Abstract
Cellular senescence is a state of stable cell growth arrest. Activation of oncogenes such as RAS in mammalian cells typically triggers cellular senescence. Oncogene-induced senescence (OIS) is an important tumor suppression mechanism, and suppression of OIS contributes to cell transformation. Oncogenes trigger senescence through a multitude of incompletely understood downstream signaling events that frequently involve protein kinases. To identify target proteins required for RAS-induced senescence, we developed a small molecule screen in primary human fibroblasts undergoing senescence induced by oncogenic RAS (H-RasG12V). Using a high-content imaging system to monitor two hallmarks of senescence, senescence-associated β-galactosidase activity expression and inhibition of proliferation, we screened a library of known small molecule kinase inhibitors for those that suppressed OIS. Identified compounds were subsequently validated and confirmed using a third marker of senescence, senescence-associated heterochromatin foci. In summary, we have established a novel high-content screening platform that may be useful for elucidating signaling pathways mediating OIS by targeting critical pathway components.
INTRODUCTION
Aberrant oncogene activation is an important driver of cellular transformation; however, initial atypical oncogene activation that occurs in primary cells typically triggers cellular senescence, a state of stable cell growth arrest 1-3. Oncogene-induced senescence (OIS) is an important tumor suppressive pathway, and suppression of OIS promotes tumorigenesis 4. For example, oncogenic RAS or BRAF triggers senescence of melanocytes, which results in formation of benign nevi and thus suppresses melanoma development 5-7. The RAS oncogene is mutated in a number of cancer types [Reviewed in 8]. Oncogenic RAS has been extensively studied in the context of OIS, where it triggers senescence by a cascade of kinases 3.
Senescent cells exhibit several distinctive morphological characteristics and molecular markers, including a large, flat morphology, decrease in cell proliferation and expression of senescence-associated β-galactosidase activity (SA-β-gal) 9 [Reviewed in 10]. Expression of SA-β-gal activity is considered a universal marker of senescent cells 11. In addition, senescence induced by oncogenic RAS is also characterized by domains of transcriptionally silenced heterochromatin, known as senescence-associated heterochromatin foci (SAHF) 12. SAHF contribute to senescence by silencing proliferation-promoting genes such as E2F target genes 13.
Inactivation of tumor suppressors such as p53 and p16 inhibits OIS 3. Due to the importance of OIS in tumor suppression, compounds/molecules that regulate OIS not only serve as useful tools in studying OIS, but may also lead to the identification of new tumor suppressors. As a result, we have developed a high-content screening assay to aid in the identification of novel OIS regulators. We utilized a kinase inhibitor library to determine compounds that inhibited OIS in the context of oncogenic RAS overexpression. In addition, the identified compounds could be studied further to help elucidate proteins involved in mediating OIS, which may lead to identification of novel tumor suppressors.
Previously, a screen for modulators of stress-induced senescence in prostate cancer has been described 14. In this study, the authors utilized percentage of SA-β-gal positive cells as a senescence output, however the screen is limited due to necessity of having to manually quantify SA-β-gal positive cells. In contrast, we report the development of a high-content imaging based screen assay. In addition, using a library of 160 well-characterized kinase inhibitors, we performed a proof-of-principle screen for inhibitors of senescence induced by oncogenic RAS.
SA-β-gal activity was utilized in a high-content screening assay in a 96-well platform as the primary measure of senescence. To eliminate artifactual hits due to decreased cell number in the absence of OIS inhibition, we quantified cell numbers using nuclear staining. Utilizing this newly developed platform, we identified 17 kinase inhibitors as suppressors of oncogenic RAS-induced senescence. We further confirmed our findings by using SAHF staining as an additional marker of senescence. All 17 inhibitors were individually validated, and 15 out of 17 these were confirmed. This study introduces a novel assay for screening of modulators of OIS, and we report the identification of several compounds as bona fide suppressors of OIS.
MATERIALS AND METHODS
Tissue Culture
Primary diploid fibroblasts (IMR90) were cultured according to the American Type Culture Collection (ATCC). Experiments were performed with IMR90 that were between 25 and 36 population doublings (PD).
Plasmids and Retrovirus
pBABE-H-RasG12V was obtained from Addgene. Retrovirus production and transduction has previously been described 15. Phoenix cells were used to facilitate retroviral packaging (Dr. Gary Nolan, Stanford University).
Screen Setup
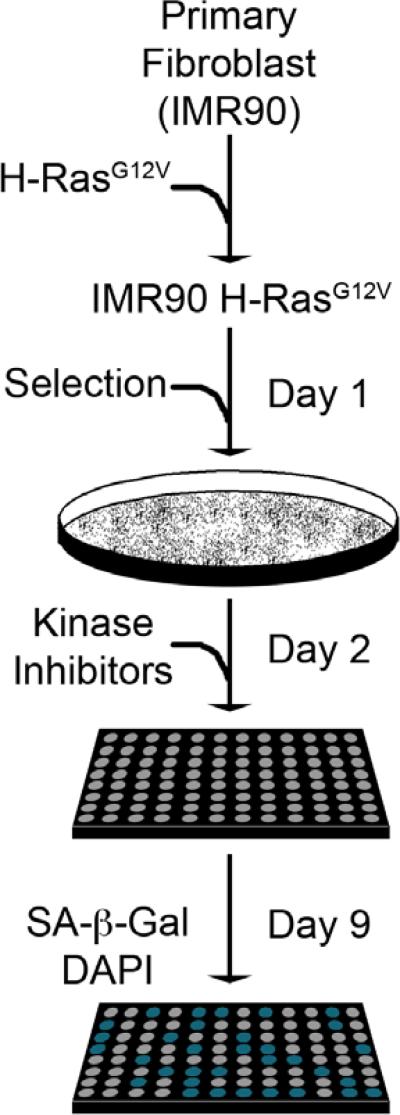
Double infections of IMR90 cells in 100 mm dishes were performed using a retrovirus encoding for pBABE-H-RasG12V (Day -1 and 0, respectively). Cells were selected with puromycin (1 μg/mL) for an additional two days and then plated into a 96-well plate (1,000 cells/well) . The initial number of cells per well (1,000) was optimized to avoid confluence-induced growth inhibition. At the end of day 2, cells were treated with kinase inhibitors (KI) by pin transfer at ~250nM. On day 9, cells were subjected to a quantitative SA-β-gal assay (described below) and stained with DAPI to visualize nuclei, which allowed for quantification of cell number.
Senescence Assay
Senescence-associated β-galactosidase (SA-β-Gal) assay has previously been described 9. Briefly, cells were fixed in 2% formaldehyde and 0.2% glutaraldehyde and washed with phosphate-buffered saline. Staining solution [40mM Na2HPO4, 150mM NaCl, 2mM MgCl2, 5mM K3Fe(CN)6, 5mM K4Fe(CN)6, 1mg/mL X-gal] was added with a multi-channel pipette, and cells were incubated for 24hrs. Cells were stained with DAPI (0.15μg/mL) to visualize nuclei.
For the screen, four fluorescence and four phase contrast images (individual image size = 2.9 megabytes) were acquired from each well of the 96-well plates with an ImageXpress Micro high-content imaging system (Molecular Devices, Sunnyvale CA). Using a 10X Pan-Fluor objective with a working distance of 15.2 mm, phase contrast images were captured (exposure time = 5 μsec) to record SA-β-gal staining. Fluorescence images were captured using a 10X Pan-Fluor objective with a Semrock filter (300W xenon lamp, Excitation 377/50 nm, Emission 447/60 nm and exposure time = 55 μsec) to record DAPI staining of cell nuclei. Note that at 10X magnification SAHF are indistinguishable. In addition, objective and filter cube change together with image-based autofocus allowing for simultaneous imaging of both channels. Images were analyzed utilizing the MultiWavelength Scoring module in MetaXpress image analysis software (Molecular Devices). Briefly, based on preliminary experiments with control and RAS-expressing cells, a threshold was determined at which to define positive beta-gal pixels. This threshold was applied to all of the phase contrast images (Figure 1, orange pixels), and the masked area for each image was systematically quantified. DAPI positive nuclei were counted with MetaXpress image analysis software by restricting counts to circular objects with a diameter greater than 1 micron. AcuityXpress software was used for informatics processing and data visualization. The four images were quantified and the values for each SA-β-gal area and nuclei number were averaged. Compared with counting percentage of SA-β-gal positive cells manually, the current approach allows for high-content screening assay to be developed. Overall, automatic image acquisition to quantification averaged 2 hours per 96-well plate.
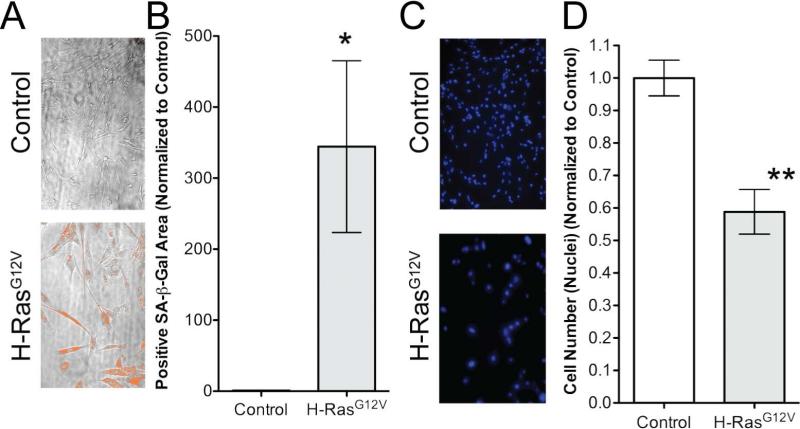
Figure 1. Screen output development.
Using the ImageXpress Micro high-content imaging system, each well was systematically imaged 8 times: 4-bright-field images and 4-fluorescence images. A) Representative bright-field images from screen. Note the “mask” (orange pixels) applied to bright-field images indicating the quantified SA-β-gal positive area. B) Quantification of SA-β-gal positive area between Control and H-RasG12V cells (*p=0.029). C) Representative fluorescence images from screen. D) Quantification of nuclei between Control and H-RasG12V cells (**p=0.0034).
We determined the average number of cells (Nctrl) and SA-β-gal area (BGPPctrl) for IMR90s transduced with pBABE-H-RasG12V treated with DMSO vehicle control, which served as a positive base value for comparison. The average number of nuclei and SA-β-gal area from each of the 160 kinase inhibitors was normalized to DMSO. These normalized values were averaged between replicates. Kinase inhibitors (KI) that inhibited senescence were identified by the following equation: KI prevented senescence if NKIx > [Nctrl + Standard Deviation of Nctrl] and at the same time BGPPKIx < [BGPPctrl - Standard Deviation of BGPPctrl].
All 17 active compounds identified as hits in the screen, along with an inactive control compound (CAS # 648449-76-7; PI3Kγ inhibitor II), were individually examined for SA-β-gal activity and SAHF formation. 300 cells from each of the groups were examined for β-galactosidase (blue) positivity within the cytosol. Utilizing total cell number and β-galactosidase positive cells the positive percentage was calculated. Cells were stained with DAPI to visualize nuclei and SAHF. 300 cells from each of the groups were examined for punctate DAPI stained dense spots, where >10 spots within the nuclear compartment were considered SAHF positive. Utilizing total cell number and SAHF positive cells the positive percentage was calculated. Cells were imaged using a Nikon Eclipse Ni fluorescence microscope as previously described 16. These experiments were performed in triplicate. Data from validation experiments is shown as a percentage of SA-β-gal or SAHF positive cells.
Statistical Analysis
Statistical significance was determined by t-test using GraphPad statistical software (La Jolla, CA). Values were considered significant if p < 0.05. Z’-Score was calculated using Microsoft Excel.
Kinase Inhibitor Library
The kinase inhibitor library, which contained 160 well-characterized kinase inhibitors, was purchased from EMD Millipore (InhibitorSelectTM). Compounds were maintained in 100% DMSO. Inhibitors were added to the growth medium of cells growing in 96-well microplates by pin transfer (Final DMSO % = 0.375%). Eight wells of cells in each plate treated with DMSO (vehicle control) were included as vehicle controls. To avoid plate location-specific effects, these DMSO wells were spread across the plate.
RESULTS
Development of a high-content screening assay for identification of small molecule suppressors of senescence induced by oncogenic RAS
Ectopic expression of activated oncogenes such as RAS is a standard approach to induce cellular senescence in a synchronized manner in primary mammalian cells 3. Thus, we ectopically expressed oncogenic H-RASG12V by retroviral transduction in primary human fibroblasts (IMR90 cells) to induce senescence. Senescence phenotypes such as SA-β-gal activity typically take several days to develop 15. SA-β-gal activity in the cytoplasm is considered a universal marker of cellular senescence 9 and can be visualized by phase contrast microscopy. In addition, a definitive marker of senescence is a decreased cell proliferation [Reviewed in 17]. We sought to develop a high-content assay for screening small molecule modulators of senescence.
In an initial pilot experiment, using MetaXpress image analysis, a threshold of SA-β-gal intensity was established to define a SA-β-gal positive pixel. This threshold was subsequently used in the large-scale screen to apply a “mask” to the four phase contrast images, and the positive pixel area per image was determined (Figure 1A, orange area). Importantly, we observed a significant and robust increase in SA-β-gal area in RAS-infected cells compared to controls (Figure 1B). We also observed a significant reduction in the number of nuclei in RAS-expressing cells compared to control cells (Figure 1C and D). The average SA-β-gal area and number of nuclei for the positive control (DMSO vehicle treated RAS-expressing cells) from the same screening plate were used as a baseline to compare with the kinase inhibitor treated cells.
It is well established that a cascade of kinases plays a role in regulating oncogenic RAS-induced senescence 18. We therefore applied our screening assay using a library of 160 well-characterized kinase inhibitors in a 96-well plate format 19. The screen was performed by treating RAS-transduced cells with 250 nM of each of the 160 kinase inhibitors or vehicle control (DMSO) (Figure 2A). This concentration was chosen based on typical concentrations at which these compounds are utilized in cell-based assays 19. Furthermore, it is known that cell density affects SA-β-gal activity. To limit the potential non-specific effects of cell density on SA-β-gal expression, a range of cell concentrations were tested during assay development to avoid cell confluence over the course of the assay. Typically, 1,000 RAS-overexpressing cells were inoculated per well. We utilized a time-point of 9 days following RAS transduction to assess OIS based on our previous work 15. Eight wells of vehicle control were included in each plate as positive controls for comparison. In addition, control virus-infected cells were included as negative controls for comparison. Cells in the 96-well plates were fixed, and markers of senescence were examined at day 9 post RAS infection. Specifically, we stained the fixed cells for SA-β-gal activity and with a fluorescent DNA dye (DAPI) to visualize nuclei, which was used as a surrogate for cell number. Four phase contrast and fluorescence images were taken of each well and systematically quantified and averaged. This assay allows us to measure the expression of a senescence marker simultaneously with cell number to reflect changes in cell proliferation in the same population of cells. To ensure that the decrease in expression of SA-β-gal activity was not due to a decrease in cell proliferation, we focused on kinase inhibitors that significantly suppressed SA-β-gal activity but did not decrease cell proliferation.
Figure 2. Senescence screen setup and analysis.
Primary fibroblasts (IMR90) were transduced with a H-RasG12V encoding retrovirus over two days (Day -1 and 0). Cells were then selected with puromycin (1 μg/mL) for two days, plated into 96-well plates (1,000 cells/well), and at the end of the second day treated with the kinase inhibitor library at a uniform concentration (250nM). Cells were incubated for seven additional days to allow for senescence and then fixed in a formaldehyde/glutaraldehyde solution. To visualize the SA-β-gal activity and determine cell number, fixed cells were incubated with X-gal and labeled with DAPI.
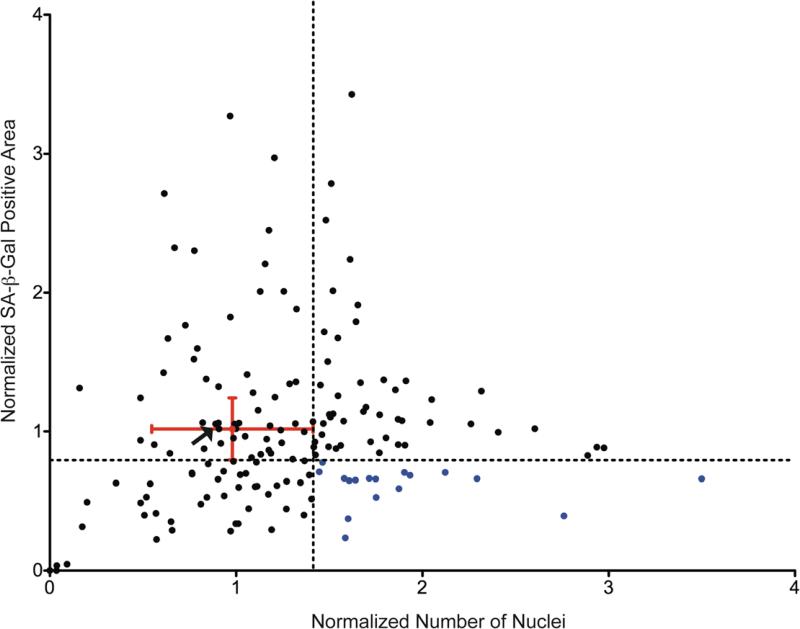
Averages of SA-β-gal area and nuclei number from biological replicate experiments were compiled and normalized to the mean of vehicle (DMSO) control wells. These data were graphed on a scatter plot (Figure 3). Hits were identified as compounds that reduced SA-β-gal area by at least one standard deviation below the mean of DMSO control and conversely increased cell number (nuclei) above one standard deviation of the DMSO control mean (Figure 3, bottom right quadrant, blue dots). According to these criteria, 17 out of the 160 compounds tested significantly suppressed SA-β-gal area while maintaining nuclei number in RAS-expressing cells (compounds are listed in Suppl. Table 1).
Figure 3. Identification of senescence suppressing kinase inhibitors.
Both the nuclei number and SA-β-gal area were normalized to DMSO-treated control wells (Red dot). The normalized numbers of nuclei (a surrogate of cell number) and SA-β-gal area were graphed. The delineation (dotted lines) separates the graph into quadrants, and placement of the lines was determined from the standard deviation of DMSO control wells (red error bars). The lower right quadrant indicates kinase inhibitors that have significantly more nuclei and less SA-β-gal area (blue dots). Arrow indicates inhibitor used as negative control in subsequent validation experiments.
Validation of identified compounds as suppressors of oncogene-induced senescence
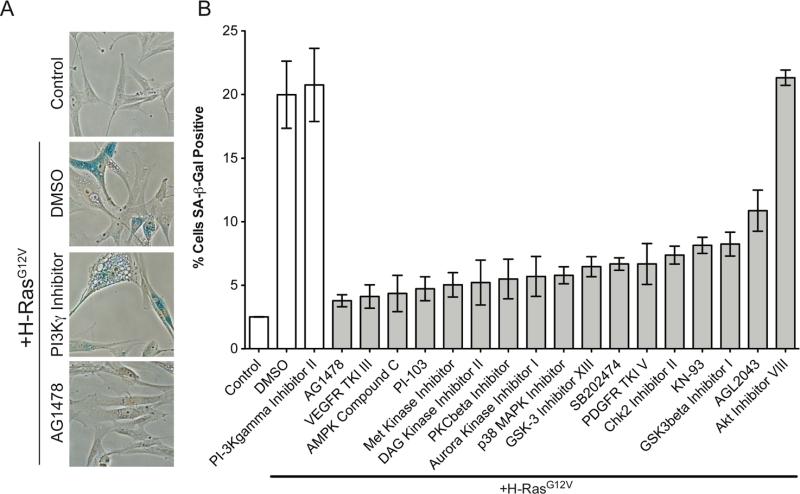
Recapitulating the experimental design of the screen, we individually treated RAS-expressing cells with DMSO vehicle control or with 250 nM of the 17 identified inhibitors. As an additional control, RAS-expressing cells were also treated with a kinase inhibitor (CAS # 648449-76-7; PI3Kγ inhibitor II) that had no effect on SA-β-gal area or cell number based on initial screening (Figure 3, arrow). These cells were then utilized in the SA-β-gal assay to examine changes in expression of SA-β-gal activity. Upon manual quantification of SA-β-gal positive cells, we discovered that 16 of 17 inhibitors significantly reduced the expression of SA-β-gal activity in our validation experiments (Figure 4 A-B and Suppl. Table 2).
Figure 4. Validation of kinase inhibitors that significantly inhibit SA-β-gal activity.
A) Primary fibroblasts (IMR90) were transduced with a control or H-RasG12V encoding retrovirus. Cells were plated into 6-well plates (15,000 cells/well), selected with puromycin (1μg/mL) for two days and at the end of the second day treated with 250nM of kinase inhibitors. Cells were incubated for seven additional days to allow for senescence and then fixed. SA-β-gal activity was visualized and nuclei were labeled with DAPI. B) Quantification of SA-β-gal positive cells. (300 cells were counted for each of the conditions). Gray bar = experimental conditions and white bars = controls. TKI = tyrosine kinase inhibitor. Average percentages and p-values are in Suppl. Table 2. Experiments were performed in triplicate (Error bars = standard error).
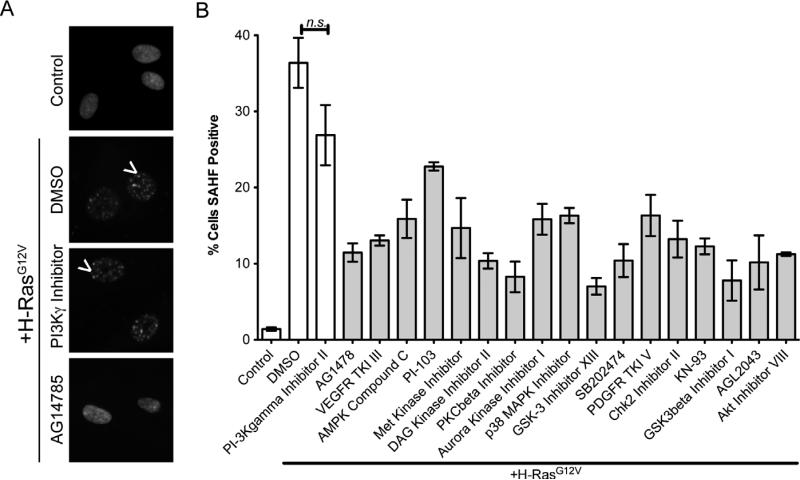
To further confirm senescence suppression by these compounds, we next assayed their effect using an independent marker of senescence, senescence-associated heterochromatin foci (SAHF), which are domains of punctate DAPI stain in the nuclei of senescent cells (Figure 5A, white arrowheads) 12. Toward this goal, RAS-expressing cells were treated with the 17 kinase inhibitors identified in the initial screen, a negative control inhibitor (CAS # 648449-76-7; PI3Kγ inhibitor II) or DMSO. After 9 days of culture, we stained the cells with DAPI to visualize the formation of SAHF. In this independent validation, we discovered that 16 of 17 inhibitors significantly inhibited oncogenic RAS-induced SAHF formation. Interestingly, the inhibitor that failed to prevent SAHF formation (CAS # 371935-74-9; PI-103) was not the same as the inhibitor that failed to reduce SA-β-gal activity (CAS# 612847-09-3; Akt Inhibitor VIII) (Figure 5 A-B and Suppl. Table 2). The discrepancy was not unexpected as there is evidence that SAHF formation is not always necessary for cells to undergo senescence 20, 21. Thus, this compound confirms that SAHF formation can be uncoupled from SA-β–gal activity and may represent a novel tool to understand this phenomenon. In addition, we also examined changes in cell number. Notably, one compound significantly decreased cell number compared to DMSO or negative control inhibitor (CAS# 371935-74-9; PI-103; data not shown), while cell number was not suppressed by the other 16 compounds. Interestingly, the compound that decreased cell number was the same compound that failed to suppress SAHF (Figure 5). These confirmation experiments reveal a false positive rate of 11.7%. In summary, we validated 15 out of the 17 compounds identified from the initial screening as bona fide suppressors of oncogenic RAS-induced senescence, demonstrating that this assay is a robust platform for identifying small molecule regulators of OIS.
Figure 5. Senescence-suppressing compounds also inhibit SAHF, an additional marker of senescence.
A) Primary fibroblasts (IMR90) were transduced with a control or H-RasG12V encoding retrovirus. Cells were plated into 6-well plates (15,000 cells/well), selected with puromycin (1μg/mL) for two days and at the end of the second day treated with 250nM of kinase inhibitors. Cells were incubated for seven additional days to allow for senescence and then fixed. Nuclei were labeled with DAPI, and nuclei containing punctate spots (arrows) were counted as SAHF positive. B) Quantification of SAHF positive cells (Cells counted >300). Order of inhibitors determined by SA-β-gal activity shown in Figure 3. Gray bars = experimental conditions and white bars = controls. TKI = tyrosine kinase inhibitor. Average percentages and p-values are in Suppl. Table 2. Experiments were performed in triplicate (Error bars = standard error).
DISCUSSION
In the current study, we describe the assay development, identification and validation of a high-content screening assay for small molecule modulators of OIS. Using a kinase inhibitor library, we have identified a list of small molecule compounds that suppress oncogenic RAS-induced senescence. We have previously demonstrated that the majority of these compounds target multiple kinases 19. Thus, understanding the mechanism of suppression of OIS by these compounds will require further identification of the relevant kinase targets. Here our identification of 15 kinase inhibitors that suppress oncogenic RAS-induced senescence validates this assay for the identification of such compounds. Significantly, we confirmed the activity of the identified inhibitors using the original screening assay (SA-β-gal activity) and further validated the compounds using an independent senescence marker, SAHF. One of the inhibitors identified (AMPK Compound C, CAS # 855405-64-3) has been reported to prevent senescence, further demonstrating the accuracy of this newly developed assay 22. Notably, none of the other identified compounds have specifically been reported to alter senescence. Follow up studies with these compounds will investigate the relationship between inhibition of a specific kinase and the OIS pathways, which will elucidate molecular mechanisms underlying RAS-induced OIS. In addition, dose-dependent effects of these compounds will also be examined. Taken together, these data establish a high-content screening assays for suppressors of senescence and identify several compounds as suppressors of oncogene-induced senescence.
The high-content screening assay reported here is unique because of the simultaneous utilization of multiple criteria to identify potential hits. Conventional single parameter screens often use two to three standard deviations to identify positive hits. Notably, only two compounds (CAS # 866405-64-3; AMPK Compound C and CAS # 120166-69-0; Diacyglycerol kinase inhibitor I) were scored as hits if two standard deviations was used as cut-off criteria for both SA-β-gal and cell number screening parameters (2 of 160 compounds, Hit rate = 1.25%). Both of the compounds were among the list of compounds confirmed in the follow-up validation assays. In addition, the identification of AMPK Compound C using two standard deviations again demonstrates the accuracy of the screen. Importantly, we confirmed 15 out of the 17 compounds identified using one standard deviation for both SA-β-gal and cell number screening parameters as cut-off. These findings support the use of the criteria described in the current assay. For single parameter based screening, Z’ score is typically used to reflect the robustness of assay. Using SA-β-gal as a single parameter, we explored Z’-score (Z’ score = 0.89) for our newly developed assay by comparing an example of compounds that do not suppress SA-β-gal (i.e. CAS # 212779-48-1; Cdc28p kinase inhibitor) with an example of compounds that efficiently suppressed SA-β-gal (i.e. CAS # 3895-92-9; PKC inhibitor) 23. This further supports the robustness of this newly developed high-content screening assay.
Compared with genetic manipulation for loss of function studies, small molecule inhibitors offer certain advantages in studying kinase function. For example, many kinases are known scaffold proteins that mediate formation of multiple protein large complexes. Genetic knockdown will lead to collapse of these protein complexes and consequently result in unintended non-specific effects. In contrast, small molecule inhibitors of these kinases offer a unique opportunity to study its kinase activity without affecting its interacting proteins. The inhibitors identified here will be invaluable tools in elucidating the role of these kinases in regulating OIS; however, a caveat to the use of small molecule inhibitors is the potential for off-target effects. Therefore future studies knocking down target kinases with shRNAs will be critical to confirm the kinases role in OIS.
The proof-of-principle kinase inhibitor screen described in this report identified compounds that suppressed senescence, which was determined by a decrease in SA-β-gal activity. In validation studies, we observed a discrepancy between kinase inhibitors that prevented SA-β-gal versus those that inhibited SAHF. Therefore, future versions of the screen will include measuring the percentage of SAHF following treatment, which in conjunction with other markers should increase reliability to identify senescent cells. In addition, the high-content screening approach utilized in this assay will also allow for future studies to examine enhancers of OIS. Taking advantage of the platform developed herein and utilizing compounds beyond kinase inhibitors may yield a more comprehensive understanding of OIS.
While senescence is known to be an important tumor suppression mechanism during tumor initiation and progression, it may also be an important mechanism for developing cancer therapeutics. For example, reactivation of the tumor suppressor p53 in murine liver and sarcoma models triggers cellular senescence and the associated tumor regression due to activation of the innate immune response 24. Likewise, we have also demonstrated that restoration of non-canonical Wnt signaling in ovarian cancer cells suppresses tumorigenesis by inducing cellular senescence 25. Thus, small molecule inhibitors that promote senescence of cancer cells could be developed as potential cancer therapeutics. Future studies will employ the high-content screening platform established in the current study to identify small molecules that promote senescence of human cancer cells such as melanoma cells, which can arise by overcoming senescence tumor suppression mechanism 26, 27. These small molecular activators of senescence would, by definition, possess anti-proliferative activity and may have value as potential anti-cancer agents.
In conclusion, we established a high-content screening platform for identifying small molecule regulators of cellular senescence and identified a list of small molecule compounds that are suppressors of senescence induced by oncogenic RAS.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by a NIH/NCI grant (R01CA160331 to R.Z.), a DOD award (OC093420 to R.Z.), and a NIH/NIGM (R01GM083025 to J.R.P.). B.G.B is an American Cancer Society Postdoctoral Fellow (PF-13-058-01-TBE). We thank Dr. Katherine Aird for critical reading of this manuscript. We thank Dr. Margret Einarson and Ms. Anna Pecherskaya at Fox Chase Cancer Center Translational Research Facility for technical assistance. Support of Core Facilities used in this study was provided by Cancer Center Support Grant (CCSG) CA010815 to The Wistar Institute.
REFERENCES
- 1.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436(7051):725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12(19):3008–19. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 4.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24(22):2463–79. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goel VK, Ibrahim N, Jiang G, Singhal M, Fee S, Flotte T, Westmoreland S, Haluska FS, Hinds PW, Haluska FG. Melanocytic nevus-like hyperplasia and melanoma in transgenic BRAFV600E mice. Oncogene. 2009;28(23):2289–98. doi: 10.1038/onc.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer J, Curtin JA, Pinkel D, Bastian BC. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J Invest Dermatol. 2007;127(1):179–82. doi: 10.1038/sj.jid.5700490. [DOI] [PubMed] [Google Scholar]
- 7.Denoyelle C, Abou-Rjaily G, Bezrookove V, Verhaegen M, Johnson TM, Fullen DR, Pointer JN, Gruber SB, Su LD, Nikiforov MA, Kaufman RJ, Bastian BC, Soengas MS. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat Cell Biol. 2006;8(10):1053–63. doi: 10.1038/ncb1471. [DOI] [PubMed] [Google Scholar]
- 8.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7(4):295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 9.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92(20):9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itahana K, Campisi J, Dimri GP. Methods to detect biomarkers of cellular senescence: the senescence-associated beta-galactosidase assay. Methods Mol Biol. 2007;371:21–31. doi: 10.1007/978-1-59745-361-5_3. [DOI] [PubMed] [Google Scholar]
- 11.Caldwell ME, DeNicola GM, Martins CP, Jacobetz MA, Maitra A, Hruban RH, Tuveson DA. Cellular features of senescence during the evolution of human and murine ductal pancreatic cancer. Oncogene. 2012;31(12):1599–608. doi: 10.1038/onc.2011.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113(6):703–16. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang R, Chen W, Adams PD. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol Cell Biol. 2007;27(6):2343–58. doi: 10.1128/MCB.02019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewald JA, Peters N, Desotelle JA, Hoffmann FM, Jarrard DF. A high-throughput method to identify novel senescence-inducing compounds. J Biomol Screen. 2009;14(7):853–8. doi: 10.1177/1087057109340314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu Z, Aird KM, Bitler BG, Nicodemus JP, Beeharry N, Xia B, Yen TJ, Zhang R. Oncogenic RAS regulates BRIP1 expression to induce dissociation of BRCA1 from chromatin, inhibit DNA repair, and promote senescence. Dev Cell. 2011;21(6):1077–91. doi: 10.1016/j.devcel.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, Pehrson JR, Berger JM, Kaufman PD, Adams PD. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8(1):19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 17.Hayflick L. The cell biology of aging. Clin Geriatr Med. 1985;1(1):15–27. [PubMed] [Google Scholar]
- 18.Sun P, Yoshizuka N, New L, Moser BA, Li Y, Liao R, Xie C, Chen J, Deng Q, Yamout M, Dong MQ, Frangou CG, Yates JR, 3rd, Wright PE, Han J. PRAK is essential for ras-induced senescence and tumor suppression. Cell. 2007;128(2):295–308. doi: 10.1016/j.cell.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 19.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29(11):1039–45. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosar M, Bartkova J, Hubackova S, Hodny Z, Lukas J, Bartek J. Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16(ink4a). Cell Cycle. 2011;10(3):457–68. doi: 10.4161/cc.10.3.14707. [DOI] [PubMed] [Google Scholar]
- 21.Di Micco R, Sulli G, Dobreva M, Liontos M, Botrugno OA, Gargiulo G, dal Zuffo R, Matti V, d'Ario G, Montani E, Mercurio C, Hahn WC, Gorgoulis V, Minucci S, d'Adda di Fagagna F. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol. 2011;13(3):292–302. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zu Y, Liu L, Lee MY, Xu C, Liang Y, Man RY, Vanhoutte PM, Wang Y. SIRT1 promotes proliferation and prevents senescence through targeting LKB1 in primary porcine aortic endothelial cells. Circ Res. 2010;106(8):1384–93. doi: 10.1161/CIRCRESAHA.109.215483. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 24.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bitler BG, Nicodemus JP, Li H, Cai Q, Wu H, Hua X, Li T, Birrer MJ, Godwin AK, Cairns P, Zhang R. Wnt5a suppresses epithelial ovarian cancer by promoting cellular senescence. Cancer Res. 2011;71(19):6184–94. doi: 10.1158/0008-5472.CAN-11-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, Larue L, Pritchard C, Marais R. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15(4):294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 27.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–4. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.