in exocrine tissue. Costaining of the β-cell marker insulin or α-cell marker glucagon with TMED6 indicated that TMED6 is expressed significantly in α cells and moderately in β cells (Fig. 3B). Further studies using confocal microscopy revealed that TMED6 appears in the cytoplasm and is not co localized with insulin in Min6 β cells or with glucagon in TC.1 α cells (Fig. 3D, C).
FIGURE 3.
Expression pattern of TMED6 in islet cells. A, Immunofluorescence staining was performed on pancreas for TMED6 (red, Alexa594). TMED6 (green, Alexa488) costained with insulin (red) and glucagon (red) on pancreatic islet (panel B) or Min6 β cells confocal views of (panel C) and TC.1 α cells (panel D). Nuclei were counterstained with DAPI.
Knockdown of TMED6 Resulted in Reduced Insulin Secretion
To determine whether TMED6 is involved in insulin secretion, we took an RNAi approach. Control and TMED6 siRNA oligonucleotides were transiently transfected into MIN6 β cells. TMED6 siRNA decreased TMED6 mRNA levels by 52%, resulting in a 35% drop in insulin secretion by Min6 β cells in culture medium containing 25 mmol/L of glucose (Fig. 4).
FIGURE 4.
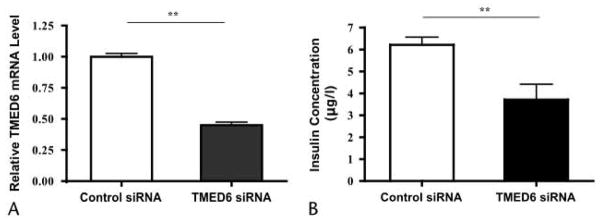
Knockdown of TMED6 resulted in decrease of insulin secretion. Min6 β cells were transfected with control (open column) and TMED6 RNAi duplexes (black column). Forty-eight hours later, TMED6 mRNA was measured by real-time quantitative PCR (A), and culture medium insulin was assayed by enzyme-linked immunosorbent assay (B). Data are expressed as mean ± SE of 4 independent experiments. **P < 0.01.
TMED6 Gene Expression Is Decreased During the Development of Diabetes in GK Rats
The GK rats developed type 2–like diabetes with moderate hyperglycemia due to impaired insulin secretion and mild to moderate insulin resistance.8,9 We confirmed by IPGTT that GK rats were insulin resistant but were not diabetic at 4 weeks of age, but were diabetic at 8 weeks of age (blood glucose: 98 vs 71 mg/dL and 135 vs 80 mg/dL at 4 and 8 weeks of age for GK and Wistar rats, respectively; Figs. 5A, B). TMED6 gene expression was determined by quantitative PCR during the development of diabetes. As shown in Figure 5C, there was no statistically significant difference in TMED6 expression between GK and Wistar control rats at 1 week of age, but TMED6 expression was significantly lower in GK rats at 4 weeks (60% of control, P < 0.01) and 8 weeks (45% of control, P < 0.01) of age. Of note, the pancreatic islet area was not significantly reduced in GK rats compared with Wistar rats (data not shown). Thus, TMED6 expression is significantly decreased in the diabetic GK rats.
FIGURE 5.
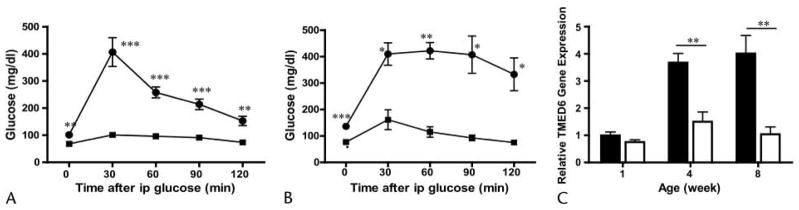
Decreased expression of TMED6 in GK rats. IPGTT of GK rats at 4 weeks (A) and 8 weeks (B) of age. *P < 0.05, **P < 0.01, ***P< 0.001. C, Quantitative PCR of TMED6 gene expression in GK (open columns) and Wistar (black columns) rats at 1, 4, and 8 weeks of age. Expression was normalized to β-actin. **P < 0.01. All comparisons are between GK and Wistar control rats and data are expressed as mean ± SE from 3 to 4 rats per group.
DISCUSSION
Pancreatic islet cells are a type of specialized endocrine cell engineered to make, process, store, and secrete hormones, especially insulin and glucagon, in large quantities. Presumably, islet cells are equipped with a unique machinery to fulfill this heavy workload, but our understanding of such machinery is limited. Toward a better understanding of islet biology at the molecular level, using a bioinformaties approach, we discovered that TMED6 is a gene that is selectively expressed in the pancreas.
TMED6 contains a transmembrane emp24 protein transport domain. Emp24 belongs to the p24 (p24/gp35L/emp/Erp) family of proteins, which play an important role in vesicle-mediated protein trafficking mediated by vesicles in the secretory pathway.7 Proteins of the p24 family are present on the membranes of secretory pathway organelles (ER, Golgi, COPI, and COPII vesicles).7,10 However, our preliminary study indicated that TMED6 was not colocalized with ER marker protein disulfide isomerase (data not shown). Nevertheless, based on the known functions of other p24 proteins, TMED6 may play a role in insulin and glucagon trafficking and secretion. Our knockdown study of TMED6 in Min6 cells supported such a role in insulin secretion. Further studies are required to determine the mechanism with which TMED6 modulates insulin secretion in cells. It is worth noting that TMED6 is expressed at higher levels in α cells than in β cells, suggesting that TMED6 may play a significant role in α-cell biology and function and warrant further studies of the gene in glucagon production and secretion.
To determine the possible relevance of TMED6 to diabetes, we examined its expression in GK rats, a type 2 diabetes model.11 The cause of diabetes in the GK model is likely a complex interaction of multiple events resulting in β-cell secretory dysfunction and/or decreased β-cell mass. GK rats are euglycemic at birth and develop progressive hyperglycemia, with most rats having glucose levels in the diabetic range by 8 weeks. The progressive decrease in TMED6 expression in GK. rats suggests that TMED6 may be implicated in the development of diabetes in GK rats.
In conclusion, we have discovered that TMED6 is selectively expressed in pancreatic α and β cells. Our preliminary functional studies in Min6 β cells indicate that the protein plays a role in insulin production/secretion and is implicated in the development of diabetes in GK rats. Based on the biological function of other p24 family members, TMED6 is likely involved in protein trafficking and processing in insulin and glucagon secretory vesicles. Future study will be focused on elucidating the unique role(s) TMED6 plays in islet biology, hormone production and the development of diabetes through targeted gene knockout and on its significance in human normal and diabetic islets.
Acknowledgments
The study was partially supported by grants from Maryland Clinical Nutrition Research Unit (P30DK072488) and the Baltimore Diabetes Research and Training Center (P60DK079637) from the National Institutes of Health.
The authors thank Drs Jianhua Shao for critical reading of the manuscript and providing Min6 A cells and Donald Steiner for providing TC.1 > cells.
Footnotes
The authors declare no conflict of interest.
References
- 1.Edlund H. Pancreatic organogenesis—developmental mechanisms and implications for therapy. Nat Rev Genet. 2002;3(7):524–532. doi: 10.1038/nrg841. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Webb G, Cao Y, et al. Contrasting patterns of expression of transcription factors in pancreatic α and β cells. Proc Natl Acad Sci U S A. 2003;100(22):12660–12665. doi: 10.1073/pnas.1735286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marselli L, Thorne J, Ahn YB, et al. Gene expression of purified β-cell tissue obtained from human pancreas with laser capture microdissection. J Clin Endocrinol Metab. 2008;93(3):1046–1053. doi: 10.1210/jc.2007-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang RZ, Blaileanu G, Hansen BC, et al. cDNA cloning, genomic structure, chromosomal mapping, and functional expression of a novel human alanine aminotransferase. Genomics. 2002;79(3):445–450. doi: 10.1006/geno.2002.6722. [DOI] [PubMed] [Google Scholar]
- 5.Shao J, Qiao L, Friedman JE. Prolactin, progesterone, and dexamethasone coordinately and adversely regulate glucokinase and cAMP/PDE cascades in MIN6 β-cells. Am J Physiol Endocrinol Metab. 2004;286(2):E304–E310. doi: 10.1152/ajpendo.00210.2003. [DOI] [PubMed] [Google Scholar]
- 6.O’Rourke CM, Davis JA, Saltiel AR, et al. Metabolic effects of troglitazone in the Goto-Kakizaki rat, a non-obese and normolipidemic rodent model of non–insulin-dependent diabetes mellitus. Metabolism. 1997;46(2):192–198. doi: 10.1016/s0026-0495(97)90301-2. [DOI] [PubMed] [Google Scholar]
- 7.Anantharaman V, Aravind L. The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biol. 2002;3(5):research0023.1–0023.7. doi: 10.1186/gb-2002-3-5-research0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitahara A, Toyota T, Kakizaki M, et al. Activities of hepatic enzymes in spontaneous diabetes rats produced by selective breeding of normal Wistar rats. Tohoku J Exp Med. 1978;126(1):7–11. doi: 10.1620/tjem.126.7. [DOI] [PubMed] [Google Scholar]
- 9.Ostenson CG, Khan A, Abdel-Halim SM, et al. Abnormal insulin secretion and glucose metabolism in pancreatic islets from the spontaneously diabetic GK rat. Diabetologia. 1993;36(1):3–8. doi: 10.1007/BF00399086. [DOI] [PubMed] [Google Scholar]
- 10.Strating JR, Martens GJ. The p24 family and selective transport processes at the ER-Golgi interlace. Biol Cell. 2009;101 (9):495–509. doi: 10.1042/BC20080233. [DOI] [PubMed] [Google Scholar]
- 11.Portha B, Lacraz G, Chavey A, et al. Islet structure and function in the GK rat. Adv Exp Med Biol. 2010;654:479–500. doi: 10.1007/978-90-481-3271-3_21. [DOI] [PubMed] [Google Scholar]