Summary
Fluorescent protein (FP) reporter alleles are useful both for identifying and purifying specific cell populations in the mouse. Here, we report the generation of mouse embryonic stem cells that contain a Pdx1 loxed cassette acceptor (Pdx1LCA) allele and the use of recombinase-mediated cassette exchange (RMCE) to derive mice that contain a Pdx1CFP (Cerulean) reporter allele. Mice with this allele exhibited cyan fluorescence within the previously well-characterized Pdx1 expression domain in posterior foregut endoderm. Immunolabeling showed that endogenous Pdx1 was co-expressed with CFP at all time points examined. Furthermore, fluorescence-activated cell sorting (FACS) was used to isolate CFP-positive cells from E11.5 and E18.5 embryonic tissues using both 405 and 445 nm lasers, although the latter resulted in a nearly 50-fold increase in emission intensity. The Pdx1CFP allele will enable the isolation of specific foregut endoderm and pancreatic cell populations, both alone and in combination with other FP reporter alleles.
Keywords for indexing: pancreas, posterior foregut endoderm
Introduction
The pancreas consists of exocrine cells, which secrete digestive enzymes into the small intestine via pancreatic ducts, and Islets of Langerhans, which contain five different types of endocrine cells that regulate blood glucose homeostasis. In mice, the pancreas arises from both dorsal and ventral evaginations from the posterior foregut endoderm beginning around embryonic day (E) 9.0 (Guz et al., 1995; Jonsson et al., 1994; Offield et al., 1996). As development proceeds, the pancreatic epithelial buds expand into the surrounding mesenchyme, partially fuse following rotation of the gut, and undergo complex expansion during which a plexus of cells is transformed into an organized series of ducts and both acinar and islet endocrine cells are formed.
Between E9.5 and approximately E12.5 the pancreatic epithelium consists of a heterogeneous population of multipotent progenitor cells (MPCs) that give rise to the endocrine, exocrine and ductal cell types found in the adult organ (Gu et al., 2002; Kawaguchi et al., 2002; Zhou et al., 2007). The allocation of undifferentiated MPCs into specific lineages is a complex process orchestrated by numerous signaling pathways (Gittes, 2009; Jensen, 2004). Pancreatic and duodenal homeobox 1 (Pdx1) plays a central role in this process and is essential for both the expansion and differentiation of pancreatic MPCs (Chiang and Melton, 2003; Jonsson et al., 1994; Krapp et al., 1998; Offield et al., 1996). Pdx1 expression begins around E8.5 in cells of the posterior foregut epithelium, some of which go on to form the pancreas. At later stages of development, Pdx1 is highly expressed in β cells where it is essential for normal insulin secretion (Melloul et al., 1993).
Numerous lines of mice expressing green fluorescent protein (GFP) (Chalfie et al., 1994; Heim et al., 1995; Shimomura et al., 1962) under control of a variety of gene loci, including Pdx1 (Micallef et al., 2005), have been reported. However, the utility of some of these reporter alleles, including that of the Pdx1GFP allele, is limited by a single spectral profile which prevents their combinatorial use. Indeed, the isolation of many distinct cell populations by fluorescence-activated cell sorting (FACS) requires the simultaneous use of two or more FPs with spectrally distinct excitation and emission profiles (Heikal et al., 2000; Nagai et al., 2002; Rizzo et al., 2004; Shaner et al., 2004).
Recently, we described a strategy for generating loxed cassette acceptor (LCA) alleles (Chen et al., 2011). Mouse embryonic stem (mES) cells containing an LCA allele allow for the use of recombinase-mediated cassette exchange (RMCE) to generate a wide variety of allelic variants. Here, we describe 1) the generation of a mES cell line containing a Pdx1LCA allele, and 2) the use of these cells to derive mice expressing Cerulean, an improved cyan fluorescent protein (CFP), under control of the Pdx1 locus. As expected, the expression of Pdx1CFP mirrors that of Pdx1 during both early and late embryogenesis. We anticipate that mES cells containing the Pdx1LCA allele will be useful for deriving additional mutant Pdx1 alleles and that the Pdx1CFP mice will be useful alone or in combination with other FP-tagged alleles for isolating specific cell populations during pancreas development.
Results and Discussion
Generation of a Pdx1LCA allele
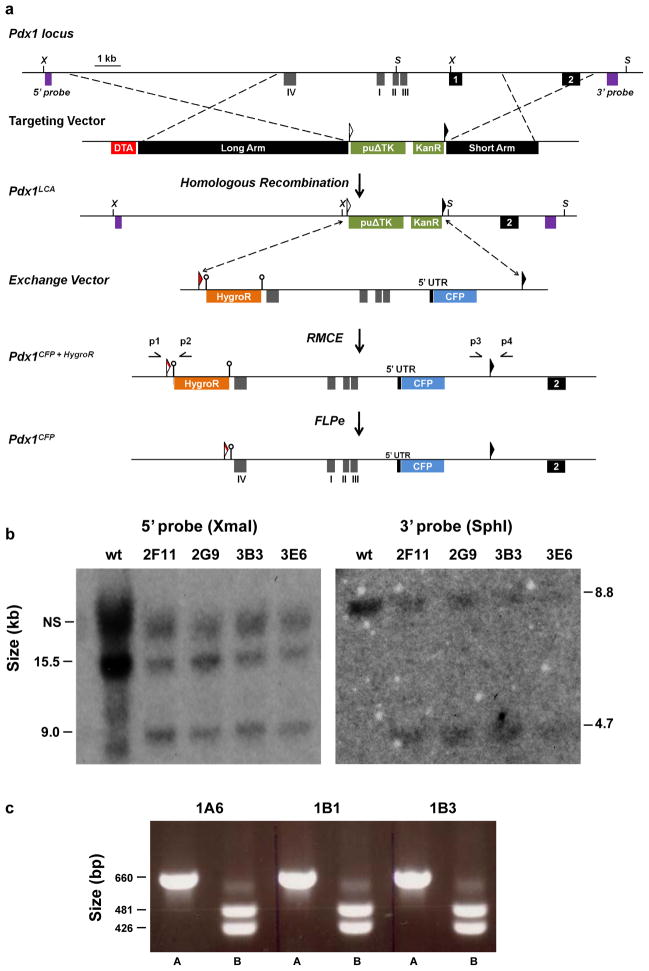
BAC recombineering and gene targeting were used to generate mES cells with a Pdx1LCA allele, as shown in Figure 1a. In this allele, an 8.6 kb region of the Pdx1 locus, containing four previously characterized conserved regulatory regions termed Areas I – IV (Gittes, 2009; Pan and Wright, 2011), as well as exon 1, were replaced with a dual positive-negative selection cassette flanked by lox71 and lox2272 sites (Araki et al., 2002; Chen et al., 2011). Southern blot analysis using probes on both the 5′ and 3′ ends of the mutated region confirmed the desired homologous recombination events (Figure 1b).
Figure 1. Overall scheme for generating the Pdx1CFP allele.
(a) Diagram of the wild type Pdx1 locus, gene targeting vector, Pdx1LCA allele, exchange vector, Pdx1CFP+HygroR allele, and Pdx1CFPallele. Homologous recombination resulted in replacement of an 8.6 kb region of the Pdx1 locus containing exon 1 and Areas I – IV with both a Pgk-driven puromycin resistance-Δthymidine kinase fusion gene (puΔTK) and an EM7-driven kanamycin resistance (KanR) gene flanked by lox71 (open triangle) and lox2272 (black triangle) sites. The exchange vector contained lox66 (red triangle) and lox2272 sites flanking a nuclear-localized CFP sequence followed by a rabbit β-globin polyadenylation sequence. A Pgk-driven hygromycin resistance (HygroR) cassette flanked by FRT sites (open circles) was used as a positive selectable marker during RMCE. Mice containing the Pdx1CFP+HygroR allele were bred with FLPe-expressing transgenic mice to remove the FRT-flanked HygroR cassette. Restriction sites: XmaI (X) and SphI (S). Primer locations: p1, p2, p3 and p4. (b) Southern blot analysis of four puromycin resistant ES cells using probes indicated in panel (a). The Pdx1LCA allele was detected by the presence of a 9.0 kb and 4.6 kb band on the 5′ and 3′ ends, respectively. NS: nonspecific band. (c) PCR analysis on both the 5′ (lane A, p1 and p2) and 3′ (lane B, p3 and p4) ends of three Pdx1CFP+HygroR exchanged clones. Properly exchanged clones were identified by 660 and 481 bp bands on the 5′ and 3′ ends, respectively.
Derivation of a Pdx1CFP allele by RMCE
To validate the functionality of the Pdx1LCA allele, we next made an exchange vector that replaced coding sequences in exon one of the Pdx1 gene with a nuclear-localized CFP (Cerulean) (Rizzo et al., 2004), thereby generating a Pdx1-null allele. A portion of the rabbit β-globin gene, containing both intronic and polyadenylation sequences, was placed downstream of the CFP coding sequences (Chen et al., 2011; Westwood et al., 1993). In addition, the exchange vector contains a Pgk-hygromycin resistance (HygroR) cassette, flanked by tandem flippase recognition target (FRT) sites. RMCE into the Pdx1LCA was achieved through the use of a staggered positive-negative selection strategy (Long et al., 2004) after co-electroporation of the exchange vector and a Cre-expression plasmid. Chimeric mice were generated by injection of an exchanged clone (Figure 1c) into E3.5 mouse blastocysts. After germline transmission, mice containing the Pdx1CFP+HygroR allele were bred with FLPe-expressing transgenic mice to remove the FRT-flanked HygroR cassette, thereby generating the Pdx1CFP allele.
Expression pattern of Pdx1CFP by whole mount fluorescence microscopy
To demonstrate the expression of CFP, we first utilized whole mount fluorescence microscopy (Figure 2). In Pdx1CFP/+ embryos, CFP expression was easily observable beginning at E9.5 in both the dorsal and ventral endoderm, consistent with the pattern previously determined from a Pdx1lacZ insertion allele (Offield et al., 1996). Between E10.5 to E11.5, CFP expression in both the dorsal and ventral pancreatic buds was brighter than that of the caudal stomach and duodenum, normal domains of Pdx1 expression. During this developmental stage, the expression domain of Pdx1 contains a subpopulation of pancreatic MPCs (Gu et al., 2002; Pan and Wright, 2011). As development proceeds, CFP expression persisted at a high level in the pancreatic epithelium and at a lower level in the caudal stomach and duodenum.
Figure 2. Whole mount fluorescent microscopy of Pdx1CFP/+ embryos.
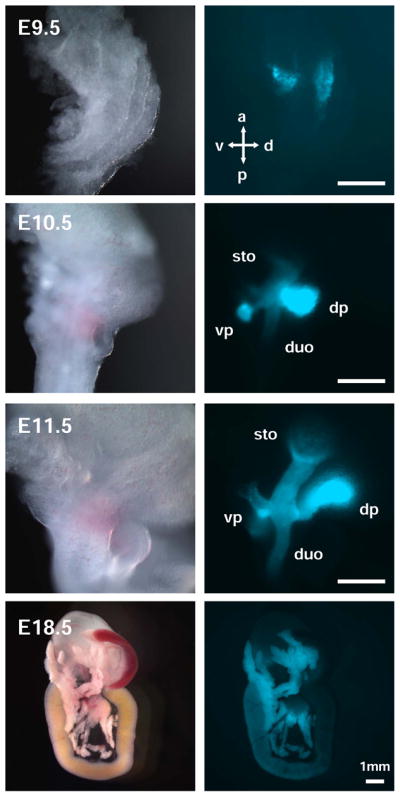
CFP fluorescence was observed in dissected Pdx1CFP/+ embryos as early as E9.5 in the dorsal and ventral endoderm, consistent with previous reports detailing the expression pattern of Pdx1. From E10.5 to E11.5, CFP expression was detected in the dorsal and ventral pancreatic buds at a higher intensity as compared to the expression visualized in the caudal stomach and duodenum. Throughout development, CFP expression persisted at higher levels in the pancreatic epithelium than in the caudal stomach and duodenum. Scale bar = 250 μm unless otherwise noted. Dorsal pancreas (dp), ventral pancreas (vp), stomach (sto), duodenum (duo), anterior (a), posterior (p), dorsal (d), ventral (v).
Expression pattern of Pdx1CFP by immunohistochemical analysis
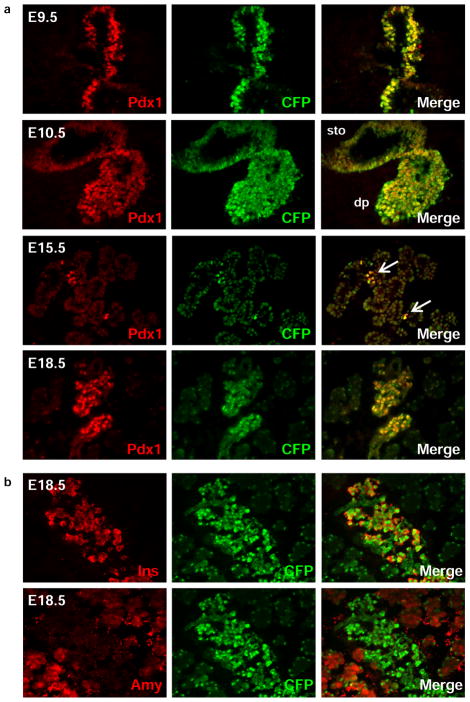
To determine whether CFP and Pdx1 were co-expressed, we performed immunohistochemical analysis using tissues from multiple developmental stages (Figure 3a and data not shown). Both Pdx1 and CFP were detected in the dorsal and ventral endodermal evaginations of the posterior foregut at E9.5. From E10.5 to E11.5, Pdx1 and CFP were detected throughout the pancreatic epithelium and expression was also observed in the stomach and duodenum. From E12.5 – E14.5, CFP immunofluorescence co-localized with Pdx1 throughout the branching epithelium, and by E15.5, high levels of Pdx1 and CFP were evident in periodic clusters throughout the epithelium with lower levels throughout the epithelium. At E18.5, high levels of Pdx1CFP expression were restricted primarily to the developing β cells as evident by the co-localization with insulin-expressing cells (Figure 3b). Additionally, lower levels of CFP expression were co-localized with amylase, indicative of Pdx1 expression in the acinar cells, again in accordance with previous reports (Figure 3b) (Guz et al., 1995; Wu et al., 1997). At each developmental stage, expression the Pdx1CFP allele paralleled that of endogenous Pdx1 expression, consistent with the Pdx1CFP allele faithfully recapitulating expression of Pdx1 (Guz et al., 1995; Leonard et al., 1993; Miller et al., 1994; Offield et al., 1996; Ohlsson et al., 1993). While perdurance of certain FP reporter alleles has been reported, (Burlison et al., 2008; Viotti et al., 2011) this was not observed at any of the time points examined in this study which may simply reflect the maintained expression of Pdx1 in numerous cell types of the pancreatic epithelium.
Figure 3. Immunofluorescent analysis of Pdx1CFP/+ embryos.
(a) Co-expression of Pdx1 and CFP at E9.5 throughout the dorsal and ventral endoderm. At E10.5, Pdx1 and CFP expression are co-localized in the pancreatic epithelium with expression also observed in other posterior foregut derivatives, such as the stomach. By E15.5, high levels of Pdx1 and CFP were evident in scattered clusters (arrows) throughout the epithelium with lower levels throughout the epithelium. At E18.5, Pdx1 and CFP are co-localized in cells displaying both high and low levels of expression. Stomach (sto), dorsal pancreas (dp). DAPI: nuclear counterstain. (b) At E18.5, Pdx1CFP expression is restricted primarily to developing β cells, as indicated by co-expression with insulin, with lower levels of CFP expression observed in the acinar cells, as indicated by co-expression with amylase.
FACS analysis of Pdx1CFP cells
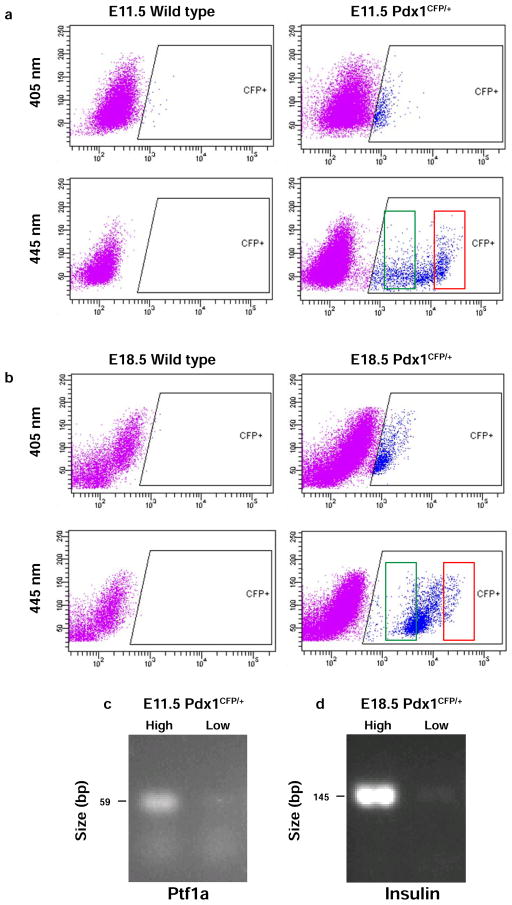
Given that the optimal excitation wavelength for Cerulean is 433 nm (Rizzo et al., 2004), we determined the effect of using either a 405 nm or 445 nm laser, both of which are commercially available, for sorting Pdx1CFP-expressing cells. As shown in Figure 4, the 445 nm laser resulted in a broader range of emission intensity using embryonic tissues at E11.5 and E18.5. Cyan fluorescence observed using the 405 nm laser was minimally distinguishable above cellular autofluorescence in wild-type embryos. However, CFP-expressing cells excited by the 445 nm laser displayed approximately 50-fold higher fluorescence intensity. At E11.5, both high- and low-intensity CFP fluorescence was observed by FACS (Figure 4a), which is similar to the high- and low-expression patterns evident by immunolabeling in the pancreatic epithelium and the stomach/duodenal epithelium, respectively. Cells exhibiting higher levels of CFP fluorescence primarily represented cells of the pancreatic epithelium as indicated by Ptf1a expression (Figure 4c). In addition, high- and low-intensity CFP fluorescence was observed at E18.5 (Figure 4b). An analysis of Insulin expression revealed that cells displaying high levels of CFP fluorescence at E18.5 were predominantly pancreatic β cells (Figure 4d).
Figure 4. Analysis of Pdx1CFP/+ embryos by fluorescence-activated cell sorting (FACS).
The emission intensity of CFP following excitation with either 405 nm or 445 nm was compared. E11.5 embryos (a) and E18.5 embryos (b) were used to isolate CFP-positive cells. The fluorescence intensity observed following excitation with 405 nm laser (top panels) is minimally distinguishable above cellular autofluorescence seen in the wild-type embryos (left panels); whereas a 445 nm laser (bottom panels) provided more optimal excitation of CFP. Both high- and low-expressing cells, indicated by red and green boxes, respectively, were isolated from both E11.5 embryos (a) and E18.5 embryos (b). (c) A higher level of Ptf1a gene expression was detected in cells showing high- versus low-intensity CFP fluorescence consistent with their origin in the pancreatic epithelium. (d) Similarly, a higher level of Insulin gene expression was observed in high- versus low-intensity CFP fluorescence cells at E18.5 consistent with them being pancreatic β cells.
Conclusions
The ability to isolate Pdx1-expressing cells by using mice containing the Pdx1CFP reporter allele at different times during pancreas development will facilitate identification and functional characterization of both gene regulatory networks and cell signaling mechanisms critical for the differentiation, growth, and maturation of the pancreas. The Pdx1CFP allele can be used in combination with green and yellow fluorescent protein reporters, such as the previously reported Ngn3GFP and Ptf1aYFP alleles, although mice that express red FPs, such as mCherry or mApple, may be preferable due to better spectral separation of cyan and red fluorescence. Finally, mouse ES cells containing the Pdx1CFP+HygroR allele may also be useful for detecting Pdx1 gene expression in cultured cells that have undergone directed differentiation.
Materials and Methods
Gene targeting vector
The gene-targeting vector used to generate the Pdx1LCA allele was made by gap repair in E. coli beginning with clone 228-B22 from the RP22 BAC library and contains long and short homology arms of 8.1 kb and 3.6 kb, respectively. The targeting replaced an 8.6 kb region of the Pdx1 gene, spanning from −6586 to +2024 bp, with a mouse phosphoglycerol kinase promoter (Pgk) driving expression of a puromycin resistance-Δthymidine kinase fusion gene (puΔTK) and a bacterial EM7 promoter driving expression of a kanamycin resistance gene (EM7-KanR). Both selectable markers were flanked with tandemly-oriented lox71 and lox2272 sites. An MC1-driven diphtheria toxin A gene (DTA) was placed outside the long homology arm to select against nonrecombinant clones.
RMCE exchange vector
To facilitate assembly of the Pdx1CFP+HygroR exchange vector, a basal exchange vector, termed Pdx1Ex1, containing the 8.6 kb of Pdx1 gene sequence absent in the Pdx1LCA allele flanked by lox61 and lox2272 sites, was generated. Coding sequences for CFP (Cerulean) tagged with 3 copies of an SV40 nuclear localization sequence (NLS) and followed by rabbit β-globin intronic and 3′ UTR sequences were amplified by PCR then cloned into the basal exchange vector. A Pgk-driven hygromycin resistance (HygroR) sequence, flanked by tandem flippase recognition target (FRT) sites, was cloned into the 5′ end of the exchange vector for positive selection following RMCE.
Gene targeting in mouse ES cells
Gene targeting was performed following standard protocols. In brief, 200 μg of the targeting vector was linearized with NotI and electroporated into 35 × 106 TL-1 mouse ES cells. Following puromycin selection, homologous recombination was verified by Southern blot analysis. Clones that had undergone the desired homologous recombination events were identified by a band of 9.0 kb after digestion with XmaI and hybridization with a 5′ probe and a band of 4.7 kb after digestion with SphI and hybridization with a 3′ probe.
RMCE
RMCE was performed using a staggered, positive-negative selection strategy as previously described (Long et al., 2004). In brief, mES cells (clone 2F11) containing the Pdx1LCA, were electroporated with equal amounts of Pdx1CFP+HygroR and pBS185, a Cre-expressing plasmid. Clones surviving selection with both hygromycin and gancyclovir were analyzed for cassette exchange by PCR using primers that spanned either the lox66/71 site on the 5′ end or the lox2272 site on the 3′ end. On the 5′ end, the combination of a 5′-TGAGATTGTATATTGCGGTGCA and 5′-ACGAGACTAGTGAGACGTGCTACT primer results in a band size of 660 bp after RMCE and on the 3′ end, use of 5′-TGAGCAATTCCAAGCAGCTGGA and 5′-ACCTTGCAGTCCTTCTGAAGT results in a 481 bp band for the exchanged allele and a 426 bp band in the wild type allele.
Mouse strains
Germline transmission was achieved following the microinjection of clone 2F11:1B1 mES cells into E3.5 C57BL/6 blastocysts. The FRT-flanked HygroR sequence was removed by breeding with Tg(ACTFLPe)9205Dym mice. The resulting Pdx1CFP allele was maintained on an outbred background. Embryos were isolated from wild-type CD-1 females crossed with Pdx1CFP/+ mice where the presence of vaginal plug at noon was considered as E0.5. Experimental protocols were approved by the Vanderbilt Institutional Animal Care and Use Committee.
Immunolabeling and imaging
Immunolabeling was performed as previously reported (Burlison et al., 2008). Primary antibodies were diluted in 1% BSA in PBS as follows: guinea pig anti-Pdx1 (C.V.E. Wright), 1:2500; chicken anti-GFP (Invitrogen), 1:1000; goat anti-amylase (Santa Cruz Biotechnology), 1:1000; guinea pig anti-insulin (Linco), 1:1000. Secondary antibodies were diluted in 1% BSA in PBS as follows: donkey anti-chicken DyLight 488 (Jackson ImmunoResearch), 1:500; donkey anti-guinea pig Cy3 (Jackson ImmunoResearch), 1:1000; donkey anti-goat Alexa Fluor 647 (Invitrogen), 1:1000. Sections were counterstained with DAPI and cover slips mounted using Aqua/Poly Mount (Polysciences, Inc.). Images were acquired using an Axioplan2 microscope (Zeiss) with a QImaging RETIGA EXi camera. Whole mount imaging of Pdx1CFP embryos was performed using a Leica MZ 16 FA stereoscope with a QImaging RETIGA 4000R camera.
FACS analysis
Pdx1CFP/+ embryos were identified by direct fluorescence and dissected tissues were dissociated using Accumax (Sigma). Following filtration through a 35 μm cell strainer (BD Biosciences) and centrifugation, cells were resuspended in FACS medium [L15 medium (Invitrogen) containing 1 mg/ml BSA, 10 mM HEPES pH 7.4, 1% penicillin/streptomycin, and 37.5 ng/ml DNase I] or FACS medium with 7-Amino-Actinomycin D (7-AAD; Invitrogen). Cells were analyzed and isolated using an Aria III (BD Biosciences). CFP was excited using either a 405 nm or 445 nm laser and emission detected with a 470 nm long pass and 510/80 bandpass filter.
RNA Isolation and semi-quantitative RT-PCR
Total RNA was isolated using TRIzol LS (Invitrogen), DNase-treated (Ambion), and column-purified (Zymo Research). Total RNA was reverse transcribed (High Capacity cDNA Archive kit; ABI), and PCR was performed using 1 ng of cDNA template. Ptf1a was detected using the following primers: 5′ – CGAATTGCCACGGATCACT and 5′ – CCCGGAAGGACGAATGG. Insulin was detected using the following primers: 5′ – CCACCCAGGCTTTTGTCAAA and 5′ – CCCAGCTCCAGTTGTTCCAC.
Acknowledgments
These studies were supported by NIH grants DK42502, DK72473 and DK89523 to MAM; CA68485 and DK58404 to the VMC Flow Cytometry Shared Resource; and CA68485 and DK20593 to the Transgenic Mouse/ES Cell Shared Resource.
We thank M. Gannon, A. Hasty, T. Labosky, and D. Piston and for critically reviewing the manuscript; K. Shelton and X. Li for assembling the targeting vector; C. Zhang for performing mES cell culture; the Vanderbilt Transgenic Mouse/ES Cell Shared Resource for performing the blastocyst microinjections; M. Southard-Smith and members of the Magnuson lab for helpful comments and technical expertise; R. Gangula for technical assistance and mouse husbandry; and D. Flaherty, B. Matlock and K. Weller for assistance with FACS.
References
- Araki K, Araki M, Yamamura K. Site-directed integration of the cre gene mediated by Cre recombinase using a combination of mutant lox sites. Nucleic Acids Res. 2002;30:e103. doi: 10.1093/nar/gnf102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlison JS, Long Q, Fujitani Y, Wright CV, Magnuson MA. Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev Biol. 2008;316:74–86. doi: 10.1016/j.ydbio.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chen SX, Osipovich AB, Ustione A, Potter LA, Hipkens S, Gangula R, Yuan W, Piston DW, Magnuson MA. Quantification of factors influencing fluorescent protein expression using RMCE to generate an allelic series in the ROSA26 locus in mice. Dis Model Mech. 2011 doi: 10.1242/dmm.006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MK, Melton DA. Single-cell transcript analysis of pancreas development. Dev Cell. 2003;4:383–393. doi: 10.1016/s1534-5807(03)00035-2. [DOI] [PubMed] [Google Scholar]
- Gittes GK. Developmental biology of the pancreas: a comprehensive review. Dev Biol. 2009;326:4–35. doi: 10.1016/j.ydbio.2008.10.024. [DOI] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Guz Y, Montminy MR, Stein R, Leonard J, Gamer LW, Wright CV, Teitelman G. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- Heikal AA, Hess ST, Baird GS, Tsien RY, Webb WW. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (Citrine) Proc Natl Acad Sci U S A. 2000;97:11996–12001. doi: 10.1073/pnas.97.22.11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Jensen J. Gene regulatory factors in pancreatic development. Dev Dyn. 2004;229:176–200. doi: 10.1002/dvdy.10460. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Ledermann B, Burki K, Berney C, Zoerkler N, Hagenbuchle O, Wellauer PK. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard J, Peers B, Johnson T, Ferreri K, Lee S, Montminy MR. Characterization of somatostatin transactivating factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol Endocrinol. 1993;7:1275–1283. doi: 10.1210/mend.7.10.7505393. [DOI] [PubMed] [Google Scholar]
- Long Q, Shelton KD, Lindner J, Jones JR, Magnuson MA. Efficient DNA cassette exchange in mouse embryonic stem cells by staggered positive-negative selection. Genesis. 2004;39:256–262. doi: 10.1002/gene.20053. [DOI] [PubMed] [Google Scholar]
- Melloul D, Ben-Neriah Y, Cerasi E. Glucose modulates the binding of an islet-specific factor to a conserved sequence within the rat I and the human insulin promoters. Proc Natl Acad Sci U S A. 1993;90:3865–3869. doi: 10.1073/pnas.90.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micallef SJ, Janes ME, Knezevic K, Davis RP, Elefanty AG, Stanley EG. Retinoic acid induces Pdx1-positive endoderm in differentiating mouse embryonic stem cells. Diabetes. 2005;54:301–305. doi: 10.2337/diabetes.54.2.301. [DOI] [PubMed] [Google Scholar]
- Miller CP, McGehee RE, Jr, Habener JF. IDX-1: a new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. Embo J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. Embo J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240:530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- Viotti M, Nowotschin S, Hadjantonakis AK. Afp::mCherry, a red fluorescent transgenic reporter of the mouse visceral endoderm. Genesis. 2011;49:124–133. doi: 10.1002/dvg.20695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood JA, Jones IM, Bishop DH. Analyses of alternative poly(A) signals for use in baculovirus expression vectors. Virology. 1993;195:90–99. doi: 10.1006/viro.1993.1349. [DOI] [PubMed] [Google Scholar]
- Wu KL, Gannon M, Peshavaria M, Offield MF, Henderson E, Ray M, Marks A, Gamer LW, Wright CV, Stein R. Hepatocyte nuclear factor 3beta is involved in pancreatic beta-cell-specific transcription of the pdx-1 gene. Mol Cell Biol. 1997;17:6002–6013. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, Melton DA. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007;13:103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]