Abstract
The mechanisms by which chromatin structure decompacts to permit access to DNA are largely unknown. Here, using a model nucleosome array system reconstituted from recombinant histone octamers we have defined the relative contribution of the individual histone octamer N-terminal tails as well as the effect of a targeted histone tail acetylation on the compaction state of the `30nm' chromatin fibre. This study goes beyond previous studies as it is based on a nucleosome array that is very long (61 nucleosomes) and contains stoichiometric concentrations of bound linker histone, which is essential for the formation of the `30nm' chromatin fibre. We find that compaction is regulated in two steps: Introduction of H4 acetylated to 30% on K16 inhibits compaction to a greater degree than deletion of the H4 N-terminal tail. Further decompaction is achieved by removal of the linker histone.
Introduction
The hierarchical packaging of eukaryotic DNA by histone proteins into chromatin represents a substantial steric barrier to all cellular processes that involve DNA. Therefore, chromatin folding (and unfolding) is necessarily part of the regulation of cellular processes such as transcription, replication, recombination and repair. Yet, the mechanisms by which chromatin decompaction and dynamics are regulated are largely unknown.
The first level of DNA compaction is achieved by the wrapping of 147bp DNA into 1.7 superhelical turns around a histone octamer, consisting of an H3-H4 tetramer and two H2A-H2B dimers 1; 2; 3. Whereas about 75% of the histone octamer mass forms the globular structure on to which the DNA is wrapped, the remaining 25% form long N-terminal tails rich in lysine and arginine residues 3. In all, eight N-terminal tails protrude from the surface of the nucleosome core where they are free to make numerous intermolecular contacts. The binding of the linker histone (H1 or H5), which is present at close to one molecule per nucleosome in the majority of eukaryotic organisms 4; 5; 6, organizes an additional 20bp of DNA to complete the nucleosome 7; 8. In the second level of compaction, an array of nucleosomes folds intramolecularly in a salt dependent manner in vitro into a compact filament with a diameter of about 30nm, the `30nm' chromatin fibre 9; 10; 11; 12; 13. X-ray diffraction and electron microscopy analyses have provided evidence for the presence of the `30nm' chromatin fibre in nuclei 14; 15; 16.
The critical role of the long and highly basic histone octamer N-terminal tails in the regulation of chromosomal processes such as transcription has become increasingly apparent with the identification of a plethora of different post-translational modifications, subsets of which correlate with regions of silenced or transcriptionally active chromatin 17; 18. Whilst it has become evident that the function of histone octamer N-terminal tails in different modification states is in the recruitment of regulatory proteins that are likely to affect chromatin structure indirectly 19; 20, in vitro studies have provided evidence that the histone octamer N-terminal tails directly modulate nucleosome array compaction 21; 22; 23. A central remaining question is whether certain histone tail modifications directly regulate chromatin compaction.
Acetylation of the histone octamer N-terminal histone tails 24, is a prevalent and reversible histone modification whose levels correlate with transcriptionally active chromatin 25; 26. The effect of acetylating lysine residues by acetyltransferases 27 is to reduce the net positive charge of the very basic histone tails and hence is expected to modulate electrostatic histone tail interactions 28 29. It is therefore believed that acetylation causes the unfolding of the chromatin fibre to permit transcription, consistent with in vitro experiments using randomly hyperacetylated histones 30. Of the many lysine residues subjected to acetylation, the specific acetylation of lysine 16 in the N-terminal tail of histone H4 (H4-K16Ac) is very frequent and is clearly functionally important in different organisms. In budding yeast, over 80% of H4 is acetylated at K16 and in vivo evidence shows that this modification has a role in maintaining or promoting gene transcription 31; 32; 33; 34. Similarly in flies, the enhancement of transcription from the male X-chromosome is also due to H4-K16Ac 35; 36. These biological observations suggest that this specific acetylation mark might play a unique role in regulating chromatin compaction. Recently, from two in vitro studies utilising a very short, reconstituted 12mer nucleosome cores arrays based on the 601 nucleosome positioning DNA sequence (177bp × 12) it has been concluded that deletion of the H4 N-terminal tail, or acetylation of H4 on K16 are sufficient to inhibit the formation of the `30nm' chromatin fibre 37 38. In the first, Richmond and colleagues 37 identified the region of the H4 N-terminal tail encompassing K16 (residues 14 to 19) as essential for salt-dependent compaction. Subsequently, Peterson and colleagues 38 showed that the compaction of the same nucleosome core array could be inhibited by H4-K16Ac. However, both of these studies suffer from the same three limitations. Firstly, the short nucleosome repeat length of 177bp chosen in the construction of these nucleosome core arrays is relatively rare in nature 39, and constrains the folding into a structure 40 that is distinct from the `30nm' chromatin fibre 13; 41. Secondly, the absence of bound linker histone in these two studies significantly limits their compaction and prevents the formation of the more relevant `30nm' chromatin fibre. Thirdly, the use of very short nucleosome arrays questions the reliability of such fibres in reflecting the compaction behaviour of native chromatin.
Here we present the results of an investigation aimed at defining the relative contribution of the linker histone and H4-K16Ac on the compaction states of chromatin. This study goes beyond previous studies as it is based on a reconstituted, model nucleosome array that is very long and contains stoichiometric concentrations of bound linker histone and hence mimics the compaction and decompaction behaviour of the `30nm' chromatin fibre. Nucleosome arrays were assembled from recombinant histone octamers containing different combinations of N-terminal tail truncations and modifications, onto a DNA array containing 61 tandem copies of 202bp 601 DNA (202bp × 61) and their compaction properties analysed using three complementary techniques: electrophoretic mobility in native agarose gels, analytical ultracentifugation and visualization by electron microscopy. We find that the decompaction of the `30nm' chromatin fibre is not a simple single-step mechanism, but a two-step mechanism: Significantly, incorporation of histone H4 partially acetylated on K16 results in a cooperative decompaction. Visualization by electron microscopy shows that acetylation leads to the loss of a compact chromatin fibre, consistent with abrogation of nucleosome–nucleosome contacts, but not the unfolding of the fibre. Further decompaction is achieved by the removal of the linker histone.
Results
Reconstitution of long nucleosome arrays containing histone octamer N-terminal tail deletions
To minimize end-effects and hence better mimic the compaction behaviour of native chromatin, we produced a very long DNA array consisting of 61 tandem copies of the 601 nucleosome positioning DNA sequence onto which the histone octamer positions uniquely 42 (202bp × 61). Chromatin fibres were assembled using our reconstitution system that produces nucleosome arrays containing stoichiometric concentrations of both histone octamer and linker histone H5 12; 43. These nucleosome arrays, at physiological salt conditions 12; 13, fold to give a highly homogeneous population of `30nm' chromatin fibres and hence provide a defined model system for analyzing the role of histone tails, their modifications and linker histone in nucleosome array compaction and in the formation of the `30nm' chromatin fibre.
Recombinant X. laevis histone octamers containing N-terminal tails deletions (Figure 1a) were produced as described previously 44. Histone octamers containing single H3 (gH3) or H4 (gH4) tail deletions, double H2A-H2B (gH2AgH2B) or H3-H4 (gH3gH4) deletions and totally tailless (gAll) (Figure 1b), as well as full length recombinant (WT) and chicken erythrocyte (CE) histone octamer were each reconstituted onto the 202 × 61 DNA array using the salt dialysis method as described previously 12; 43. The point of saturation for each histone octamer variant was established empirically from titrations with increasing protein concentrations (Supplementary Figure 1, online). Prior to the analysis of the role of the various histone octamer N-terminal tails in nucleosome array compaction, we tested whether deletion of the tails would affect linker histone H5 binding. The analysis of H5 binding to reconstituted mononucleosomes containing CE, WT, gH2AgH2B and gH3gH4 histone octamers, shows that linker histone binding is unaffected by the absence of specific histone tails (Supplementary Figure 2, online). The linker histone concentration required to obtain saturation in binding and used for all the nucleosome arrays containing various N-terminal tail deletions was determined from a titration of the 202bp × 61 array reconstituted with WT histone octamer (not shown).
Figure 1. Recombinant histone octamers containing different N-terminal tail deletions.

(a) Schematic representation of N-terminal histone octamer tail deletion. The position of the truncation point in each histone N-terminal tail is indicated with an arrow and the number of the residue.
(b) SDS-PAGE analysis of the histone composition of histone octamers containing multiple histone octamer deletions. Purified chicken erythrocyte (CE) (Lane 1) and WT recombinant (Lane 2) histone octamers are shown for reference. Tailless histones are indicated above the lanes (Lanes 3–5) by the prefix “g” which refers to the globular nature of the resulting histones.
(c) SDS-PAGE analysis of the histone composition of histone octamers containing single histone tail deletions (Lanes 2–3).
Histone H4 N-terminal tail is essential for the compaction of nucleosome arrays into the `30nm' chromatin fibre
First, to obtain a quantitative measure of the role the histone octamer N-terminal tails in the formation of the `30nm' chromatin fibre, we compared the compaction properties of nucleosome arrays containing gH2AgH2B or gH3gH4 histone octamers in the presence of linker histone H5. Arrays containing CE, WT and gAll histone octamers were reconstituted side-by-side to provide standards of full compaction. All arrays were folded by dialysis into a buffer containing 1.6mM MgCl2 which, as we have shown previously, gives rise to the same degree of compaction as 120mM NaCl and results in the formation of fully folded `30nm' chromatin fibres12; 13.
The folding properties of the various nucleosome arrays were first analysed by electrophoresis in native agarose gels (Figure 2a). In these gels, the migration rate of a nucleosome array reflects the degree of compaction, with a tightly folded nucleosome array migrating significantly faster than its unfolded form due to a significant reduction in the Stokes radius. In order to maintain the solution compaction state of each folded nucleosome array during electrophoresis, it was necessary to fix the chromatin samples with glutaraldehyde. Each nucleosome array was subjected to electrophoretic analysis both in its fixed and unfixed form. The unfixed chromatin samples unfold in the gel-loading buffer and the resulting migration rate instead reflects the protein composition of the nucleosome array. Comparison of the unfixed arrays containing CE, WT, gH3gH4, gH2AgH2B, gAll histone octamers (Figure 2a, lanes 3, 5, 7, 9 and 11 respectively) and gH3 and gH4 histone octamers (Figure 2b, lanes 4 and 6), shows they have nearly identical gel migration properties and hence are equally saturated with histones (Supplementary Figure 1, online). Importantly, the sharpness of the bands is indicative of highly homogeneous populations of nucleosome arrays, rather than a mixture of subsaturated populations (Figure 2a).
Figure 2. The H4 N-terminal tail is the most important in nucleosome array compaction.
(a) Deletion of the H3-H4 tails results in partial inhibition of compaction. Electrophoretic analysis in native 0.9% agarose of the compaction state of 202bp × 61 nucleosome arrays fully reconstituted with histone octamers containing combinations of N-terminal tail deletions as indicated above the lanes: Chicken erythrocyte (CE); wild type recombinant (WT); H3-H4 tailless (gH3gH4); H2A-H2B-tailless (gH2AgH2B); and completely tailless (gAll) histone octamers. The presence of the linker histone H5 is indicated (+). Arrays were folded in 1.6mM Mg2Cl and 20mM TEA pH7.4 and analysed in both the unfixed (−) or fixed state (+). Fixation was by 0.1% (v/v) glutaraldehyde. The migration in the gel reflects the compaction state of the nucleosome array: the faster migration the more compact the fibre.
(b) Deletion of the H4 tail has the most crucial effect on compaction. Electrophoretic analysis in native 0.9% agarose of the compaction state of 202bp × 61 nucleosome arrays fully reconstituted with different histone octamers as indicated above the lanes: tailless H3 (gH3) and tailless H4 (gH4). The presence of the linker histone H5 is indicated (+). The nucleosome arrays were reconstituted, folded and fixed as in (a).
Comparison of the migration rates of the various fixed nucleosome arrays reveals clear differences in their compaction states. The arrays containing full-length chicken erythrocyte (CE) and recombinant histone octamers (WT) (Figure 2a, lanes 4 and 6) show an identical and dramatic increase in migration rate, corresponding to the maximal compaction resulting from the formation of a fully folded `30nm' chromatin fibre (Figure 3a)13. In contrast, arrays lacking all N-terminal tails (gAll) migrate only slightly faster than the unfolded nucleosome arrays (Figure 2a, lanes 11 and 12) reflecting the inability of these arrays to undergo a salt-dependent compaction. Nucleosome arrays containing gH2AgH2B histone octamers migrate slightly faster than the nucleosome arrays containing full-length histone octamers (Figure 2a, lanes 10 and 4), demonstrating that the N-terminal tails of H2A and H2B appear to be entirely dispensable for compaction. In contrast, arrays containing gH3gH4 histone octamers migrate at a significantly slower rate, intermediate between the fully compacted WT and CE nucleosome arrays and the fully gAll decompacted nucleosome arrays (Figure 2a, lanes 4, 6, 7, 8 and 12), suggesting that these arrays are only partially folded, consistent with previous observations derived from native chromatin in which various tails had been removed by trypsin digest21; 22; 23.
Figure 3. Deletion of the H4 N-terminal leads to decompaction of the `30nm' chromatin fibre.

Electron micrographs showing the effects of different N-terminal histone octamer tail deletions on the compaction state of the chromatin fibres. The electron micrographs of the 202bp × 61 nucleosome arrays with combinations of N-terminal tail deletions analysed by gel electrophoresis in Figure 2. Nucleosome arrays were folded as described in Figure 2b and fixed gently in 0.1% (v/v) glutaraldehyde and stained with 2% (w/v) uranyl acetate. In the background, surrounding the folded chromatin fibres, individual nucleosomes formed from excess histones and competitor DNA are seen. For each nucleosome array three representative examples are shown. The bar corresponds to 100nm.
We next investigated which of the H3 or H4 N-terminal tail is most important for compaction. Histone octamers containing these tail deletions were reconstituted into nucleosome arrays and folded as described above. The native gel electrophoretic analysis clearly shows that whilst fixed gH3 nucleosome arrays have the same migration rate as WT arrays (Figure 2b, lanes 3 and 5), deletion of the H4 N-terminal tail (gH4) has a dramatic effect resulting in a much slower migration rate than that of fully compacted WT nucleosome arrays (Figure 2b, lanes 3 and 7). In summary, the systematic deletion of the histone octamer N-terminal tails together with the electrophoretic analysis of their compaction state has pinpointed the H4 N-terminal tail as being of key importance in the compaction of nucleosome arrays into the `30nm' chromatin fibre containing the linker histone.
To directly visualise the effects of the N-terminal tail deletions on the morphology of the chromatin fibres, we analysed each of the folded nucleosome arrays by electron microscopy (Figure 3). The resulting electron micrographs of negatively stained samples show that nucleosome arrays containing WT, gH2AgH2B and gH3 histone octamers, all show fully compact, rod-shaped chromatin fibres with an uniform diameter of about 34nm13 (Figure 3). This observation confirms that the N-terminal tails of H2A, H2B and H3 are dispensable for compaction into the `30nm' chromatin fibre. In contrast, electron micrographs of fibres containing gH4 histone octamers show a population of decompacted and irregular fibres (Figure 3). The gH4 fibres are morphologically most similar to fibres containing gH3gH4 histone octamers and gAll histone octamers, which also form irregular assemblies of nucleosomes (Figure 3). Thus, the EM analysis confirms the crucial importance of the H4 N-terminal in the formation of fully compacted `30nm' chromatin fibre. However, it is evident that unfolding resulting from the deletion of the H4 tail is only partial, as we do not observe well resolved nucleosomes in the electron micrographs (Figure 3), consistent with the intermediate electrophoretic mobility of the gH4 containing nucleosome array in gels (Figure 2b, lane 7).
Partial acetylation of K16 in the N-terminal tail of histone H4 inhibits the formation of the `30nm' chromatin fibre
Having established the key role of the H4 N-terminal tail in nucleosome array compaction, we next tested the effect of H4-K16 acetylation, a ubiquitous mark of transcriptionally active chromatin32; 33; 35; 36, on the compaction state of the `30nm' chromatin fibre. To test whether acetylation functions by the resulting reduction in charge simply weakening electrostatic interactions28, we also prepared an H4-K16Q point mutant. Histone octamers acetylated uniquely at H4-K16 were produced using the highly specific enzymatic activity of a recombinant form of the Drosophila histone acetyltransferase MOF complex45. The MOF complex (MOF-MSL1-MSL3) was overexpressed using recombinant baculoviruses in insect cells and purified to homogeneity using affinity purification45. From acetylation trials using [3H] Acetyl CoA and a defined concentration of mononucleosome cores containing WT recombinant hitone octamer, it was determined that H4-K16 could be acetylated to a maximum level of approximately 30% (see Methods). A control reaction using nucleosome cores reconstituted with the H4-K16Q point mutant gave no acetylation signal, demonstrating that acetylation was specific for H4-K16 (data not shown). The histone octamer acetylated at H4-K16 was recovered from mononucleosomes and used in reconstitutions of the 202bp × 61 DNA arrays side by side with H4-K16Q and WT recombinant histone octamers (all in the presence of the linker histone H5). The three nucleosome arrays were folded, fixed and their relative compaction state analysed by native gel electrophoresis as described above (Figure 2). Strikingly, arrays saturated with histone octamers containing only 30% H4-K16Ac show almost no salt-dependent compaction (Figure 4a, lanes 4 and 5). In other words, 30% H4-K16Ac appears to disrupt folding to the same extent as the complete removal of all N-terminal histone tails (Figure 2). The disruption caused by 30% H4-K16Ac can be seen clearly in the electron micrographs of negatively stained samples (Figure 4b). Significantly, since the nucleosome array containing the H4-K16Q mutation (100% mutation), which should mimic the reduction in charge offered by acetylation is fully folded, the inhibition in compaction by H4-K16Ac cannot simply by attributed to the change in charge of the H4 N-terminal tail (Figure 4a, lanes 3 and 7).
Figure 4. Partial acetylation of H4-K16 disrupts the `30nm' chromatin fibre.
(a) Electrophoretic analysis of the effect of 30% H4-K16Ac on the compaction state of the `30nm' chromatin fibre. The 202bp × 61 DNA arrays were reconstituted with wild type recombinant histone octamer (WT), 30% acetylated H4-K16Ac histone octamer or the histone octamer containing the H4-K16Q mutation. The presence of the linker histone H5 is indicated (+). Arrays were folded in 1.6mM Mg2Cl and 20mM TEA pH7.4 and analysed in both their unfixed (−) or fixed state (+). Fixation was by 0.1% (v/v) glutaraldehyde.
(b) EM analysis of the effect of 30% H4-K16Ac on the compaction state of the `30nm' chromatin fibre. Nucleosome arrays were folded as described in (a) and fixed gently in 0.1% (v/v) glutaraldehyde and stained with 2% (w/v) uranyl acetate. The electron micrographs show four representative examples of the WT and H4-K16Ac chromatin fibres as indicated. In the background, surrounding the folded chromatin fibres, individual nucleosomes formed from excess histones and competitor DNA are seen. The bar corresponds to 100nm.
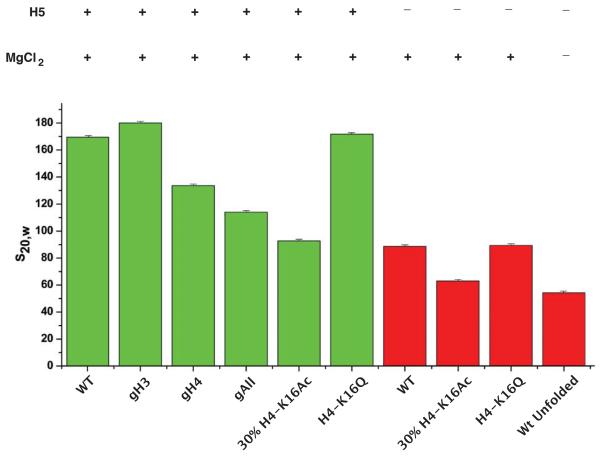
To obtain a quantitative measure of the inhibition of compaction resulting from 30% H4-K16Ac and to assess the relative contribution of linker histone H5 in nucleosome array compaction, we carried out sedimentation velocity analysis. The sedimentation rates of the nucleosome array reconstituted with various tailless and full-length histone octamers were also determined to act as references for fully compact, partially compact and completely unfolded nucleosome arrays. After reconstitution each nucleosome array was folded side-by-side in the same 1.6mM MgCl2 folding buffer and analysed without fixation. Comparison of the sedimentation coefficients of the various 202bp × 61 nucleosome arrays containing the linker histone H5 (Figure 5, green histograms) show varying degrees of compaction, that correlate rather well with the electrophoretic migration rates of the same, but fixed, samples in gels (Figure 2). Nucleosome arrays containing WT, gH3 or H4-K16Q histone octamers, all sediment at approximately the same rate: 170S, 180S and 173S respectively. The close correspondence of these sedimentation coefficients provides a reference value for the fully compacted `30nm' chromatin fibre, and illustrates the reproducibility of the nucleosome array reconstitutions and folding. Whilst the removal of the H4 tail (gH4) results in a sedimentation coefficient of 133S, a value that is intermediate between that of the WT (170S) and completely tailless gAll arrays (115S), the H4-K16Ac containing array has a sedimentation coefficient of 92S. This demonstrates that acetylation H4-K16 at a level of only 30% inhibits compaction to a greater extent than deletion of the H4 tail (133S), indicating a cooperative effect on decompaction.
Figure 5. `30nm' chromatin fibre compaction is additive involving both histone tails and the linker histone: a unique role for H4-K16Ac in fibre decompaction.
Summary of the sedimentation coefficients for the 202bp × 61 nucleosome arrays acetylated at K16 of H4 as well as different combinations of histone octamer N-terminal tail deletions folded in the presence (green histograms) and absence (red histograms) of the linker histone H5. Reconstituted arrays were folded side-by-side in the same folding buffer containing 1.6mM Mg2+ and the sedimentation analysis was performed without the prior fixation of the nucleosome arrays. The composition of the different nucleosome arrays is indicated below the histograms. The presence of the linker histone H5 is indicated (+) as well as whether the nucleosome arrays were folded in MgCl2 (+) or not (−). The error bar represents the 95% confidence interval in the fit of to the g(s*) distribution, indicating the high homogeneity of the individual samples.
The analysis of the folded 202bp × 61 nucleosome arrays reconstituted in the absence of linker histone H5 (Figure 5, red histograms) shows that the H4-K16Ac is not the sole determinant of chromatin compaction. In the absence of linker histone, the nucleosome core array containing WT recombinant histone octamer has a sedimentation coefficient of 89S, a value approximately half of that of fully folded nucleosome arrays in the presence of H5 (170S). Nevertheless, also in the absence of linker histone 30% H4-K16Ac causes further reduction in sedimentation coefficient to 64S, a value close to that of the fully unfolded nucleosome array analysed in the absence of folding buffer (53S). Taken together, these sedimentation velocity results show that decompaction of the `30nm' chromatin fibre is regulated in two steps: one is by acetylation of K16 in the N-terminal tail of histone H4, the other is by the linker histone.
Discussion
The biologically relevant starting point for understanding the regulatory mechanisms that determine the compaction state of the `30nm' chromatin fibre is an analysis of the folding behaviour of chromatin arrays containing the linker histone. As such, our results go beyond previous studies using model nucleosome core arrays that did not include the linker histone in the reconstitutions37; 38. Our conclusions are particularly reliable because they are derived from the use of three independent but complementary analytical techniques.
The conclusion that the N-terminal tail of H4 is the most important of all the histone octamer N-terminal tails in the formation of a fully compact `30nm' chromatin fibre was derived from the observation that whereas the removal of the H2A, H2B and H3 N-terminal tails failed to perturb the salt-dependent compaction of the nucleosome arrays (Figures 2 and 3), the deletion of the H4 N-terminal tail alone results in a significant reduction in the level of compaction of the folded fibre from (170S to 133S) (Figure 5). Our findings are in agreement with those of Richmond and colleagues37, derived from the analysis of the role of the individual histone octamer N-terminal tails in the salt-dependent compaction properties of the 177bp x×12 nucleosome core array lacking the linker histone. This suggests that the H4 N-terminal tail plays the same role in nucleosome array compaction, regardless of whether or not the nucleosomes are arranged in the bone fide `30nm' chromatin fibre generated using our reconstitution method including the linker histone12; 13, or in the less compact structures lacking the linker histone40. In both structures, a key interaction driving compaction is the tight nucleosome-nucleosome packing and it would seem that the H4 N-terminal tail regulates this interaction regardless of the specific architecture of the fibre.
Significant for understanding the mechanism by which chromatin fibre decompaction results in transcriptional activation is our finding that acetylation of a single lysine residue (K16) in the N-terminal tail of histone H4 causes a significant loss of compaction of folded nucleosome arrays (Figures 4 and 5). Although for the nucleosome arrays folded in the presence of linker histone the loss in compaction is dramatic, from 170S to 92S, in the absence of linker histone it is smaller but still significant, from 89S to 64S (Figure 5). The latter result is consistent with earlier findings by Petersen and colleagues38 who also observed a similar H4-K16Ac dependent effect for the 177bp × 12 nucleosome core array lacking the linker histone. The interaction affected by acetylation is likely to be the one observed in nucleosome core crystals, where the stretch of amino acids from the H4 N-terminal tail containing K16 makes very specific contacts with the acidic patch on the H2A-H2B dimer of a neighbouring nucleosome3; 46. Therefore, the most logical explanation for the effects of acetylation we observe, is that acetylation disrupts this specific interaction and hence nucleosome-nucleosome interactions. This is consistent with the observation that the high levels of H4-K16 acetylation in budding yeast and a high basal level of transcription correlate with a decondensed state of the genome34; 47. Similarly, the transcriptional upregulation of the male X-chromosome (dosage compensation) in flies involves the specific acetylation of H4-K16 by the MOF complex35, which correlates with a change in the morphology of the X-chromosome48.
The finding that the partial acetylation of H4-K16 has a greater effect in disrupting compaction than the removal of the entire H4 N-terminal tail (Figure 5) is intriguing and suggests not only that the effect of acetylation is cooperative, but also that it is somehow actively disruptive to chromatin folding. The apparent cooperative effect can be explained if the disruption of a few nucleosome-nucleosome contacts is sufficient for the fibre to open up. However, this does not explain the size of the effect (from 170S to 92S) (Figure 5). One possible explanation is that H4-K16Ac somehow interferes with the binding of the linker histone49. This appears to be unlikely because the nucleosome array reconstituted with partially acetylated H4-K16Ac in the absence of linker histone, reach a significantly lower level of compaction (64S) than the same arrays reconstituted with linker histone (92S) (Figure 5). Furthermore, the dominant destabilising effect is highly specific for H4-K16Ac and cannot be accounted for by a weakened electrostatic interaction28; 37, since the array reconstituted with the H4-K16Q mutant histone octamer is fully compacted (173S) (Figure 5). One possibility that could explain the size of the disruptive effect is that acetylation of K16 modifies the secondary structure of the H4 N-terminal tail that is though to be α-helical {Baneres, 1997 #874}, causing both a disruption of nucleosome-nucleosome interactions as well as actively preventing nucleosome array folding.
Although H4-K16 acetylation as been called a central switch for controlling higher-order chromatin structure by inhibiting the formation of the `30nm' chromatin fibre38; 50, the results presented here show that the compaction or folding (and hence decompaction) of nucleosome arrays into the `30nm' chromatin fibre is not a simple single-step mechanism, but a two-step mechanism involving both H4-K16 acetylation and the linker histone. Whilst our results are consistent with the finding that the single H4-K16Ac provides the mechanism for disrupting nucleosome-nucleosome contacts, in the presence of native-like concentrations of the linker histone the decompaction resulting from acetylation is partial, from 170S to 92S, as the fully unfolded nucleosome array has a sedimentation coefficient of 53S (Figure 5). Therefore, H4-K16 acetylation is one of two major determinants of chromatin fibre compaction. The observation that the WT nucleosome array lacking the linker histone has approximately half the sedimentation coefficient (89S) of the otherwise identical arrays containing H5 (170S), taken together with the observation that arrays containing H4-K16Ac are even less compact in the absence of H5 (64S), show that the linker histone is the second major determinant of chromatin compaction. Further investigations are required to establish whether the partial decompaction resulting from H4-K16 acetylation is sufficient for establishing transcriptionally active chromatin regions, or whether this is coupled to the eviction or modulation of linker histone binding 51,52.
Materials and Methods
601 DNA array
The DNA array is based on the Widom 601 DNA nucleosome positioning sequence42 and contains 61 tandem copies of 202bp of the 601 DNA fragment. Monomeric DNA fragments were ligated together in a tandem arrangement to form multimers and then cloned into pUC18as as previously described12; 13; 43. Plasmids were grown in DH5α E. coli cells. The 601 DNA array was excised by digestion with EcoRV and purified. Mixed sequence competitor DNA (crDNA), about 147bp in length, was obtained from nucleosome core preparations12; 13. The DNA was extracted using phenol/chloroform and precipitated with ethanol.
Histone octamer and linker histone
Wild type (WT) and tailless recombinant X. laevis H2A, H2B, H3 and H4 histones were expressed in E. coli and purified as described previously44. Recombinant histone octamers were reconstituted from purified histones by refolding an equimolar mixture of each of the four denatured histone by dialysis against 2M NaCl buffer. The intact histone octamers were fractionated from histone tetramers and hexamers by size exclusion chromatography as described44. Native linker histone H5 and histone octamer were purified from chicken erythrocyte nuclei as described previously12.
Reconstitution and folding of nucleosome arrays
The 202bp × 61 DNA arrays was reconstituted by the salt dialysis method at 25μg/ml DNA and the molar input ratios of histone octamer (Supplementary Figure 1, online) and linker histones H5 were empirically determined as previously described12. Mixed sequence crDNA (~147bp) was added in all reconstitutions at a crDNA:601 DNA array mass ratio of 1:1 to prevent super-saturation of the 601 DNA arrays with excess histone octamer, ensuring that one histone octamer was bound per 601 DNA repeat. The H5 input level giving the optimal population of homogeneous and fully condensed `30nm' fibres, was then used in the side-by-side reconstitution of the arrays with histone octamers containing combinations of N-terminal tail deletions. Following reconstitution, chromatin arrays were folded by dialysed into 1.6mM MgCl2, 10mM TEA pH7.4. The reconstitution and folding of nucleosome arrays was monitored by electrophoresis in native agarose gels and by electron microscopy of negatively stained samples12.
MOF expression and purification
The Drosophila melanogaster acetyltransferase MOF and its interaction partners MSL1 and MSL3 were co-expressed using recombinant baculoviruses in SF9 insect cells as described previously45. Large-scale purification of the MOF-MSL complex was performed using the pellet from a 500ml liquid SF9 cell culture and purified using an N-terminally FLAG-tagged MSL-1 construct as previously described45. The concentration of the MOF-complex was estimated from a comparison of the intensity of MSL protein bands against that from a known quantity of pure histone H5 after SDS-PAGE separation and Coomassie Blue staining.
Acetylated histone octamer preparation and purification
The acetylation reaction was carried out on nucleosomes to ensure substrate specificity for H4-K16 as described previously45. Mononucleosomes were prepared using the salt dialysis reconstitution method from WT recombinant histone octamer and an equimolar concentration of a ~150bp DNA fragment of mixed sequence DNA generated from micrococcal nuclease digestion of chicken erythrocyte chromatin. The acetylation reaction was carried out by incubating 2mg of reconstituted mononucleosomes in a reaction volume of 10ml containing 5μg/ml purified MOF complex, 2.45μM cold acetyl CoA, 0.245μM [3H] Acetyl CoA, 50mM Hepes pH 7.6, 50mM KCl, and 0.5mM EDTA, for 12 hours at 26°C. The acetylated histone octamers were recovered by increasing the salt concentration to 2M NaCl followed by fractionation on Sephacryl S200 (GE Healthcare) gel filtration column equilibrated in 2M NaCl and 20mM Tris/HCl pH 7.4 to separate the modified histone octamer from the MOF complex, nucleosomal DNA and free [3H] Acetyl CoA. The % acetylation was estimated by comparing the scintillation counts of the acetylated histone octamer at a known concentration, against a standard curve generated from different concentrations of [3H] Acetyl CoA.
Native gel electrophoresis
Native gel electrophoresis was carried out in 0.9% agarose gels using 17×19cm flatbed gels and 0.2x TB (18mM Tris/borate pH 8.3) electrophoresis buffer and a running voltage of 20v/cm. Visualization of the nucleosome arrays was either by ethidium bromide staining or, in experiments in which the DNA was 5' labelled [32P], by standard phosphoimaging techniques of the dried gel.
Electron Microscopy
For visualisation in negative stain, chromatin samples at a concentration of 50μg/ml were gently fixed on ice in 0.1% (v/v) glutaraldehyde for 30mins. 4μl drops were applied to a continuous carbon layer covering an air-discharge treated 200-mesh copper/palladium grid. After one minute, grids were washed with 40μl 2% (w/v) uranyl acetate, blotted and left to air-dry. Images were recorded on a Philips 208S microscope at an operating voltage of 80 keV at 56'000X direct magnification at −1 to −2μm defocus.
Analytical Ultracentrifugation
Sedimentation coefficients were determined using a Beckman XL-A analytical ultracentrifuge equipped with scanner optics. Optical density was measured at 260nm with an initial absorbance between 0.8 and 1.2. Sedimentation analysis was carried out for 2 hours at 5°C at 14'000 rpm in 12mm double-sector cells and a Beckman AN60 rotor. Sedimentation coefficients were determined by the time-derivative method described by Stafford53 to calculate a differential apparent sedimentation coefficient distribution g(s*) using John Philo's Dcdt+ data analysis program (version 2.05)54. g(s*) values were not mass corrected according to linker histone composition or histone tail deletion due to their relatively small mass contribution (never greater than 9%) and as the buoyant mass is unpredictable on account of disordered linker and core histone tails. Sedimentation coefficients were corrected to S20,W using a partial specific volume of 0.69 ml/g for chromatin arrays as calculated previously for native rat liver chromatin55. Solvent viscosity and solvent density were corrected according to buffer composition.
Supplementary Material
Acknowledgements
We thank Tony Crowther for helpful discussions. We thank Peter Becker for providing recombinant baculoviruses for the expression of the MOF-MSL1-MSL2 complex. We thank Louise Fairall for help and John Widom for providing the 601 DNA sequence. We thank Sara Sandin for advice with electron microscopy and discussions and Jo Bulter for advice on analytical ultracentrifugation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–54. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 2.Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984;311:532–7. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 3.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 4.Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–9. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 5.Bates DL, Thomas JO. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 1981;9:5883–94. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas JO. Histone H1: location and role. Curr Opin Cell Biol. 1999;11:312–7. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 7.Noll M, Kornberg RD. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977;109:393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- 8.An W, Leuba SH, van Holde K, Zlatanova J. Linker histone protects linker DNA on only one side of the core particle and in a sequence-dependent manner. Proc Natl Acad Sci U S A. 1998;95:3396–401. doi: 10.1073/pnas.95.7.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976;73:1897–901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–27. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Widom J, Klug A. Structure of the 300A chromatin filament: X-ray diffraction from oriented samples. Cell. 1985;43:207–13. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- 12.Huynh VA, Robinson PJ, Rhodes D. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol. 2005;345:957–68. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 13.Robinson PJJ, Fairall L, Huynh VAT, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fibre: Evidence for a compact, interdigitated structure. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langmore JP, Schutt C. The higher order structure of chicken erythrocyte chromosomes in vivo. Nature. 1980;288:620–2. doi: 10.1038/288620a0. [DOI] [PubMed] [Google Scholar]
- 15.Marsden MP, Laemmli UK. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979;17:849–58. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- 16.Andersson K, Mahr R, Bjorkroth B, Daneholt B. Rapid reformation of the thick chromosome fiber upon completion of RNA synthesis at the Balbiani ring genes in Chironomus tentans. Chromosoma. 1982;87:33–48. doi: 10.1007/BF00333508. [DOI] [PubMed] [Google Scholar]
- 17.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 20.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–83. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 21.Allan J, Harborne N, Rau DC, Gould H. Participation of core histone “tails” in the stabilization of the chromatin solenoid. J Cell Biol. 1982;93:285–97. doi: 10.1083/jcb.93.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fletcher TM, Hansen JC. Core histone tail domains mediate oligonucleosome folding and nucleosomal DNA organization through distinct molecular mechanisms. J Biol Chem. 1995;270:25359–62. doi: 10.1074/jbc.270.43.25359. [DOI] [PubMed] [Google Scholar]
- 23.Krajewski WA, Ausio J. Modulation of the higher-order folding of chromatin by deletion of histone H3 and H4 terminal domains. Biochem J. 1996;316(Pt 2):395–400. doi: 10.1042/bj3160395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allfrey VG, Mirsky AE. Structural Modifications of Histones and their Possible Role in the Regulation of RNA Synthesis. Science. 1964;144:559. doi: 10.1126/science.144.3618.559. [DOI] [PubMed] [Google Scholar]
- 25.Allegra P, Sterner R, Clayton DF, Allfrey VG. Affinity chromatographic purification of nucleosomes containing transcriptionally active DNA sequences. J Mol Biol. 1987;196:379–88. doi: 10.1016/0022-2836(87)90698-x. [DOI] [PubMed] [Google Scholar]
- 26.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–45. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 27.Brownell JE, Allis CD. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–84. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 28.Clark DJ, Kimura T. Electrostatic mechanism of chromatin folding. J Mol Biol. 1990;211:883–96. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- 29.Sun J, Zhang Q, Schlick T. Electrostatic mechanism of nucleosomal array folding revealed by computer simulation. Proc Natl Acad Sci U S A. 2005;102:8180–5. doi: 10.1073/pnas.0408867102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–38. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durrin LK, Mann RK, Kayne PS, Grunstein M. Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell. 1991;65:1023–31. doi: 10.1016/0092-8674(91)90554-c. [DOI] [PubMed] [Google Scholar]
- 32.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–83. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 33.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–7. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 34.Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, Smith DL. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Anal Biochem. 2003;316:23–33. doi: 10.1016/s0003-2697(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 35.Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–75. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 36.Smith ER, Allis CD, Lucchesi JC. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J Biol Chem. 2001;276:31483–6. doi: 10.1074/jbc.C100351200. [DOI] [PubMed] [Google Scholar]
- 37.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 38.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 39.Widom J. A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc Natl Acad Sci U S A. 1992;89:1095–9. doi: 10.1073/pnas.89.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–41. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 41.Robinson PJ, Rhodes D. Structure of the `30 nm' chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–43. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 43.Huynh V. Reconstitution of the 30nm Chromatin Fibre. University of Cambridge; 2001. [Google Scholar]
- 44.An W, Palhan VB, Karymov MA, Leuba SH, Roeder RG. Selective requirements for histone H3 and H4 N termini in p300-dependent transcriptional activation from chromatin. Mol Cell. 2002;9:811–21. doi: 10.1016/s1097-2765(02)00497-5. [DOI] [PubMed] [Google Scholar]
- 45.Morales V, Straub T, Neumann MF, Mengus G, Akhtar A, Becker PB. Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. Embo J. 2004;23:2258–68. doi: 10.1038/sj.emboj.7600235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davey CA, Richmond TJ. DNA-dependent divalent cation binding in the nucleosome core particle. Proc Natl Acad Sci U S A. 2002;99:11169–74. doi: 10.1073/pnas.172271399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mizzen CA, Allis CD. Transcription. New insights into an old modification. Science. 2000;289:2290–1. doi: 10.1126/science.289.5488.2290. [DOI] [PubMed] [Google Scholar]
- 48.Bashaw GJ, Baker BS. Dosage compensation and chromatin structure in Drosophila. Curr Opin Genet Dev. 1996;6:496–501. doi: 10.1016/s0959-437x(96)80073-6. [DOI] [PubMed] [Google Scholar]
- 49.Baneres JL, Essalouh L, Jariel-Encontre I, Mesnier D, Garrod S, Parello J. Evidence indicating proximity in the nucleosome between the histone H4 N termini and the globular domain of histone H1. J Mol Biol. 1994;243:48–59. doi: 10.1006/jmbi.1994.1629. [DOI] [PubMed] [Google Scholar]
- 50.Shia WJ, Pattenden SG, Workman JL. Histone H4 lysine 16 acetylation breaks the genome's silence. Genome Biol. 2006;7:217. doi: 10.1186/gb-2006-7-5-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Holde K, Zlatanova J. What determines the folding of the chromatin fiber? Proc Natl Acad Sci U S A. 1996;93:10548–55. doi: 10.1073/pnas.93.20.10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodcock CL. Chromatin architecture. Curr Opin Struct Biol. 2006 doi: 10.1016/j.sbi.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 53.Stafford WF., 3rd Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal Biochem. 1992;203:295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 54.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–46. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 55.Butler P, Thomas J. Changes in chromatin folding in solution. J. Mol. Biol. 1980;140:505–529. doi: 10.1016/0022-2836(80)90268-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.