Abstract
By combining experimental neuron models and mathematical tools, we developed a “systems” approach to deconvolve cellular mechanisms of neurodegeneration underlying the most common known cause of Parkinson's disease (PD), mutations in leucine-rich repeat kinase 2 (LRRK2). Neurons ectopically expressing mutant LRRK2 formed inclusion bodies (IBs), retracted neurites, accumulated synuclein, and died prematurely, recapitulating key features of PD. Degeneration was predicted from the levels of diffuse mutant LRRK2 that each neuron contained, but IB formation was neither necessary nor sufficient for death. Genetic or pharmacological blockade of its kinase activity destabilized LRRK2 and lowered its levels enough to account for the moderate reduction in LRRK2 toxicity that ensued. By contrast, targeting synuclein, including neurons made from PD patient-derived induced pluripotent cells, dramatically reduced LRRK2-dependent neurodegeneration and LRRK2 levels. These findings suggest that LRRK2 levels are more important than kinase activity per se in predicting toxicity and implicate synuclein as a major mediator of LRRK2-induced neurodegeneration.
Keywords: LRRK2, mechanisms, Parkinson's disease, single cell, synuclein
Introduction
Parkinson's disease (PD) is a progressive neurodegenerative disorder resulting in motor and cognitive dysfunction. No disease-modifying treatment exists. Mutations in leucine-rich repeat kinase 2 (LRRK2), the most common genetic cause of PD (Paisán-Ruíz et al., 2004; Zimprich et al., 2004; Clark et al., 2006), lead to a dominantly inherited phenotype that is similar to idiopathic PD (Haugarvoll et al., 2008). The mechanisms of mutant LRRK2-induced neurodegeneration are elusive. If identified, they might point to novel therapeutic targets.
Increased kinase activity is a prevailing hypothesis for mutant LRRK2-mediated toxicity. Of the known LRRK2 mutations that cause PD, the G2019S mutation consistently enhances LRRK2 kinase activity, but whether this is a pathogenic mechanism common to all LRRK2 mutants is unclear (Greggio et al., 2006; West et al., 2007). Alternatively, because the pathogenic mutations are nonconservative, their harmful effects may be mediated by misfolded LRRK2. Misfolded proteins tend to accumulate and aggregate within cells (Chiti and Dobson, 2006). In vitro, LRRK2 aggregates and forms inclusion bodies (IBs) that correlate with mutant-LRRK2-induced toxicity and kinase activity (Greggio et al., 2006; MacLeod et al., 2006). Although LRRK2 IBs are an indicator of protein misfolding, their role and importance, if any, in disease progression are unknown.
Misfolded proteins may cause toxicity by adopting toxic conformations or by overloading the protein-homeostasis machinery (Gidalevitz et al., 2006). Overexpressing the C-terminus of Hsp70 interacting protein or downregulating Hsp90, both of which interact with LRRK2, reduce mutant LRRK2-induced toxicity in a kinase-independent manner and are each associated with lower steady-state levels of mutant LRRK2 (Wang et al., 2008; Ko et al., 2009). Changes in LRRK2 levels correlate with changes in kinase activity, as genetic and pharmacological manipulations that inhibit kinase activity reduce LRRK2 stability (Lin et al., 2009; Herzig et al., 2011). If LRRK2-related toxicity depends on physiologic LRRK2 levels, toxicity might therefore be better explained by changes in LRRK2 levels than by kinase activity itself.
Independent of kinase activity, the PD-associated protein α-synuclein accumulates in cells with elevated LRRK2 levels (Lin et al., 2009; Sanchez-Danes et al., 2012). PD patients with LRRK2 mutations often display synuclein pathology (Biskup and West, 2009), and symptoms occur earlier if specific genetic variants in the synuclein gene (SNCA; Botta-Orfila et al., 2012) are present along with LRRK2 mutations. However, whether α-synuclein is required for LRRK2-mediated degeneration is unknown. Given its role in disrupting protein-degradation pathways (Cuervo et al., 2004; Alegre-Abarrategui et al., 2009; Gomez-Suaga et al., 2011), α-synuclein might mediate LRRK2 toxicity by disrupting protein homeostasis and changing LRRK2 levels.
Typically, “snap-shot” cellular approaches are used to identify disease-associated events. However, often a cellular event is measured from populations of cells, masking the relationship between the event and cell death (e.g., does it improve or exacerbate cell death). In contrast, following thousands of single cells throughout the course of neurodegeneration enables both the identification of disease-associated events and the determination of their prognostic value for cell death. Using an imaging platform, we longitudinally tracked primary neurons expressing LRRK2 throughout their lives. LRRK2-associated parameters (e.g., levels, IB formation, and kinase activity) were captured within each neuron and linked to when the neuron died. Using multivariate survival models, cell-to-cell variability was harnessed to establish quantitative and temporal relationships between LRRK2-related events and cell death. By combining experimental and statistical models (Pastrana, 2012; Skibinski and Finkbeiner, 2013), we established a basic cellular “systems” model of LRRK2-mediated neurodegeneration that led to several significant insights.
Materials and Methods
Plasmids.
LRRK2 was derived from pAcGFP-LRRK2 (Greggio et al., 2006; from Mark Cookson, NIH) and subcloned into pGW1-CMV (British Biotechnologies). Y1699C, G2019S, and D1994A were generated using Quikchange Site-Directed Mutagenesis Kit (Stratagene). Wild-type and mutant LRRK2 were cloned into pGW1-CMV (Arrasate et al., 2004). All constructs were verified by sequencing. In cotransfection experiments with pGW1-Venus-LRRK2, we used pGW1-monomeric red fluorescent protein (mRFP) or pGW1-mApple as a survival marker. The huntingtin (Htt) constructs (Htt586) with 17 or 136 polyglutamine repeats were cloned into pGW1-GFP from full-length Htt constructs JM031 and JM032 (gift from Ray Truant McMaster University, ON Canada). Knockdown of α-synuclein was achieved using MISSION Sigma-Aldrich pLKO.1 constructs. To track Map2-expressing human cells, mApple was cloned into a plasmid that put its expression under the control of the human Map2 promoter (System Biosciences).
Cell culture and transfections.
For Western blot analysis, HEK293 cells were grown in DMEM medium containing 10% calf serum, 2 mm glutamine, and penicillin/streptomycin (100 U ml−1/100 μg·ml−1). Primary rat cortical neurons were prepared from rat pup cortices at embryonic days (E)20–E21, cultured at 0.6 × 106 cells/ml. At 5 d in vitro (DIV) neurons were transfected with plasmids by the calcium phosphate method as described (Finkbeiner et al., 1997; Saudou et al., 1998) or by Lipofectamine 2000. For survival analysis, neurons in 24- or 96-well plates were cotransfected with pGW1-RFP and pGW1-Venus-LRRK2 at a 1:12 molar ratio (0.5–1 μg of DNA per well). Postnatal (P) rat midbrain cultures were prepared from the ventral mesencephalon of P0–P1 rats (Mena et al., 1997). For survival analysis, postnatal neurons (100,000/well) in 96-well plates were transfected using Lipofectamine (0.8 μg of DNA per well). To prepare cortical cultures from mice, cortices were taken from E21 αβγ-synuclein triple knock-out (TKO) mice (Nakamura et al., 2011) and wild-type littermates (Ctrl). For confocal analysis, cortical and midbrain neurons were transfected and fixed 24–48 h post-transfection. To assess the detergent-resistance of IBs, neurons were fixed with 1% paraformaldehyde in PBS for 10 min at 37°C, rinsed twice with PBS, and treated with 1–2.5% Triton X-100 and 1–2.5% SDS for 20 min at 37°C (Kazantsev et al., 1999). Neurons were then rinsed with PBS and imaged by fluorescence microscopy.
iPSC differentiation to DA neurons.
Control-induced pluripotent cell (iPSC) lines derived from PD patients harboring a homozygous G2019S mutation (ND35367) were obtained from the NINDS iPSC repository at Coriell Institute. Reprogramming and characterization of the control iPSC line (normal female, 40-y-old; CRL-2524; ATCC) were as reported (Park et al., 2008; Bilican et al., 2012). Neural stem cells (NSCs) were generated from the control and PD iPSC lines, lifted off into suspension and treated with dual SMAD inhibitors, SB431542 and LDN193189, for 2–3 d. NSCs were grown in suspension as spherical aggregates as reported (HD iPSC Consortium, 2012). For DA neuron differentiation, NSCs were cultured on PA6 stromal cells in serum-free insulin/transferring/selenium medium with ascorbic acid (ITS + AA), as described (Park et al., 2005). ITS + AA medium was supplemented with bFGF and purmorphamine. Cells were harvested from the PA6 cells and plated onto fibronection/poly-l-ornithine (FN)-coated plates and cultured in ITS + AA supplemented with bFGF, FGF8a, and purmorphamine. In the final stage of precursor differentiation, cells were dissociated into single cells using Accutase and plated onto FN-coated 96-well plates (0.1 × 106 cells/well). The cells were then cultured in neurobasal media, N2, B27, ascorbic acid, BDNF, GDNF, and cAMP (adapted from Xi et al., 2012). Terminally differentiated cultures were transfected with the mApple-Map2 reporter.
Immunocytochemistry.
Cortical and postnatal rat midbrain neurons and human differentiated DA neurons were grown on 96-well plates or 12 mm coverslips. Neurons were transfected at 4 DIV or 6–20 d into the final stage of precursor differentiation. At 48 h post-transfection, immunocytochemistry was performed as described (Saudou et al., 1998) and labeled with at least one of the following, anti-tyrosine hydroxylase (1:1500, Pel-Freez), anti-MAP2 (1:200, Millipore), anti-LRRK2 antibody (1:2000, Novus Biologicals; or 1:100, Cell Signaling Technology), and anti-α-synuclein (1:500, BD Biosciences). LRRK2 100–500, LRRK2 phospho (pS935), and LRRK2 phospho (pS910) were provided by Dr. Dario Alessi (MRC-PPU, Dundee University, Dundee Scotland) and purchased from Epitomics. Primary antibody staining was followed by secondary antibody with either, anti-rabbit Cy3 (1:500), anti-mouse Cy5 (1:200), anti-rabbit Cy5 (1:200), or anti-mouse Cy5 (1:500; Jackson Immunochemical).
Robotic microscope imaging system and image analysis.
For neuronal survival analysis, images of neurons were taken at 12–24 h intervals after transfection with an automated imaging platform as described (Arrasate et al., 2004; Arrasate and Finkbeiner, 2005; Daub et al., 2009). Measurements of LRRK2 expression, IB formation, and neuron survival were obtained from files generated with automated imaging. Digital images were analyzed with MetaMorph, ImagePro, and original proprietary programs that were written in MATLAB or pipeline pilot. These custom-based algorithms were then used to capture and analyze neurons in each group in a high throughput and unbiased manner. Live transfected neurons were selected for analysis based on fluorescence intensity and morphology. Neurons were only selected if they had extended processes at the start of the experiment. The abrupt loss of cotransfected mRFP was used to estimate the survival time of the neuron (Arrasate et al., 2004). Data from each experimental group were analyzed in an identical manner so there was little need for blinding. The expression of Venus-tagged versions of LRRK2 was estimated by measuring Venus fluorescence intensity over a region of interest that corresponded to the cell soma, using the fluorescence of cotransfected mRFP as a morphology marker (Arrasate et al., 2004). The Venus intensity values were background-subtracted by using an adjacent area of the image. The fold difference in levels of overexpressed Venus-LRRK2 to endogenous LRRK2 was determined by measuring LRRK2 (labeled with anti-rabbit Cy5 antibodies) staining intensity in Venus-LRRK2- or Venus-transfected cells. Total neuronal process length and number were calculated from traces of neurons coexpressing mRFP and LRRK2. Traces of neuronal processes were made with Fiji imaging software using the simple neurite tracer plug-in.
Statistical analysis.
With longitudinal single cell data collected by tracking individual neurons over time, survival analysis was used to accurately determine differences in longevity between populations of cells (Jager et al., 2008). For longitudinal survival analysis, the survival time of a neuron was defined as the time point at which a cell was last seen alive. Statview software was used to construct Kaplan–Meier curves from the survival data. Survival functions were fitted to these curves and used to derive cumulative hazard (or risk-of-death) curves that describe the instantaneous risk-of-death for individual neurons in the cohort being tracked. Differences in the cumulative risk-of-death curves were assessed with the log rank test. We used Cox proportional hazards regression analysis (Cox analysis) to generate hazard ratios that quantified the relative risk-of-death between two cohorts of neurons or the predictive values of variables such as the presence of a PD-associated mutation, changes in LRRK2 levels or IB formation on the risk-of-death (Miller et al., 2010, 2011). As IB formation is a time-dependent variable, time-dependent Cox analysis (Dekker et al., 2008) was used to measure the contribution of IB formation to neuron death. The sign and size of the coefficient of the cellular feature describe, respectively, its predictive relationship to the fate of the cell and the magnitude of its effect. Hazard ratios and their respective p values were generated using the coxph function in the survival package for R statistical software (Andersen and Gill, 1982). All Cox models were analyzed for violations of proportional hazards and for outlier data points using cox.zph and dfbeta functions in R, respectively. Images of cells that were out of the focal plan of Venus fluorescence were excluded from analysis for IB formation (this was <1–5% of cells under analysis). Selection of IB formation was based on expression intensity of Venus-LRRK2 and localization. For risk of IB formation, death can preclude IB formation making it a competing effect of IB formation. To control for this, hazard ratios and p values for the risk-of-IB formation were determined with a multivariate regression analysis using the semiparametric proportional hazards model (Fine and Gray, 1999), that incorporates the presence of competing risk events (competing risk analysis). The analysis was performed using the crr package for R statistical software (Scrucca et al., 2010). The cumulative risk-of-IB formation curves are the estimated cumulative subdistribution hazard for IB formation and are derived from the competing risk regression model. Significance was derived from the competing risks proportional hazard regression model.
Cox analysis was used to generate hazard ratios for the relative risk-of-death of neurons expressing mutant LRRK2 or Venus compared with wild-type LRRK2 or Ctrl. In Table 1, time-dependent Cox analysis was used to determine the predictive nature of IB formation on risk-of-death. Time of IB formation was included as a time-dependent covariate, whereas Venus-LRRK2 fluorescence was included as a baseline covariate in the regression model. Violations of proportional hazards by cox.zph were accommodated by including a time-interaction term for IB formation as a covariate, which measures how the effect of IB formation on risk-of-death varies with time. In Table 2, hazard ratios for the relative risk-of-IB formation were determined with a multivariate regression analysis using the semiparametric proportional hazards model (Fine and Gray, 1999) that incorporates the presence of competing risk events. The analysis was performed using the crr package for R statistical software (Scrucca et al., 2010). Covariates in the regression model included Venus-LRRK2 fluorescence. In several models, the date of the experiment was included as a stratification variable. Any models that included date as a nominal covariate gave comparable results. Simple linear regression was used to correlate Venus-LRRK2 expression and LRRK2 staining. To compare differences across two groups, the groups were statistically compared using an unpaired t test or a Mann–Whitney test. To compare differences across groups, for example when comparing average LRRK2 levels or neurite length, groups were statistically compared using one-way ANOVA for repeated measures and Fisher's PLSD post hoc tests. Levels of endogenous α-synuclein in cohorts were determined by normalizing against α-synuclein levels in a cohort of neurons expressing only Venus or Ctrl. To compare non-normal data across multiple groups, the Kruskal–Wallis test was used.
Table 1.
Cox proportional hazards analysis of the effects of expression level and IB formation of LRRK2 on neuronal survival
| Group | LRRK2 related variable | HR | 95% CI | p value |
|---|---|---|---|---|
| Wild-type LRRK2 (n = 279) | IB formation | 0.73 | (0.332–1.62) | n.s. |
| IB formation: time | 1.01 | (1.003–1.02) | 0.0065 | |
| Venus-LRRK2 fluorescence (a.u.) | 2.75 | (1.637–4.63) | 0.0001 | |
| G2019S LRRK2 (n = 305) | IB formation | 0.39 | (0.142–1.09) | n.s. |
| IB formation: time | 1.01 | (1.001–1.03) | 0.03 | |
| Venus-LRRK2 fluorescence (a.u.) | 4.03 | (1.549–10.48) | 0.0043 | |
| Y1699C LRRK2 (n = 116) | IB formation | 0.48 | (0.163–1.40) | n.s. |
| IB formation: time | 1.01 | (0.997–1.03) | n.s. | |
| Venus-LRRK2 fluorescence (a.u.) | 5.75 | (2.439–13.57) | <0.0001 | |
| Venus (n = 182) | Venus fluorescence (a.u.) | 1 | (1.0–1.0) | n.s. |
CI, Confidence interval; IB, inclusion body; n, number of neurons; a.u., arbitrary unit; n.s., not significant; p > 0.05.
Table 2.
Cox proportional hazards analysis of the effects of kinase activity and α-synuclein on LRRK2-mediated, toxicity, or IB formation before and after adjustment for Venus-LRRK2 levels
| Group | Risk | HR | 95% CI | p value | Adjusted for Venus-LRRK2 levels |
||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | p value | |||||
| G2019S D1994A (n = 283) vs G2019S (n = 181) | Death | 0.752 | (0.59–0.95) | 0.018 | 0.863 | (0.66–1.12) | 0.275 |
| IB formation | 0.521 | (0.23–1.17) | 0.11 | 1.35 | (0.45–4.07) | 0.59 | |
| Y1699C D1994A (n = 260) vs Y1699C (n = 354) | Death | 0.807 | (0.66–0.99) | 0.044 | 0.895 | (0.69–1.17) | 0.409 |
| IB formation | 0.360 | (0.2–0.753) | 0.007 | 0.529 | (0.25–1.11) | 0.093 | |
| G2019S/0.05uM LRRK2-IN1 (n = 292) vs G2019S/vehicle (n = 295) | Death | 0.874 | (0.7–1.09) | 0.24 | 0.926 | (0.74–1.16) | 0.508 |
| G2019S/0.1uM LRRK2-IN1 (n = 345) vs G2019S/vehicle (n = 295) | Death | 0.785 | (0.63–0.98) | 0.04 | 0.856 | (0.68–1.08) | 0.182 |
| G2019S/0.5uM LRRK2-IN1 (n = 321) vs G2019S/vehicle (n = 295) | Death | 0.738 | (0.57–0.96) | 0.023 | 0.835 | (0.64–1.09) | 0.180 |
| G2019S/sh_α-SYN (n = 160) vs G2019S/sh_scr (n = 185) | Death | 0.757 | (0.59–0.97) | 0.028 | 0.792 | (0.6–1.02) | 0.07 |
| Y1699C/sh_α-SYN (n = 147) vs Y1699C/sh_scr (n = 93) | Death | 0.716 | (0.51–0.98) | 0.042 | 0.738 | (0.53–1.02) | 0.07 |
CI, Confidence interval; n, number of neurons.
Western blot analysis.
In experiments to confirm expression of full-length LRRK2, HEK293 cells were transiently transfected with pGW1-Venus, pGW1-Venus-wild-type-LRRK2, pGW1-Venus-Y1699C-LRRK2, or pGW1-Venus-G2019S LRRK2. In experiments looking at the influence of mutations within the kinase domain, HEK293 cells were transfected with pGW1-Venus-D1994A-LRRK2 constructs with or without PD-associated mutations. In experiments that confirmed that Venus-LRRK2 constructs were kinase active, HEK293 cells were transiently transfected with wild-type or mutant pGW1-Venus-LRRK2 or pGW1-LRRK2. At 24 h post-transfection, cells were harvested.
In experiments that confirmed that the kinase inhibitor LRRK2-IN1 was active, HEK293 cells were transiently transfected with pGW1-Venus-LRRK2 G2019S. At 24 h post-transfection, cells transfected with pGW1-Venus-G2019S were treated with DMSO or 1 μm LRRK2-IN1 (kindly provided by Dr. Dario Alessi, MRC-PPU, Dundee University, Dundee Scotland) and harvested 2 h later. In all experiments, lysates were prepared using RIPA buffer (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 0.5% deoxycholate, 1% NP-40 and 0.1% SDS) with protease inhibitors (Roche). Western analysis was carried by loading equal amounts of protein per treatment condition, determined by Bradford assay (Bio-Rad). The samples were electrophoresed through a 4–12% gradient Bis-Tris gel (NuPage) and transferred to PVDF (Thermo Scientific). After Western blotting, membranes were blocked with 5% nonfat milk and subsequently incubated in rabbit polyclonal antibody to LRRK2 (1:1000, overnight at 4°C; Cell Signaling Technology), anti-GFP (1:50,000, 30 min at 37°C; Abcam), β-actin, (1:20000, for 1 h at RT, Sigma-Aldirch), rabbit monoclonal anti-LRRK2 (100-500), anti-LRRK2 P-ser935 or anti-LRRK2 P-ser910 (all 1:20,000, overnight at 4°C, were kindly provided by Dr. Dario Alessi and Epitomics). Immunoreactivity was measured by probing with horseradish peroxidase-conjugated anti-rabbit (1:5000, Jackson ImmunoResearch Laboratory) and anti-mouse IgG (1:5000, Calbiochem) followed by exposure to ECL detection reagents (PerkinElmer).
Results
Mutant LRRK2 in primary neurons recapitulates features of PD
We developed a neuronal model that recapitulates key features of PD and the progression of LRRK2-induced degeneration in individual neurons. To visualize cells expressing LRRK2 in real time, we generated a Venus-tagged full-length LRRK2 construct (Venus-LRRK2; Fig. 1A,B). By immunocytochemistry (Fig. 1B), Venus fluorescence levels correlated significantly with LRRK2 levels in Venus-LRRK2-expressing neurons (Fig. 1C) but not in neurons expressing only Venus (Fig. 1D). Primary neurons had fivefold more transfected LRRK2 than endogenous LRRK2 (Fig. 1E).
Figure 1.
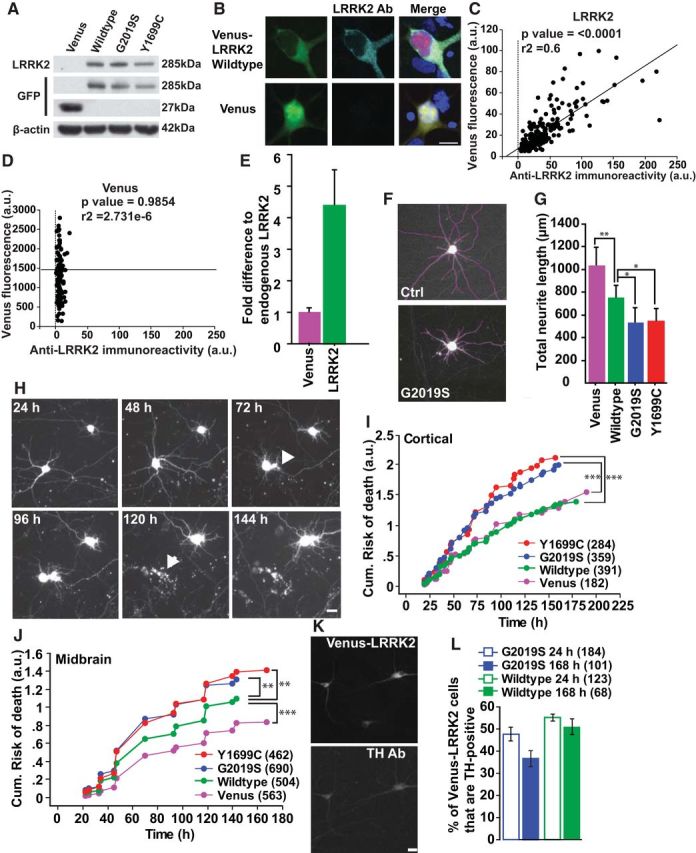
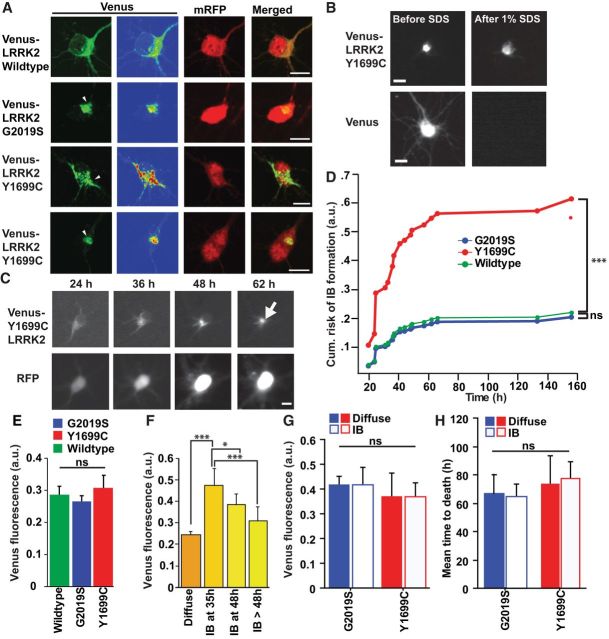
A neuron model that recapitulates key features of PD, including neurite degeneration and cell loss. A, Representative Western blots of transfected HEK293 cell lysates show expression of full-length Venus-tagged LRRK2 blotted with antibodies raised against LRRK2 or GFP (recognizes Venus). β-Actin was blotted as a loading control. B, Confocal images of neurons overexpressing wild-type Venus-LRRK2 and stained with LRRK2 antibody. Cells were fixed 48 h post-transfection at 7 DIV. C, Linear regression analysis showing that single-neuron levels of diffuse wild-type Venus-LRRK2 fluorescence correlate with measurements taken by immunocytochemistry with a LRRK2 antibody. More than 150 neurons, two experiments combined, linear regression analysis, p < 0.0001, r2 = 0.6. D, Linear regression analysis of fluorescence measurements from neurons expressing Venus alone. There is no correlation with LRRK2 immunofluorescence. More than 150 neurons, two experiments combined, linear regression analysis, p = n.s. and r2 = 2.731e−6. E, Histogram showing that neurons transfected with wild-type Venus-LRRK2 have LRRK2 levels approximately fivefold greater than neurons expressing just Venus based on measurements taken by immunocytochemistry with a LRRK2 antibody (>50 neurons). F, Representative traces (purple lines) of neuronal processes from images of neurons cotransfected with Venus or mutant Venus-LRRK2 and mRFP. Venus-only transfected neurons were used as controls (Ctrl). At 48 h post-transfection, mRFP images were used to trace and quantify the total number and lengths of neuronal processes. G, Average total neurite length was significantly lower in neurons transfected with the PD-associated LRRK2 mutants than in neurons expressing wild-type Venus-LRRK2 or Venus alone. Neurons expressing wild-type LRRK2 also had shorter neurites than Venus-expressing neurons. More than 65 neurons in each group, two experiments combined, one-way ANOVA with Fisher's PLSD post hoc tests, **p < 0.001, *p < 0.05; F = 13.2. Error bars are 95% confidence intervals. H, Longitudinal imaging of two neurons expressing mRFP and wild-type Venus-LRRK2. The top neuron remains alive throughout the imaging period. The bottom neuron underwent neurite degeneration at 72 h (arrowhead) and then died by 120 h (arrow). Neurons were 5 DIV at initiation of tracking. Scale bars, 10 μm. I, J, Primary cortical (I) or postnatally derived midbrain (J) neurons expressing LRRK2 mutants G2019S and Y1699C have a significantly greater cumulative risk-of-death than neurons expressing wild-type LRRK2. Neurons per group specified in brackets on the figures, ≥3 experiments combined, log rank test, **p < 0.001, ***p < 0.0001. K, Representative images of midbrain neurons transfected with wild-type Venus-LRRK2 and stained with TH 48 h later. Neurons are 5–6 DIV at time of transfection. Scale bar, 10 μm. L, Histogram showing the percentage of LRRK2-expressing neurons that are TH-positive at 24 or 168 h post-transfection. Neurons expressing mutant LRRK2 G2019S trended toward a lower percentage of TH-positive neurons at 7 d. The number of neurons in each group ranged from 65 to 184, three independent experiments combined, one-way ANOVA with Fisher's PLSD post hoc tests; p = 0.173, F = 1.805. Error bars are 95% confidence intervals.
Neuronal processes degenerate early in PD (Li et al., 2009). To determine whether LRRK2-expressing neurons display this PD phenotype, we examined two mutations: the most common PD-associated mutation, G2019S (Kay et al., 2006) and Y1699C. G2019S, Y1699C, or wild-type Venus-LRRK2 constructs were cotransfected into primary cortical neurons with mRFP. As reported in vivo (MacLeod et al., 2006), G2019S or Y1699C-LRRK2 expression led to shorter total neurite lengths in neurons than wild-type Venus-LRRK2 or Venus alone. Wild-type Venus-LRRK2 expression was associated with shorter neurite lengths than Venus alone (Fig. 1F,G).
To determine whether mutant LRRK2 recapitulates cortical and midbrain degeneration, primary neurons were transfected with Venus-LRRK2 and mRFP. Approximately 24 h after transfection, automated imaging (Arrasate et al., 2004) was used to collect consecutive images of fluorescent neurons. Using this approach hundreds of neurons, expressing Venus LRRK2 and mRFP were tracked individually every 12–24 h for 7 d (Fig. 1H). The mRFP signal was used to track the cell's morphology throughout its lifetime. Abrupt loss of signal corresponded to a loss of membrane integrity, and the neuron's death was recorded at that time (Fig. 1H). Time-of-death data from hundreds of neurons were then analyzed in a Kaplan–Meier survival model. In cortical neurons expressing Venus or wild-type Venus-LRRK2 alone, the cumulative risk-of-death was minimal. However, neurons expressing G2019S or Y1699C had a significantly greater risk-of-death and were 50% more likely to die than controls (Fig. 1I; hazard ratio (HR) 1.5; p < 0.0001; 95% CI 1.3–1.8 for G2019S and 1.3–1.9 for Y1699C). In postnatally derived ventral midbrain neurons cotransfected with Venus-LRRK2 constructs and mRFP, midbrain neurons expressing Y1699C or G2019S had a significantly higher risk-of-death than wild-type-LRRK2-expressing neurons (Fig. 1J; HR 1.3; p = 0.0007; 95% CI 1.1–1.5 for G2019S and HR1.3; p = 0.0009; 95% CI 1.3–1.9 for Y1699C). However, compared with cortical neurons, ventral midbrain neurons were more susceptible to death after transfection with wild-type Venus-LRRK2 than Venus alone (Venus ctrl compared with wild-type Venus-LRRK2; HR 1.0; p = not significant (n.s.); 95% CI 0.9–1.2 for cortical neurons and HR 0.8; p = 0.004; 95% CI 0.67–0.93 for midbrain neurons). To determine whether TH-positive neurons were dying specifically from LRRK2 expression, we compared the percentage of TH-positive neurons in LRRK2-transfected midbrain neurons at 24 and 168 h post-transfection (Fig. 1K,L). Neurons expressing wild-type LRRK2 had similar percentages of TH-positive neurons at both times, indicating that TH-positive neurons expressing LRRK2 were dying, but at a rate similar to TH-negative neurons (Fig. 1L). Neurons expressing mutant G2019S LRRK2 trended toward a lower percentage of TH-positive neurons at 168 h post-transfection (Fig. 1L), suggesting that TH-positive neurons might be more vulnerable to mutant G2019S LRRK2.
IB formation is not required for mutant LRRK2-induced toxicity
PD-associated mutations in LRRK2 lead to misfolding and IB formation (Smith et al., 2005; MacLeod et al., 2006; Higashi et al., 2007), common features of PD. However, the importance of LRRK2 IB formation in PD pathogenesis is unknown. In primary neurons, Venus-LRRK2 is diffusely distributed in the cytosol and excluded from the nucleus, as reported (West et al., 2005; Fig. 2A). Yet, in a subset of neurons, wild-type, mutant Y1699C, or G2019S LRRK2 formed heterogeneous intracellular IBs that were insoluble in 1% SDS/Triton X-100. Venus expressed alone was solubilized by SDS (Fig. 2A,B). Immunocytochemistry failed to observe any colocalization of LRRK2 inclusions with α-synuclein (data not shown). We then used our longitudinal imaging platform to track LRRK2-Venus IB formation in real time (Fig. 2C). As death can preclude IB formation, to correctly estimate the risk-of-IB formation, death must be treated as a competing effect. Neurons expressing Y1699C Venus-LRRK2 were at greater risk of IB formation than neurons transfected with wild-type or G2019S Venus-LRRK2 (Fig. 2D). The relative increase in IB formation in Y1699C-expressing neurons was not due to different LRRK2 levels, as the expression of Venus-LRRK2 before IB formation was similar for both G2019S and Y1699C (Fig. 2E). In other models of neurodegeneration, the intracellular level of a disease protein predicts IB formation (Arrasate et al., 2004; Barmada et al., 2010). To determine whether this is true for LRRK2, Venus-LRRK2 levels were measured in neurons 24 h after transfection and before IB formation. Neurons were split into cohorts based on whether and when an IB formed. Neurons that developed IBs earlier started with significantly higher LRRK2 levels than those that formed IBs later or not at all (Fig. 2F).
Figure 2.
IB formation depends on the PD-associated LRRK2 mutation and LRRK2 levels, but is not required for LRRK2-dependent cell death. A, Confocal images of primary cortical neurons overexpressing wild-type Venus-LRRK2 show diffuse cytosolic LRRK2 expression. Expression of PD-associated mutants G2019S or Y1699C LRRK2 leads to the formation of heterogeneous cytosolic IBs (arrowheads). Neurons were fixed at 48 h post-transfection at 7 DIV. Scale bars, 10 μm. B, Detergent-resistance assay shows that IBs were resistant to treatment with 1% SDS/1% Triton X-100. Scale bar, 10 μm. C, Longitudinal imaging of a neuron expressing mRFP and mutant Y1699C Venus-LRRK2, undergoing IB formation (arrow). Neurons were 5 DIV at initiation of tracking. Scale bars, 10 μm. D, Cumulative risk-of-IB formation is greater in neurons expressing Y1699C Venus-LRRK2 than those expressing wild-type Venus-LRRK2 or G2019S Venus-LRRK2. More than 110 neurons per group, ≥3 experiments combined, using a semiparametric proportional hazards model, that incorporates the presence of competing risk events, ***p < 0.0001. E, Neurons expressing wild-type, G2019S, or Y1699C Venus-LRRK2 had similar Venus-LRRK2 levels. Number of neurons for Y1699C = 121, G2019S = 308, and wild-type = 283, three experiments combined, Fisher's PLSD post hoc tests, not significant (ns). Error bars are 95% confidence intervals. F, Neurons that did not form IBs had significantly lower LRRK2 levels than neurons that formed LRRK2 IBs by 35, 48, or after 48 h. More than 32 neurons in each group, three experiments combined, one-way ANOVA with Fisher's PLSD post hoc tests, *p < 0.01, ***p < 0.0001; F = 36.71. Error bars are 95% confidence intervals. G, Venus-LRRK2 levels are matched in cohorts of neurons expressing G2019S and Y1699C Venus-LRRK2. H, In neurons expressing G2019S or Y1699C Venus-LRRK2, the average time-to-death in neurons that form an IB at 36–48 h is similar to the average time-of-death in neurons without an IB. More than 40 neurons per group, three experiments combined, one-way ANOVA with Fisher's PLSD post hoc tests. Error bars are 95% confidence intervals.
To determine whether LRRK2 IB formation was necessary for LRRK2-mediated toxicity, we identified neurons that formed inclusions at 36–48 h and compared their mean time to death to neurons with similar LRRK2 levels (Fig. 2G) but that did not form an IB at 36–48 h. If IB formation is necessary for toxicity, neurons with IBs should die earlier than those without. However, we found no difference in mean time to death in neurons with or without IBs (Fig. 2H), even though the levels of LRRK2 were the same in both groups.
Time-dependent Cox proportional hazard (CPH) analysis was used to assess whether LRRK2 IBs contribute to toxicity, act as a cellular coping response or are an incidental event during the disease progression. CPH analysis is predominantly used in clinical studies to determine the relationship between an explanatory variable(s), such as IBs, and a specified endpoint (e.g., cell death; Fleming and Lin, 2000). One advantage of CPH analysis is the ability to control potentially confounding effects by including the potentially confounding effect (e.g., LRRK2 level) as another explanatory variable simultaneously in the CPH model. A second advantage is that all neurons can be included in the CPH analysis, regardless of their LRRK2 level or if/when they formed an IB, which increases the power and sensitivity of the analysis.
To determine the role of IBs during LRRK2-mediated toxicity in the CPH model, we included both the LRRK2 level and if/when an IB forms in each neuron. We found that, in neurons expressing mutant or wild-type LRRK2, IBs did not significantly enhance or reduce mutant LRRK2-induced toxicity (Table 1), suggesting that IB formation is an incidental event during neurodegeneration.
Diffuse LRRK2 has a dose-dependent toxic effect in neurons
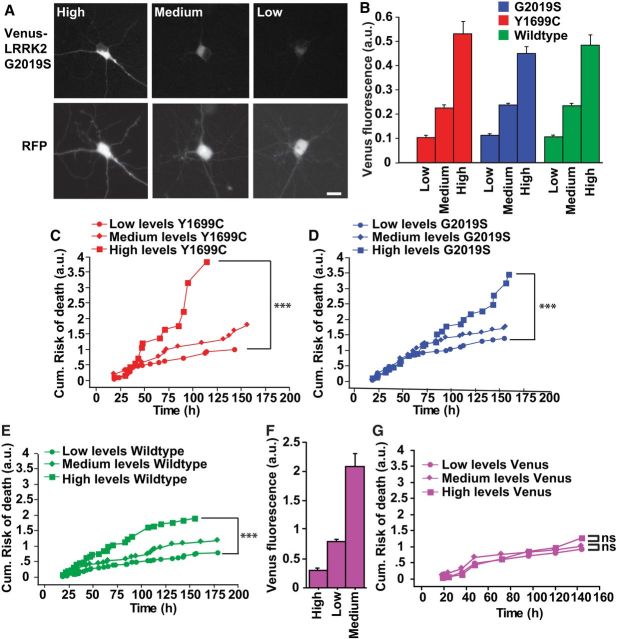
In other neurodegenerative diseases, intracellular toxic species occur outside IBs (Arrasate et al., 2004; Barmada et al., 2010). To determine whether diffuse forms of mutant LRRK2 are responsible for neurodegeneration, we measured Venus-LRRK2 levels in live neurons at 20–24 h post-transfection. Cohorts that expressed low, medium or high levels of Venus-LRRK2 were followed prospectively (Fig. 3A,B). Neurons with high levels of mutant LRRK2 died at a greater rate than those with low levels (Fig. 3C,D). Wild-type LRRK2 also exhibited dose-dependent toxicity, which is consistent with the discovery of wild-type LRRK2 as a risk locus for PD (Satake et al., 2009; Fig. 3E). No relationship was noted between control Venus expression levels and survival, indicating that these effects were specific to LRRK2 (Fig. 3F,G). To validate that LRRK2 levels were proportional to cell death and ensure no user bias in selecting terciles of expression, all cells were included simultaneously into a Cox analysis. This independent and complementary approach, demonstrated that the level of LRRK2 expressed in the cell was proportional to the cell's risk of death. No relationship was found in cells expressing Venus alone (Table 1). The average levels of Venus-LRRK2 were similar in populations of cells expressing mutant or wild-type LRRK2 (Fig. 2E), suggesting elevated steady-state mutant LRRK2 levels cannot explain mutant-LRRK2-induced toxicity.
Figure 3.
Higher levels of venus-LRRK2 are more toxic. A, Representative images of primary neurons expressing different levels of G2019S Venus-LRRK2. In each neuron, a quantitative measure of LRRK2 is made using the Venus-LRRK2 fluorescence. Scale bar, 10 μm. B, Average levels of Venus-LRRK2 after neurons were split into separate cohorts expressing low, medium, or high LRRK2 levels. Error bars are 95% confidence intervals. C–E, Neurons expressing higher levels of Y1699C Venus-LRRK2, G2019S Venus-LRRK2, or wild-type Venus-LRRK2 have a greater cumulative risk-of-death than neurons expressing lower levels. Number of neurons for Y1699C = 157, G2019S = 345 neurons, and wild-type = 400 neurons, ≥3 experiments combined, log rank test, ***p < 0.0001. F, Cohorts of neurons expressing low, medium, and high levels of Venus. Error bars are 95% confidence intervals. G, In neurons expressing higher, medium, or low levels of Venus there is no difference in the cumulative risk-of-death. Number of neurons, >180, ≥3 experiments, log-rank test.
Kinase-inactivating mutations modify toxicity by reducing LRRK2 levels
LRRK2 mutations are thought to provide a toxic gain of function to LRRK2 that includes, at least for G2019S, increased kinase activity. To test this, neurons were transfected with versions of wild-type, G2019S, and Y1699C LRRK2 with the D1994A mutation, which blocks kinase activity (Lee et al., 2010; Fig. 4A,B). Automated imaging was used to track individually transfected neurons, and their time of death was recorded. As reported (Greggio et al., 2006; Smith et al., 2006; Lee et al., 2010), blocking kinase activity modestly but significantly reduced Y1699C or G2019S toxicity (Fig. 4C–E). However, when we measured Venus-LRRK2 levels within individual neurons, mutations within the kinase domain also led to reduced steady-state levels of LRRK2 (Fig. 4F). If LRRK2 levels and kinase activity correlate with mutant-LRRK2-induced toxicity, but mutations inhibiting kinase activity reduce LRRK2 levels, which is the primary cause of mutant LRRK2-induced toxicity?
Figure 4.
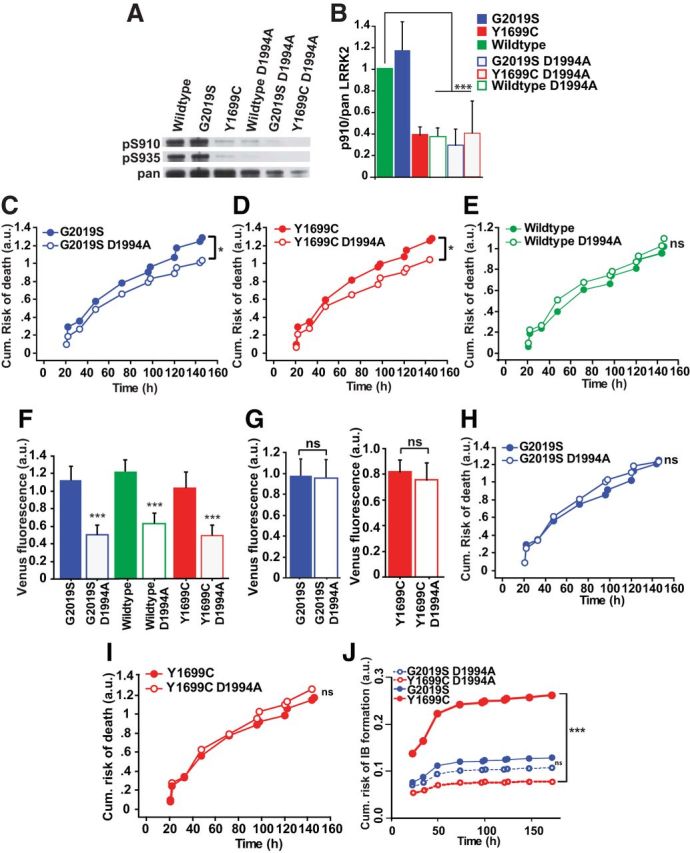
Kinase-inactivating mutations reduce mutant LRRK2-induced toxicity by reducing LRRK2 levels. A, Representative Western blots of lysates from HEK293 cells transfected with wild-type, G2019S, or Y1699C Venus-LRRK2 constructs with or without the D1994A mutation and immunoblotted for the detection of LRRK2 phosphorylation at Ser910 (top) and Ser935 (middle), and total LRRK2 (pan LRRK2, bottom). B, Quantification of phosphorylated LRRK2 at Ser910 normalized to total pan LRRK2. Three independent experiments combined, Mann–Whitney test, wild-type versus wild-type D1994A (S910 ***p = 0.0495; Z = 1.964), G2019S vs G2019S D1994A (S910 ***p = 0.0495, Z = 1.964), Y1699C versus Y1699C D1994A (S910 p = 0.8273; Z = 0.218). Error bars indicate the SD. C, D, Before controlling for LRRK2 levels, cumulative risk-of-death curves show that the D1994A mutation led to a significant reduction in toxicity in neurons expressing the mutant G2019S and Y1699C Venus-LRRK2; number of neurons for G2019S = 181, G2019S D1994A = 283, Y1699C = 354, and Y1699C D1994A = 260, three experiments combined, log rank test, *p < 0.01. E, Graph showing no significant difference in the survival of neurons expressing wild-type Venus-LRRK2 with or without the D1994A mutation. Number of neurons for wild-type = 392 and wild-type D1994A = 277, three experiments combined, log rank test. F, Average expression levels of LRRK2 constructs with or without the D1994A mutation in primary neurons. Number of neurons as in C, D, three experiments combined, one-way ANOVA with Fisher's PLSD post hoc tests, ***p < 0.0001; F = 22.91. Error bars are 95% confidence intervals. G, Neurons expressing mutant Y1699C or G2019S Venus-LRRK2 with or without the D1994A mutation were matched for LRRK2 levels. Number of neurons for G2019S = 170, G2019S D1994A = 138, Y1699C = 297, and Y1699C D1994A = 166, three experiments combined. H, I, After matching for LRRK2 levels, cumulative risk-of-death curves show neurons expressing Y1699C and G2019S with or without D1994A exhibit no significant difference in toxicity. Number of neurons as in G, three experiments combined, log rank test. J, Before controlling for LRRK2 levels, cumulative risk-of-IB formation curves show that the D1994A mutation significantly reduces the risk-of-IB formation of mutant Y1699C, but not mutant G2019S. Number of neurons for G2019S and G2019S D1994A (449) and for Y1699C and Y1699C D1994A (565), three experiments combined, using a semiparametric proportional hazards model that incorporates the presence of competing risk events, ***p < 0.0001.
To dissect out the independent contributions of kinase activity and levels over the course of LRRK2-mediated neurodegeneration, cohorts of neurons expressing either kinase-active or dead constructs were matched for similar LRRK2 levels. When LRRK2 levels were matched, cells expressing kinase dead constructs no longer showed a reduction in toxicity (Fig. 4G–I). In a complementary approach that avoids any confound of excluding low or high-expressing LRRK2 neurons, all neurons were incorporated into a multivariate Cox model. By simultaneously incorporating LRRK2 level and kinase activity into the model, we could determine their independent contributions to neuron death. Before incorporating LRRK2 levels into the Cox model, kinase-dead versions reduce neuronal toxicity; however, once LRRK2 levels were incorporated into the model, any difference in toxicity disappeared (Table 2). Thus, the reduction in LRRK2 levels fully accounted for the improvement in survival that resulted from the disruption of LRRK2 kinase activity.
As reported (Greggio et al., 2006; Kett et al., 2012), blocking kinase activity significantly reduced the risk-of-IB formation in neurons expressing Y1699C-LRRK2 but not in G2019S-LRRK2-expressing neurons (Fig. 4J). As LRRK2 levels influence IB formation (Fig. 2F) and are modulated by kinase activity (Fig. 4F), the reduction in IB formation for Y1699C-LRRK2 could be due to changes in LRRK2 levels or kinase activity. Before adjusting for LRRK2 levels, blocking kinase activity reduced the risk-of-IB formation in neurons expressing Y1699C-LRRK2. When we controlled for LRRK2 levels, blocking kinase activity had no effect (Table 2).
Pharmacologically inhibiting LRRK2 kinase activity modulates LRRK2 stability and localization
To exclude any confounding effects of kinase-inactivating mutations, we tested whether LRRK2 kinase activity is required for mutant LRRK2-induced toxicity with acute pharmacological inhibition. LRRK2-IN1, one of the most selective and potent known LRRK2 kinase inhibitors, dephosphorylates LRRK2 at Ser910 and Ser935 by an unknown regulatory feedback loop (Deng et al., 2011). By Western blot analysis in HEK293 cells (Fig. 5A–C) and by immunocytochemistry in neurons (Fig. 5D), we found that treating G2019S Venus-LRRK2-expressing cells with LRRK2-IN1 dephosphorylated Ser910 and Ser935 in a dose-dependent manner. As LRRK2-IN1 concentrations >1 μm were toxic (data not shown), lower doses (0.05, 0.1, or 0.5 μm) were tested. In cells transfected with G2019S LRRK2, LRRK2-IN1 reduced G2019S-LRRK2-induced toxicity (Fig. 5E; Table 2).
Figure 5.
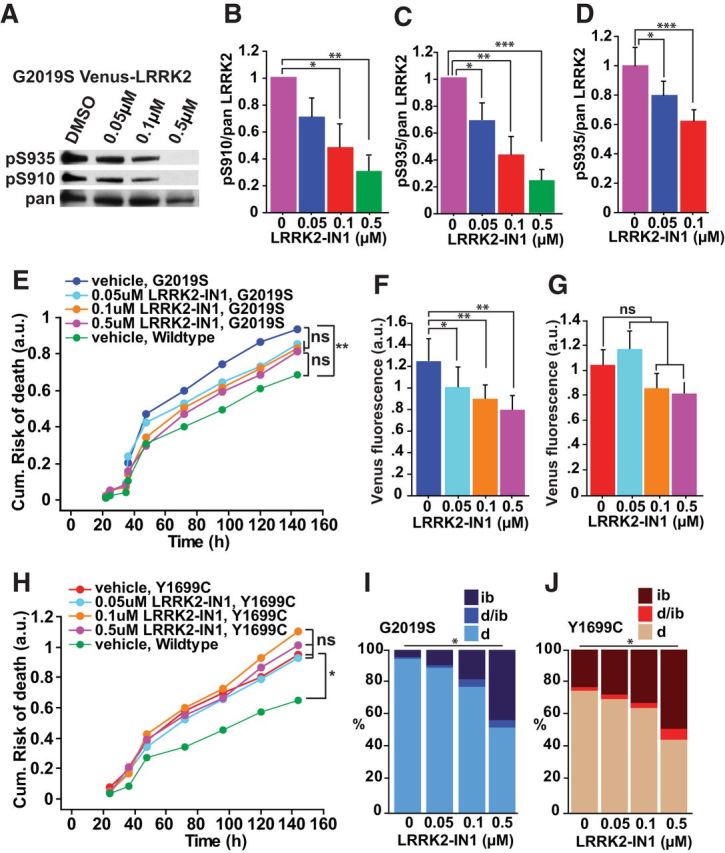
Pharmacological inhibition of LRRK2 kinase activity reduces LRRK2 toxicity by modulating IB formation and LRRK2 levels. A, Western blot of HEK293 cell lysates transfected with G2019S Venus-LRRK2, treated with DMSO or increasing concentrations of LRRK2-IN1 for 2 h, and extracted for immunoblotting to detect LRRK2 phosphorylation at Ser910 (top) and Ser935 (middle), and total LRRK2 (bottom). B, C, Western blot analysis was used to take measurements of LRRK2 phosphorylation at Ser910 and Ser935 normalized against total LRRK2. Three experiments combined, Mann–Whitney test, 0 μm versus 0.05 μm (S910 and S935 p = 0.0495; Z = 1.964), 0 μm versus 0.1 μm (S910 and S935 p = 0.0495; Z = 1.964), 0 μm versus 0.5 μm (S910 and S935 p = 0.025; Z = 2.236). Error bars indicate SD of the mean. D, Immunocytochemistry measurements of LRRK2 phosphorylation at Ser935 normalized against total LRRK2, from individual rat cortical neurons overexpressing G2019S Venus-LRRK2. More than 30 neurons per group, one-way ANOVA with Fisher's PLSD post hoc tests, *p < 0.01, ***p < 0.0001; F = 14.579. Error bars indicate 95% confidence intervals. E, Application of increasing concentrations of LRRK2-IN-1 trends towards a reduction in the cumulative risk-of-death of neurons expressing mutant G2019S LRRK2. Number of neurons for vehicle = 295, 0.05 μm = 292, 0.1 μm = 345 and 0.5 μm = 321, three experiments combined, log rank test, **p < 0.01. F, G, Average Venus-LRRK2 levels measured in neurons with and without LRRK2-IN1 treatment. F, Increasing doses of LRRK2-IN1 caused a dose-dependent reduction in G2019S-Venus-LRRK2 levels and G, no significant difference was found with Y1699C Venus-LRRK2. Number of neurons same as E, H, three experiments combined, one-way ANOVA with Fisher's PLSD post hoc tests, G2019S (*p < 0.05, **p < 0.01; F = 4.659) and Y1699C (not significant (ns); F = 1.256). Error bars are 95% confidence intervals. H, Application of increasing concentrations of LRRK2-IN1 caused no difference in risk of death for mutant Y1699C-expressing neurons. Number of neurons for vehicle = 274, 0.05 μm = 222, 0.1 μm = 238, and 0.5 μm = 231, three experiments combined, log rank test, *p < 0.05. I, J, Bar graphs showing the percentage of cells with diffuse mutant G2019S or Y1699C LRRK2 expression (d), an IB (ib) or that do not have a clearly defined IB, but show signs of an IB precursor (d/ib) at 24 h post-transfection. Increasing concentrations of LRRK2-IN increase the percentage of neurons with IBs for both mutants. Number of neurons is same as above in E, H, three experiments combined, Kruskal–Wallis test, G2019S (*p = 0.042, H = 8.231) and Y1699C (*p = 0.038, H = 8.426).
As reported for LRRK2 kinase inhibitors (Herzig et al., 2011), increasing LRRK2-IN1 concentrations decreased the steady-state levels of G2019S LRRK2 (Fig. 5F). Because LRRK2 concentration is proportional to cell death (Fig. 3C–E), Cox analysis was used to test the contribution of LRRK2 steady-state levels in reducing G2019S-induced toxicity mediated by LRRK2-IN1 exposure. Before adjusting for LRRK2 levels, LRRK2-IN1 reduced mutant LRRK2-induced toxicity, but not after LRRK2 levels were controlled (Table 2). In the case of Y1699C, increasing LRRK2-IN1 concentrations did not reduce toxicity and only trended toward reducing LRRK2 levels (Fig. 5G,H), again supporting the importance of LRRK2 concentration in predicting neuron death.
As reported (Dzamko et al., 2010; Deng et al., 2011), increasing LRRK2-IN1 concentrations caused increased LRRK2 IB formation for G2019S and Y1699C LRRK2 (Fig. 5I,J). In the case of G2019S LRRK2, the redistribution of LRRK2 from a diffuse form into IBs after LRRK2-IN1 application associated with a lowering of diffuse LRRK2 levels and a reduction in LRRK2-mediated toxicity. These results suggest that the redistribution of LRRK2 into IBs might be a viable coping response for the cell to deal with diffuse LRRK2.
By genetic and pharmacological approaches, LRRK2 levels, kinase activity and IB formation are interrelated variables. The ability to incorporate kinase activity and LRRK2 levels simultaneously into our model enabled us to show that changes in toxicity and IB formation are better explained by effects on LRRK2 levels than kinase activity per se.
Removing endogenous synuclein alleviates mutant LRRK2-induced toxicity
We then turned our attention to another feature of PD-associated toxicity: α-synuclein accumulation (Lin et al., 2009), a cause of PD (Singleton et al., 2003; Satake et al., 2009; Nakamura et al., 2011) that correlates with neuronal deficits in the substantia nigra (SN; Kovacs et al., 2008) and is toxic in a dose-dependent manner (Nakamura et al., 2011). Indeed, expression of G2019S- or Y1699C-LRRK2 significantly increased endogenous cytoplasmic α-synuclein levels (Fig. 6A,B) as reported (Lin et al., 2009; Sanchez-Danes et al., 2012), suggesting that mutant-LRRK2-induced toxicity may require α-synuclein.
Figure 6.
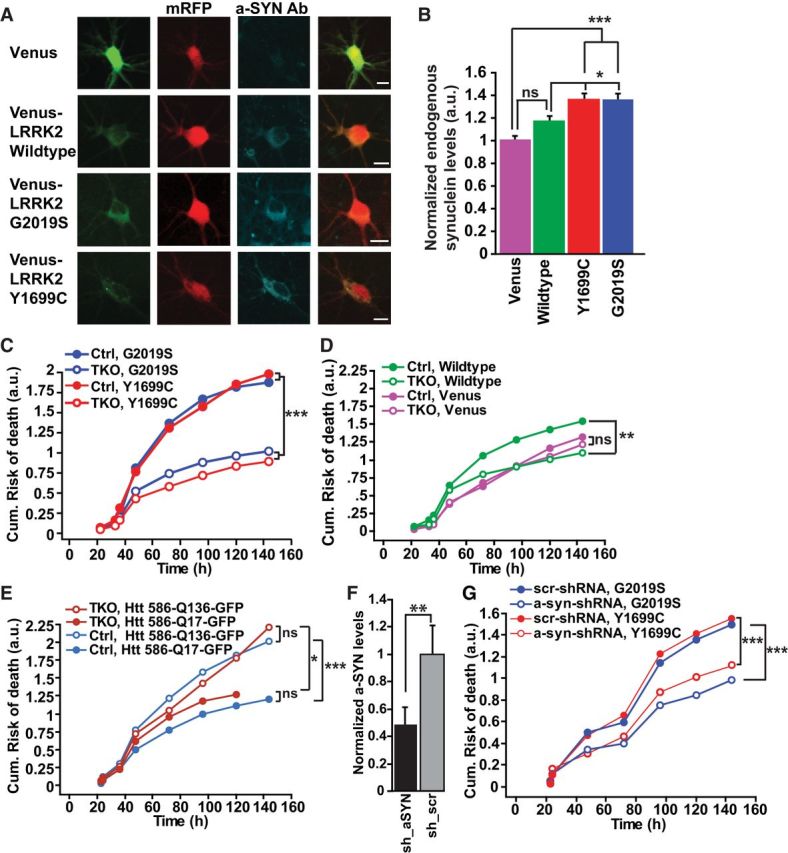
Synuclein is required for mutant LRRK2-induced toxicity. A, Representative images of primary cortical neurons transfected with Venus, wild-type, or mutant Venus-LRRK2, fixed 24 h later and stained with α-synuclein. Neurons are 7 DIV. Scale bar, 10 μm. B, Average levels of endogenous α-synuclein (determined by anti-α-synuclein immunocytochemistry) in cortical neurons transfected with wild-type, Y1699C, or G2019S Venus-LRRK2. Values are normalized relative to α-synuclein levels in neurons expressing a Venus control. More than 100 neurons per group, four experiments combined, one-way ANOVA with Fisher's PLSD post hoc tests, *p < 0.05, ***p < 0.0001; F = 7.707. Error bars are 95% confidence intervals. C, Cumulative risk-of-death curves show that TKO neurons expressing Y1699C or G2019S had significantly lower toxicity than Ctrl neurons. More than 245 neurons per group, three experiments combined, log rank test, ***p > 0.0001. D, Cumulative risk-of-death curves show that TKO neurons expressing wild-type Venus-LRRK2 had significantly lower toxicity than Ctrl neurons. Ctrl and TKO neurons transfected with Venus control had no difference in survival. More than 180 neurons per group, three experiments combined, log rank test, **p < 0.001. E, Ctrl and TKO neurons transfected with mutant Htt586-Q136-GFP show no difference in toxicity in Ctrl or TKO neurons. More than 120 neurons per group, two experiments combined, log rank test, *p < 0.01, ***p > 0.0001. F, Quantification of shRNA knockdown of endogenous α-synuclein levels. Primary rat cortical neurons were transfected with shRNA against α-synuclein or shRNA scrambled control. Forty-eight hours after transfection cells were fixed and stained with α-synuclein. Neurons were 7 DIV when fixed. More than 34 neurons per group, two experiments combined, unpaired t test, **p < 0.001; t = 3.688. Error bars are 95% confidence intervals. G, Cumulative risk-of-death curves show that in rodent neurons knockdown of α-synuclein by transfection of shRNA constructs (a-syn-shRNA) significantly reduce mutant LRRK2 toxicity, compared with neurons transfected with scrambled shRNA (scr-shRNA). More than 150 neurons per group, three experiments combined, log rank test, ***p < 0.0001.
Given the functional redundancy of α-, β-, and γ-synuclein (Chandra et al., 2004; Anwar et al., 2011), we began investigating a role of synuclein in LRRK2-mediated toxicity by transfecting Venus-LRRK2 into primary cortical neurons from αβγ-synuclein TKO and wild-type matched Ctrl mice (Nakamura et al., 2011). In contrast to Ctrl neurons, toxicity induced by overexpressing G2019S and Y1699C LRRK2 was reduced in TKO neurons (Fig. 6C; HR 0.634; p < 0.0001, CI 0.519–0.774 for G2019S and HR 0.506; p < 0.0001; CI 0.407–0.630 for Y1699C). Toxicity from wild-type Venus-LRRK2 was also reduced in TKO neurons (Fig. 6D). However, TKO or Ctrl neurons transfected with control Venus showed no difference in baseline survival. Furthermore, removing endogenous synuclein did not protect neurons from toxicity induced by mutant huntingtin expression, another toxic aggregation-prone protein, suggesting that the benefits of synuclein loss are specific to LRRK2 (Fig. 6E).
Next, we investigated whether removing α-synuclein alone was sufficient to reduce mutant LRRK2 toxicity. ShRNA-mediated knockdown of α-synuclein, confirmed by immunohistochemistry (Fig. 6F), was used to reduce α-synuclein levels in rat neurons transfected with LRRK2. Longitudinal imaging revealed that reduction of α-synuclein alone reduced mutant-LRRK2-mediated toxicity (Fig. 6G; HR 0.747; p = 0.001, CI 0.627–0.889 for G2019S and HR 0.699; p < 0.0001; CI 0.587–0.833).
LRRK2 patient-derived human neurons display α-synuclein-dependent toxicity
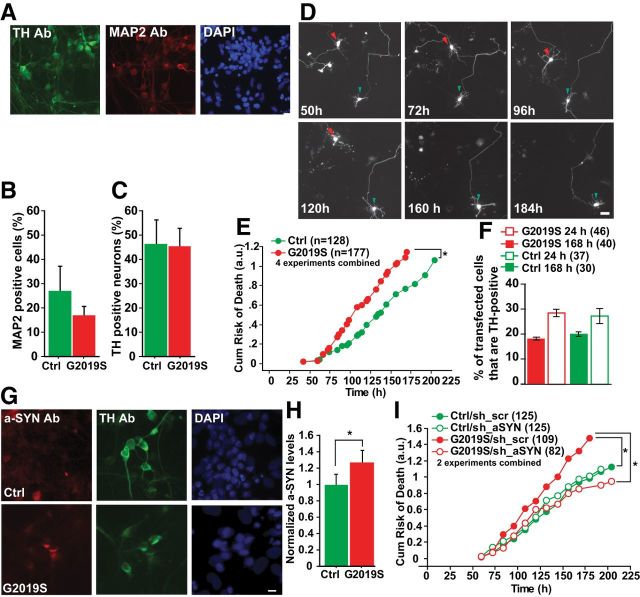
To confirm the physiological relevance of our findings, neurons were differentiated from G2019S-LRRK2 PD patient-derived and iPSCs, where LRRK2 expression is under the control of its endogenous promoter. Differentiated cells were fixed and stained for microtubule-associated protein 2 (MAP2), TH, and DAPI (Fig. 7A). The percentage of neurons in the culture was calculated as the percentage of cells with MAP2 and DAPI overlap (Fig. 7B). The percentage of dopaminergic neurons was calculated as the percentage of MAP2-positive cells with both MAP2 and TH staining (Fig. 7C). In parallel, differentiated cells transfected with a mApple-Map2 fluorescent reporter were imaged and followed for at least 7 d (Fig. 7D). Tracked cells displayed neuronal features, including at least two projecting processes. In parallel, separate cultures were fixed, and 30–40% of these morphologically similar cells stained positive for TH (data not shown). Human cells harboring the G2019S mutation had a higher cumulative risk-of-death than control cells for G2019S without the application of exogenous stressors (Fig. 7E; HR 1.4; p = 0.018; 95% CI 1.1–1.9). To determine whether the neurons that we were tracking were TH-positive neurons and that they were specifically dying, we compared the percentage of transfected neurons that were TH-positive at 24 and 168 h post-transfection. In human cells harboring the G2019S mutation and control cells, there was a trend toward a lower percentage of TH-positive neurons at 168 h post-transfection (Fig. 7F), indicating that TH neurons were indeed dying and potentially at a faster rate than TH-negative neurons.
Figure 7.
LRRK2 patient-derived neurons display α-synuclein-dependent toxicity. A, Representative images from immunocytochemistry of neurons differentiated from iPSCs from a control individual, costained with TH, MAP2, and DAPI. Scale bar, 20 μm. B, The percentage of neurons in the culture was calculated as the percentage of cells with MAP2 and DAPI overlap. C, The percentage of dopaminergic neurons was calculated as the percentage of MAP2-positive cells with colocalization of both MAP2 and TH staining. D, Longitudinal tracking of single neurons differentiated from iPSCs and transfected with a mApple-Map2 fluorescent reporter. The time of death is captured for each neuron that is followed. The cell indicated with a red arrowhead dies by 160 h. Degeneration of neurites was observed at 120 h and complete soma loss observed at 160 h. In contrast the neuron indicated by the green arrowhead, lives throughout the length of the experiment. Scale bar, 20 μm. E, Neurons derived from patients harboring the LRRK2 mutation G2019S had a greater risk-of-death than controls. Cells per group specified in brackets on the figure, four experiments combined, log rank test, *p < 0.05. F, Histogram showing the percentage of mApple-transfected neurons that are TH-positive at 24 or 168 h post-transfection. TH-positive neurons expressing mutant LRRK2 G2019S trended toward a lower percentage of TH-positive neurons at 6 d. Approximately 30–40 neurons in each group, two independent experiments combined, one-way ANOVA with Fisher's PLSD post hoc tests, p = 0.246; F = 1.563. Error bars are 95% confidence intervals. G, Representative images of TH-positive neurons derived from G2019S LRRK2 and control iPSCs indicate elevated endogenous α-synuclein staining in G2019S LRRK2 patient cells. Scale bar, 10 μm. H, Average levels of endogenous α-synuclein (determined by anti-α-synuclein immunocytochemistry) in TH-positive neurons are significantly higher in neurons differentiated from G2019S LRRK2 iPSCs compared with control iPSCs. Values are normalized relative to α-synuclein levels in control TH neurons. More than 100 neurons per group, three experiments combined, unpaired t test, *p < 0.01; t = 2.692. Error bars are 95% confidence intervals. I, Reducing α-synuclein levels in neurons derived from patients harboring the LRRK2 mutation G2019S caused a significant reduction in risk-of-death compared with those with a scrambled control shRNA. Reducing α-synuclein levels in neurons derived from unaffected individuals showed no significant difference in survival compared with those with a scrambled control shRNA. Brackets indicate cells per group, two experiments combined, log rank test, *p < 0.05.
In addition to recapitulating mutant-LRRK2-induced cell loss, DA neurons derived from G2019S iPSCs accumulated more α-synuclein than control neurons (Fig. 7G,H). As in primary rodent neurons, removal of α-synuclein reduced toxicity in iPSC-derived neurons with the G2019S LRRK2 mutation (Fig. 7I; HR 0.676; p = 0.03; 95% CI 0.5–0.96). α-Synuclein knockdown did not affect survival in an independent control line (Fig. 7I; HR 1; p = 0.83; 95% CI 0.7–1.5).
Removal of endogenous synuclein reduces levels of diffuse mutant LRRK2
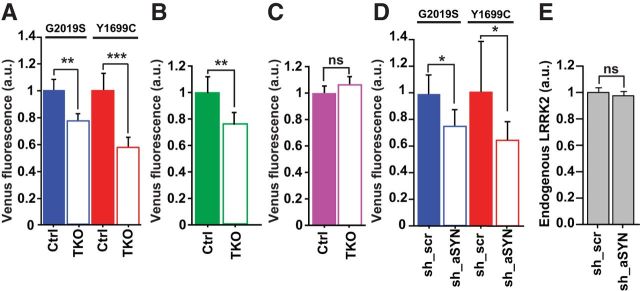
In rodent and human cells, synuclein is required for LRRK2-mediated toxicity. As α-synuclein accumulation disrupts proteostasis and its removal enhances it (Cuervo et al., 2004; Fornai et al., 2005), synuclein might mechanistically mediate mutant LRRK2 toxicity by modulating LRRK2 levels. In TKO neurons overexpressing LRRK2, mutant and wild-type LRRK2 levels were lower than in Ctrl neurons (Fig. 8A,B). Removing synuclein was specific to LRRK2: there was no difference in Venus levels in neurons expressing only Venus (Fig. 8C). Knocking down α-synuclein in rat neurons overexpressing mutant LRRK2 was also associated with a reduction in mutant LRRK2 levels (Fig. 8D), indicating that the removal of α-synuclein alone was sufficient to modulate LRRK2 levels. We were unable to detect an effect of knockdown of α-synuclein caused on endogenous levels of LRRK2 (Fig. 8E).
Figure 8.
Loss of α-synuclein reduces LRRK2 levels. A, Venus-LRRK2 levels of the Y1699C and G2019S LRRK2 mutants were significantly lower in TKO neurons than in Ctrl mouse neurons. Number of neurons for G2019S = 204, G2019S TKO = 251, Y1699C = 160, Y1699C TKO = 194, three experiments combined, one-way ANOVA with Fisher's PLSD post hoc tests, **p < 0.01, ***p < 0.0001; F = 8.674. Error bars are 95% confidence intervals. B, Histogram showing quantification of Venus-LRRK2 fluorescence in Ctrl and TKO neurons cotransfected with wild-type Venus-LRRK2. TKO neurons had significantly lower levels of Venus-LRRK2 as measured by Venus fluorescence. More than 271 neurons per group, three experiments combined, unpaired t test, **p < 0.002, t = 3.179. Error bars are 95% confidence intervals. C, In Ctrl and TKO neurons cotransfected with Venus, levels of Venus were unchanged. More than 188 neurons per group, three experiments combined, unpaired t test, p = 0.11, t = 1.6. Error bars are 95% confidence intervals. D, Venus-LRRK2 levels were also significantly lower in rat neurons with knockdown of endogenous α-synuclein (sh_aSYN) versus scrambled shRNA (sh_scr). Number of neurons G2019S/sh_scr = 185, G2019S/sh_aSYN = 160, Y1699C/sh_scr = 93, Y1699C/sh_aSYN = 147, two experiments combined, one-way ANOVA with Fisher's PLSD post hoc tests, *p < 0.05; F = 3.841. Error bars are 95% confidence intervals. E, Endogenous LRRK2 levels were unchanged in rat neurons with knockdown of endogenous α-synuclein (sh_aSYN) versus those with scrambled shRNA (sh_scr). Number of neurons sh_aSYN = 189, sh_scr = 180, three experiments combined, one-way ANOVA, p = 0.153; F = 2.056. Error bars are 95% confidence intervals.
As removing α-synuclein reduces toxicity and levels of LRRK2, and given LRRK2 levels' role as a powerful predictor of neuron death (Figs. 3–5), the benefit of removal might be explained by α-synuclein's effect on LRRK2 levels. Before controlling for LRRK2 levels, cells overexpressing mutant LRRK2 with knockdown of α-synuclein had lower toxicity. However, once LRRK2 levels were controlled for, there was no difference in toxicity between cells with or without α-synuclein (Table 2). By controlling for LRRK2 levels, we show that α-synuclein is required for LRRK2-induced toxicity and its effect is mediated by modulating LRRK2 levels.
Discussion
Our basic cellular systems model of PD led to several novel and significant findings about the role of LRRK2-related events during neurodegeneration. Using an imaging platform that longitudinally tracks primary neurons throughout their lifetime, we established rodent and human primary neuron models of LRRK2 that recapitulate features of PD-associated LRRK2 neurodegeneration, including neurite retraction, α-synuclein accumulation, and mutant LRRK2-induced neuron loss. In contrast to previous studies (Byers et al., 2011; Nguyen et al., 2011; Liu et al., 2012; Reinhardt et al., 2013), in our human PD model, we demonstrated neuronal loss without an exogenous cell stressor. Cellular features associated with LRRK2-mediated toxicity, including LRRK2 levels, IB formation, and kinase activity, were tracked and linked to cell fate. Using statistical survival tools, we built a predictive, quantitative model of LRRK2 that measured the roles and importance of these cellular features during neurodegeneration, independently of each other.
The cellular concentrations of diffuse mutant and wild-type LRRK2 strongly predicted cell death. Past studies relied on Western blotting to measure LRRK2 levels, which were often performed in cell lines that are substantially different from neurons (LePage et al., 2005; Tsvetkov et al., 2010). Furthermore, given LRRK2's ubiquitous expression and linkage to a subset of diseases (Franke et al., 2010), its function and metabolism may be cell-type-specific. Finally, measuring protein levels by population-based assays lacks single-cell resolution. Cell-to-cell variation in the LRRK2 dose is treated as a technical rather than biological variation, reducing sensitivity to detect phenotypes (e.g., cell death) across cell populations. In contrast, we measured the LRRK2 dose in each neuron and related it to that cell's fate, harnessing cell-to-cell variability to enhance the sensitivity and power to resolve dose–response relationships.
Measuring LRRK2 levels to predict cell death was critical for investigating the role of LRRK2 kinase activity during neurodegeneration. Genetic and pharmacologic blockade of kinase activity reduced mutant LRRK2 toxicity and LRRK2 levels. However, unlike previous studies (Greggio et al., 2006; Lin et al., 2009; Lee et al., 2010; Herzig et al., 2011), our systems biology approach enabled us to measure the independent contributions of LRRK2 levels and kinase activity during cell death, even though they are intertwined and a change in one affects the other. Critically, we show that disrupting kinase activity reduced mutant LRRK2 toxicity by affecting LRRK2 levels rather than kinase activity per se.
We also found neurons with higher LRRK2 levels have a greater propensity to form IBs. However, when LRRK2-IN1 was used to inhibit LRRK2 kinase activity, IB formation increased, and LRRK2 levels decreased. LRRK2-IN1 disrupts the binding of LRRK2 to 14-3-3 protein isoforms by dephosphorylating Ser910 and Ser935 leading to increased LRRK2 aggregation (Deng et al., 2011). Reducing the 14-3-3-LRRK2 interaction destabilizes the LRRK2 structure (Nichols et al., 2010), promoting sequestration of diffuse LRRK2 into IBs and reducing diffuse LRRK2 levels elsewhere in the cell. Unlike G2019S, LRRK2-IN1 did not reduce Y1699C-mediated toxicity. As the Y1699C mutation itself impairs the binding of LRRK2 to 14-3-3 (Nichols et al., 2010) and has a greater propensity to form IBs, LRRK2-IN1 causes a smaller increase in IB formation than the G2019S mutant, leading to lower reductions in LRRK2 levels. Although LRRK2 levels were a stronger predictor of toxicity for both mutants, the distinct differences of the mutants for IB formation and 14-3-3 binding indicate mechanistic differences between the PD-associated mutants that may be relevant for therapeutic development.
In the absence of LRRK2-IN1, LRRK2 IBs neither promoted nor retarded mutant LRRK2 toxicity. This may be due to their inefficiency in reducing toxic diffuse LRRK2 levels elsewhere in the cell. In other neurodegenerative disorders, IBs are well formed and help sequester the diffuse toxic species into IBs, so that the IBs act as a coping response (Arrasate et al., 2004). Therefore, in addition to therapeutics that clear LRRK2, ones that help promote more efficient sequestration of toxic diffuse mutant LRRK2 into more compact IBs may be a viable therapeutic strategy against LRRK2-mediated toxicity.
Elevated levels of mutant LRRK2 also caused the accumulation of the PD-associated protein α-synuclein. We found removing endogenous synuclein significantly reduced mutant LRRK2-induced toxicity and suggested a reciprocal link between α-synuclein and LRRK2 in PD-related neurodegeneration. To demonstrate that these key observations were due specifically to the PD-associated mutations in LRRK2, complementary experiments were performed in human dopaminergic neurons with the endogenous LRRK2 G2019S mutation and in murine neurons with ectopic expression of mutant LRRK2. In murine neurons, the genetic background was identical in neurons expressing wild-type or mutant LRRK2, so that any increased risk of death could be attributed to the PD-causing mutation in LRRK2. Similarly, the reduction of LRRK2 G2019S-induced neurodegeneration by synuclein knockdown in human dopaminergic neurons was supported with an analogous murine experiment in which the genetic background was fully controlled. Furthermore, the knockdown of α-synuclein in human dopaminergic neurons with the LRRK2 G2019S mutation only rescued survival back to levels of the control line, and had no effect on survival of the control line, consistent with the idea that the increased risk of death in the LRRK2 G2019S line is a LRRK2 mutation-dependent effect.
How the removal of α-synuclein facilitates LRRK2-induced neurodegeneration is unclear. Evidence from LRRK2 and synuclein knock-out models indicates that these two proteins have at least some normal functions that are independent of each other, at least in some cell types. For example, α-synuclein KO mice display cognitive impairments (Kokhan et al., 2012) and subtle evidence of abnormalities in neurotransmission (Abeliovich et al., 2000; Cabin et al., 2002), whereas LRRK2 knock-out mice showed neither dopaminergic degeneration nor alterations in dopamine dynamics (Andres-Mateos et al., 2009; Hinkle et al., 2012; Tong et al., 2012) but display age-dependent alterations in autophagic activity in the kidneys. In addition, we observed no detectable change in endogenous LRRK2 levels with knockdown of α-synuclein.
However, this does not go against the possibility that during neurodegeneration, triggered by PD causing mutations in LRRK2, the actions of LRRK2 become critically linked to α-synuclein, at least in neurons. For example, even though α-synuclein knock-out mice are resistant to MPTP-induced toxicity (Dauer et al., 2002) and LRRK2 KO mice show no response to this toxin (Andres-Mateos et al., 2009), LRRK2 and MPTP might act on independent pathways to cause neurodegeneration but both require synuclein.
In the disease state, α-synuclein could mediate LRRK2-mediated toxicity by modulating LRRK2 levels, which are toxic in a dose-dependent manner (Fig. 3). Without α-synuclein, mutant LRRK2 levels were significantly lower. Because α-synuclein disrupts protein homeostasis pathways (Snyder et al., 2003; Cuervo et al., 2004; Fornai et al., 2005) and removing α-synuclein bolsters them (Fornai et al., 2005), synuclein might hinder degradation and handling of mutant LRRK2, thereby leading to LRRK2's accumulation and toxicity. However, the change in LRRK2-mediated toxicity before and after controlling for LRRK2 levels in the absence of α-synuclein, although significant, was small. The lowering of LRRK2 levels is likely one of multiple contributing mechanisms by which synuclein removal protects neurons from LRRK2 toxicity. As α-synuclein is toxic in a dose-dependent manner (Singleton et al., 2003; Nakamura et al., 2011), LRRK2-mediated toxicity may directly result from α-synuclein build-up. However, the increase in α-synuclein by LRRK2 is probably insufficient to fully explain the toxicity of mutant LRRK2.
Alternatively, mutant LRRK2 and α-synuclein might lead to neurodegeneration, via a synergistic mechanism. The presence of genetic variants within the 3′UTR of SNCA that increase α-synuclein levels in the SN (Fuchs et al., 2008) cause an earlier onset of PD in patients harboring LRRK2 mutations (Botta-Orfila et al., 2012). A synergistic relationship between these two key PD-associated proteins may explain the variable penetrance of LRRK2 mutations (Healy et al., 2008) and why synuclein pathology is common in LRRK2 patients (Biskup and West, 2009).
With α-synuclein's role in protein homeostasis disruption (Snyder et al., 2003; Cuervo et al., 2004; Fornai et al., 2005) and as LRRK2 itself modulates α-synuclein degradation (Orenstein et al., 2013) and α-synuclein-mediated toxicity (Lin et al., 2009), a synergistic relationship between LRRK2 and α-synuclein may converge on dysfunction of protein homeostasis within cells. The accumulation of misfolded proteins causes proteome instability (Gupta et al., 2011), resulting in global cellular consequences. This might explain the widespread cellular dysfunction in vesicular trafficking, neurotransmitter release, cytoskeletal dynamics, protein degradation (Greggio et al., 2011), and mitochondrial dynamics (Nakamura et al., 2011; Wang et al., 2012), observed independently in α-synuclein and LRRK2-associated neurodegeneration.
As the cellular environment may be important for LRRK2's physiological role (Lewis and Manzoni, 2012), cell-type specificity may contribute critically to the interplay between these proteins. In the hindbrain, LRRK2 modulated α-synuclein neuropathology (Lin et al., 2009), but it was not required for α-synuclein-mediated degeneration (Daher et al., 2012). Understanding how and where the functional pathways of LRRK2 and α-synuclein converge will provide greater insight into the common mechanisms underlying neurodegeneration in PD.
Footnotes
This work was supported with a gift from the DeClerq family in memory of Betty Brown. The Gladstone Institutes received support from a National Center for Research Resources Grant RR18928. G.S. is supported by the Hillblom Foundation. K.N. is supported by the Burroughs-Wellcome Medical Scientist Fund Career Award, NINDS (1K08NS062954-01A1) and award no. P30NS069496 from the National Institute of Neurological Disorders and Stroke. M.R.C. is supported in part by the Intramural Research Program of the NIH, National Institute on Aging. S.F. is supported by Grants U24 NS078370, 3R01 NS039074, 2R01 NS04549, 2P01 AG0224074, CIRM RB4-06079 research Grant, the Koret/Taube Center, and the Hellman Family Foundation. We thank members of the Finkbeiner lab for useful discussions, A.L. Lucido, S. Ordway, and G. Howard for editorial assistance, and K. Nelson for administrative assistance. Dr. Dario Alessi (MRC-PPU, Dundee University, Dundee Scotland) who kindly provided LRRK2-IN1 and rabbit monoclonal anti-LRRK2 (100-500), anti-LRRK2 P-ser935, or anti-LRRK2 P-ser910.
The authors declare no competing financial interests.
References
- Abeliovich A, Schmitz Y, Fariñas I, Choi-Lundberg D, Ho WH, Castillo PE, Shinsky N, Verdugo JM, Armanini M, Ryan A, Hynes M, Phillips H, Sulzer D, Rosenthal A. Mice lackingα-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron. 2000;25:239–252. doi: 10.1016/S0896-6273(00)80886-7. [DOI] [PubMed] [Google Scholar]
- Alegre-Abarrategui J, Christian H, Lufino MM, Mutihac R, Venda LL, Ansorge O, Wade-Martins R. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet. 2009;18:4022–4034. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen PK, Gill RD. Cox's regression model for counting processes, a large sample study. Ann Statistics. 1982;10:1100–1120. doi: 10.1214/aos/1176345976. [DOI] [Google Scholar]
- Andres-Mateos E, Mejias R, Sasaki M, Li X, Lin BM, Biskup S, Zhang L, Banerjee R, Thomas B, Yang L, Liu G, Beal MF, Huso DL, Dawson TM, Dawson VL. Unexpected lack of hypersensitivity in LRRK2 knock-out mice to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) J Neurosci. 2009;29:15846–15850. doi: 10.1523/JNEUROSCI.4357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar S, Peters O, Millership S, Ninkina N, Doig N, Connor-Robson N, Threlfell S, Kooner G, Deacon RM, Bannerman DM, Bolam JP, Chandra SS, Cragg SJ, Wade-Martins R, Buchman VL. Functional alterations to the nigrostriatal system in mice lacking all three members of the synuclein family. J Neurosci. 2011;31:7264–7274. doi: 10.1523/JNEUROSCI.6194-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M, Finkbeiner S. Automated microscope system for determining factors that predict neuronal fate. Proc Natl Acad Sci U S A. 2005;102:3840–3845. doi: 10.1073/pnas.0409777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Barmada SJ, Skibinski G, Korb E, Rao EJ, Wu JY, Finkbeiner S. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilican B, Serio A, Barmada SJ, Nishimura AL, Sullivan GJ, Carrasco M, Phatnani HP, Puddifoot CA, Story D, Fletcher J, Park IH, Friedman BA, Daley GQ, Wyllie DJ, Hardingham GE, Wilmut I, Finkbeiner S, Maniatis T, Shaw CE, Chandran S. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc Natl Acad Sci U S A. 2012;109:5803–5808. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskup S, West AB. Zeroing in on LRRK2-linked pathogenic mechanisms in Parkinson's disease. Biochim Biophys Acta. 2009;1792:625–633. doi: 10.1016/j.bbadis.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta-Orfila T, Ezquerra M, Pastor P, Fernández-Santiago R, Pont-Sunyer C, Compta Y, Lorenzo-Betancor O, Samaranch L, Martí MJ, Valldeoriola F, Calopa M, Fernández M, Aguilar M, de Fabregas O, Hernández-Vara J, Tolosa E. Age at onset in LRRK2-associated PD is modified by SNCA variants. J Mol Neurosci. 2012;48:245–247. doi: 10.1007/s12031-012-9820-7. [DOI] [PubMed] [Google Scholar]
- Byers B, Cord B, Nguyen HN, Schüle B, Fenno L, Lee PC, Deisseroth K, Langston JW, Pera RR, Palmer TD. SNCA triplication Parkinson's patient's iPSC-derived DA neurons accumulate α-synuclein and are susceptible to oxidative stress. PloS One. 2011;6:e26159. doi: 10.1371/journal.pone.0026159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabin DE, Shimazu K, Murphy D, Cole NB, Gottschalk W, McIlwain KL, Orrison B, Chen A, Ellis CE, Paylor R, Lu B, Nussbaum RL. Synaptic vesicle depletion correlates with attenuated synaptic responses to prolonged repetitive stimulation in mice lacking α-synuclein. J Neurosci. 2002;22:8797–8807. doi: 10.1523/JNEUROSCI.22-20-08797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Fornai F, Kwon HB, Yazdani U, Atasoy D, Liu X, Hammer RE, Battaglia G, German DC, Castillo PE, Südhof TC. Double-knock-out mice for alpha- and beta-synucleins: effect on synaptic functions. Proc Natl Acad Sci U S A. 2004;101:14966–14971. doi: 10.1073/pnas.0406283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Clark LN, Wang Y, Karlins E, Saito L, Mejia-Santana H, Harris J, Louis ED, Cote LJ, Andrews H, Fahn S, Waters C, Ford B, Frucht S, Ottman R, Marder K. Frequency of LRRK2 mutations in early- and late-onset Parkinson disease. Neurology. 2006;67:1786–1791. doi: 10.1212/01.wnl.0000244345.49809.36. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- Daher JP, Pletnikova O, Biskup S, Musso A, Gellhaar S, Galter D, Troncoso JC, Lee MK, Dawson TM, Dawson VL, Moore DJ. Neurodegenerative phenotypes in an A53T α-synuclein transgenic mouse model are independent of LRRK2. Hum Mol Genet. 2012;21:2420–2431. doi: 10.1093/hmg/dds057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub A, Sharma P, Finkbeiner S. High-content screening of primary neurons: ready for prime time. Curr Opin Neurobiol. 2009;19:537–543. doi: 10.1016/j.conb.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer W, Kholodilov N, Vila M, Trillat AC, Goodchild R, Larsen KE, Staal R, Tieu K, Schmitz Y, Yuan CA, Rocha M, Jackson-Lewis V, Hersch S, Sulzer D, Przedborski S, Burke R, Hen R. Resistance of α-synuclein null mice to the parkinsonian neurotoxin MPTP. Proc Natl Acad Sci U S A. 2002;99:14524–14529. doi: 10.1073/pnas.172514599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker FW, de Mutsert R, van Dijk PC, Zoccali C, Jager KJ. Survival analysis: time-dependent effects and time-varying risk factors. Kidney Int. 2008;74:994–997. doi: 10.1038/ki.2008.328. [DOI] [PubMed] [Google Scholar]
- Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, Lee JD, Patricelli MP, Nomanbhoy TK, Alessi DR, Gray NS. Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2. Nat Chem Biol. 2011;7:203–205. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzamko N, Deak M, Hentati F, Reith AD, Prescott AR, Alessi DR, Nichols RJ. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14-3-3 binding and altered cytoplasmic localization. Biochem J. 2010;430:405–413. doi: 10.1042/BJ20100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine J, Gray R. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. doi: 10.1080/01621459.1999.10474144. [DOI] [Google Scholar]
- Finkbeiner S, Tavazoie SF, Maloratsky A, Jacobs KM, Harris KM, Greenberg ME. CREB: a major mediator of neuronal neurotrophin responses. Neuron. 1997;19:1031–1047. doi: 10.1016/S0896-6273(00)80395-5. [DOI] [PubMed] [Google Scholar]
- Fleming TR, Lin DY. Survival analysis in clinical trials: past developments and future directions. Biometrics. 2000;56:971–983. doi: 10.1111/j.0006-341X.2000.0971.x. [DOI] [PubMed] [Google Scholar]
- Fornai F, Schlüter OM, Lenzi P, Gesi M, Ruffoli R, Ferrucci M, Lazzeri G, Busceti CL, Pontarelli F, Battaglia G, Pellegrini A, Nicoletti F, Ruggieri S, Paparelli A, Südhof TC. Parkinson-like syndrome induced by continuous MPTP infusion: convergent roles of the ubiquitin-proteasome system and α-synuclein. Proc Natl Acad Sci U S A. 2005;102:3413–3418. doi: 10.1073/pnas.0409713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J, Tichopad A, Golub Y, Munz M, Schweitzer KJ, Wolf B, Berg D, Mueller JC, Gasser T. Genetic variability in the SNCA gene influences α-synuclein levels in the blood and brain. FASEB J. 2008;22:1327–1334. doi: 10.1096/fj.07-9348com. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Gomez-Suaga P, Luzon-Toro B, Churamani D, Zhang L, Bloor-Young D, Patel S, Woodman PG, Churchill GC, Hilfiker S. Leucine-rich repeat kinase 2 regulates autophagy through a calcium-dependent pathway involving NAADP. Hum Mol Genet. 2011;21:511–525. doi: 10.1093/hmg/ddr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, Ahmad R, Miller DW, Kesavapany S, Singleton A, Lees A, Harvey RJ, Harvey K, Cookson MR. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Disease. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Greggio E, Bisaglia M, Civiero L, Bubacco L. Leucine-rich repeat kinase 2 and α-synuclein: intersecting pathways in the pathogenesis of Parkinson's disease? Mol Neurodegener. 2011;6:6. doi: 10.1186/1750-1326-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Kasturi P, Bracher A, Loew C, Zheng M, Villella A, Garza D, Hartl FU, Raychaudhuri S. Firefly luciferase mutants as sensors of proteome stress. Nat Methods. 2011;8:879–884. doi: 10.1038/nmeth.1697. [DOI] [PubMed] [Google Scholar]
- Haugarvoll K, Haugarvoll K, Rademakers R, Kachergus JM, Nuytemans K, Ross OA, Gibson JM, Tan EK, Gaig C, Tolosa E, Goldwurm S, Guidi M, Riboldazzi G, Brown L, Walter U, Benecke R, Berg D, Gasser T, Theuns J, Pals P, et al. Lrrk2 R1441C parkinsonism is clinically similar to sporadic Parkinson disease. Neurology. 2008;70:1456–1460. doi: 10.1212/01.wnl.0000304044.22253.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HD iPSC Consortium. Induced pluripotent stem cells from patients with Huntington's disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy DG, Falchi M, O'Sullivan SS, Bonifati V, Durr A, Bressman S, Brice A, Aasly J, Zabetian CP, Goldwurm S, Ferreira JJ, Tolosa E, Kay DM, Klein C, Williams DR, Marras C, Lang AE, Wszolek ZK, Berciano J, Schapira AH, et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson's disease: a case-control study. Lancet Neurol. 2008;7:583–590. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig MC, Kolly C, Persohn E, Theil D, Schweizer T, Hafner T, Stemmelen C, Troxler TJ, Schmid P, Danner S, Schnell CR, Mueller M, Kinzel B, Grevot A, Bolognani F, Stirn M, Kuhn RR, Kaupmann K, van der Putten PH, Rovelli G, et al. LRRK2 protein levels are determined by kinase function and are crucial for kidney and lung homeostasis in mice. Hum Mol Genet. 2011;20:4209–4223. doi: 10.1093/hmg/ddr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi S, Biskup S, West AB, Trinkaus D, Dawson VL, Faull RL, Waldvogel HJ, Arai H, Dawson TM, Moore DJ, Emson PC. Localization of Parkinson's disease-associated LRRK2 in normal and pathological human brain. Brain Res. 2007;1155:208–219. doi: 10.1016/j.brainres.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Hinkle KM, Yue M, Behrouz B, Dächsel JC, Lincoln SJ, Bowles EE, Beevers JE, Dugger B, Winner B, Prots I, Kent CB, Nishioka K, Lin WL, Dickson DW, Janus CJ, Farrer MJ, Melrose HL. LRRK2 knock-out mice have an intact dopaminergic system but display alterations in exploratory and motor coordination behaviors. Mol Neurodegener. 2012;7:25. doi: 10.1186/1750-1326-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager KJ, van Dijk PC, Zoccali C, Dekker FW. The analysis of survival data: the Kaplan-Meier method. Kidney Int. 2008;74:560–565. doi: 10.1038/ki.2008.217. [DOI] [PubMed] [Google Scholar]
- Kay DM, Zabetian CP, Factor SA, Nutt JG, Samii A, Griffith A, Bird TD, Kramer P, Higgins DS, Payami H. Parkinson's disease and LRRK2: frequency of a common mutation in U.S. movement disorder clinics. Mov Disord. 2006;21:519–523. doi: 10.1002/mds.20751. [DOI] [PubMed] [Google Scholar]
- Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc Natl Acad Sci U S A. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kett LR, Boassa D, Ho CC, Rideout HJ, Hu J, Terada M, Ellisman M, Dauer WT. LRRK2 Parkinson disease mutations enhance its microtubule association. Hum Mol Genet. 2012;21:890–899. doi: 10.1093/hmg/ddr526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HS, Bailey R, Smith WW, Liu Z, Shin JH, Lee YI, Zhang YJ, Jiang H, Ross CA, Moore DJ, Patterson C, Petrucelli L, Dawson TM, Dawson VL. CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proc Natl Acad Sci U S A. 2009;106:2897–2902. doi: 10.1073/pnas.0810123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokhan VS, Afanasyeva MA, Van'kin GI. α-Synuclein knockout mice have cognitive impairments. Behav Brain Res. 2012;231:226–230. doi: 10.1016/j.bbr.2012.03.026. [DOI] [PubMed] [Google Scholar]
- Kovacs GG, Milenkovic IJ, Preusser M, Budka H. Nigral burden of α-synuclein correlates with striatal dopamine deficit. Mov Disord. 2008;23:1608–1612. doi: 10.1002/mds.22207. [DOI] [PubMed] [Google Scholar]
- Lee BD, Shin JH, VanKampen J, Petrucelli L, West AB, Ko HS, Lee YI, Maguire-Zeiss KA, Bowers WJ, Federoff HJ, Dawson VL, Dawson TM. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson's disease. Nat Med. 2010;16:998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LePage KT, Dickey RW, Gerwick WH, Jester EL, Murray TF. On the use of neuro-2a neuroblastoma cells versus intact neurons in primary culture for neurotoxicity studies. Crit Rev Neurobiol. 2005;17:27–50. doi: 10.1615/CritRevNeurobiol.v17.i1.20. [DOI] [PubMed] [Google Scholar]
- Lewis PA, Manzoni C. LRRK2 and human disease: a complicated question or a question of complexes? Sci Signal. 2012;5:pe2. doi: 10.1126/scisignal.2002680. [DOI] [PubMed] [Google Scholar]
- Li LH, Qin HZ, Wang JL, Wang J, Wang XL, Gao GD. Axonal degeneration of nigra-striatum dopaminergic neurons induced by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine in mice. J Int Med Res. 2009;37:455–463. doi: 10.1177/147323000903700221. [DOI] [PubMed] [Google Scholar]
- Lin X, Parisiadou L, Gu XL, Wang L, Shim H, Sun L, Xie C, Long CX, Yang WJ, Ding J, Chen ZZ, Gallant PE, Tao-Cheng JH, Rudow G, Troncoso JC, Liu Z, Li Z, Cai H. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant α-synuclein. Neuron. 2009;64:807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GH, Liu GH, Qu J, Suzuki K, Nivet E, Li M, Montserrat N, Yi F, Xu X, Ruiz S, Zhang W, Wagner U, Kim A, Ren B, Li Y, Goebl A, Kim J, Soligalla RD, Dubova I, Thompson J, et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012;491:603–607. doi: 10.1038/nature11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Mena MA, Khan U, Togasaki DM, Sulzer D, Epstein CJ, Przedborski S. Effects of wild-type and mutated copper/zinc superoxide dismutase on neuronal survival and l-DOPA-induced toxicity in postnatal midbrain culture. J Neurochem. 1997;69:21–33. doi: 10.1046/j.1471-4159.1997.69010021.x. [DOI] [PubMed] [Google Scholar]
- Miller J, Arrasate M, Shaby BA, Mitra S, Masliah E, Finkbeiner S. Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into huntington's disease molecular pathogenesis. J Neurosci. 2010;30:10541–10550. doi: 10.1523/JNEUROSCI.0146-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, Miller J, Arrasate M, Brooks E, Libeu CP, Legleiter J, Hatters D, Curtis J, Cheung K, Krishnan P, Mitra S, Widjaja K, Shaby BA, Lotz GP, Newhouse Y, Mitchell EJ, Osmand A, Gray M, Thulasiramin V, Saudou F, et al. Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat Chem Biol. 2011;7:925–934. doi: 10.1038/nchembio.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Nemani VM, Azarbal F, Skibinski G, Levy JM, Egami K, Munishkina L, Zhang J, Gardner B, Wakabayashi J, Sesaki H, Cheng Y, Finkbeiner S, Nussbaum RL, Masliah E, Edwards RH. Direct membrane association drives mitochondrial fission by the Parkinson disease-associated protein α-synuclein. J Biol Chem. 2011;286:20710–20726. doi: 10.1074/jbc.M110.213538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, Gujar P, Kee K, Schüle B, Dolmetsch RE, Langston W, Palmer TD, Pera RR. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8:267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RJ, Dzamko N, Morrice NA, Campbell DG, Deak M, Ordureau A, Macartney T, Tong Y, Shen J, Prescott AR, Alessi DR. 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson's disease-associated mutations and regulates cytoplasmic localization. Biochem J. 2010;430:393–404. doi: 10.1042/BJ20100483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein SJ, Kuo SH, Tasset I, Arias E, Koga H, Fernandez-Carasa I, Cortes E, Honig LS, Dauer W, Consiglio A, Raya A, Sulzer D, Cuervo AM. Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci. 2013;16:394–406. doi: 10.1038/nn.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisán-Ruíz C, Jain S, Evans EW, Gilks WP, Simón J, van der Brug M, López de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Martí-Massó JF, Pérez-Tur J, Wood NW, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Park CH, Minn YK, Lee JY, Choi DH, Chang MY, Shim JW, Ko JY, Koh HC, Kang MJ, Kang JS, Rhie DJ, Lee YS, Son H, Moon SY, Kim KS, Lee SH. In vitro and in vivo analyses of human embryonic stem cell-derived dopamine neurons. J Neurochem. 2005;92:1265–1276. doi: 10.1111/j.1471-4159.2004.03006.x. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Pastrana E. A handle on neurodegenerative disease complexity. Nat Methods. 2012;9:21. doi: 10.1038/nchembio.1130. [DOI] [PubMed] [Google Scholar]
- Reinhardt P, Schmid B, Burbulla LF, Schöndorf DC, Wagner L, Glatza M, Höing S, Hargus G, Heck SA, Dhingra A, Wu G, Müller S, Brockmann K, Kluba T, Maisel M, Krüger R, Berg D, Tsytsyura Y, Thiel CS, Psathaki OE, et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12:354–367. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Sanchez-Danes A, Richaud-Patin Y, Carballo-Carbajal I, Jimenez-Delgado S, Caig C, Mora S, Di Guglielmo C, Ezquerra M, Patel B, Giralt A, Canals JM, Memo M, Alberch J, Lopez-Barneo J, Vila M, Cuervo AM, Tolosa E, Consiglio A, Raya A. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson's disease. EMBO Mol Med. 2012;4:380–395. doi: 10.1002/emmm.201200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, Kawaguchi T, Tsunoda T, Watanabe M, Takeda A, Tomiyama H, Nakashima K, Hasegawa K, Obata F, Yoshikawa T, Kawakami H, Sakoda S, Yamamoto M, Hattori N, Murata M, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/S0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- Scrucca L, Santucci A, Aversa F. Regression modeling of competing risk using R: an in depth guide for clinicians. Bone Marrow Transplant. 2010;45:1388–1395. doi: 10.1038/bmt.2009.359. [DOI] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, et al. α-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Skibinski G, Finkbeiner S. Longitudinal measures of proteostasis in live neurons: features that determine fate in models of neurodegenerative disease. FEBS Lett. 2013;587:1139–1146. doi: 10.1016/j.febslet.2013.02.043. [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Moore DJ, Liang Y, West AB, Dawson VL, Dawson TM, Ross CA. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc Natl Acad Sci U S A. 2005;102:18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- Snyder H, Mensah K, Theisler C, Lee J, Matouschek A, Wolozin B. Aggregated and monomeric α-synuclein bind to the S6′ proteasomal protein and inhibit proteasomal function. J Biol Chem. 2003;278:11753–11759. doi: 10.1074/jbc.M208641200. [DOI] [PubMed] [Google Scholar]
- Tong Y, Giaime E, Yamaguchi H, Ichimura T, Liu Y, Si H, Cai H, Bonventre JV, Shen J. Loss of leucine-rich repeat kinase 2 causes age-dependent bi-phasic alterations of the autophagy pathway. Mol Neurodegener. 2012;7:2. doi: 10.1186/1750-1326-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov AS, Miller J, Arrasate M, Wong JS, Pleiss MA, Finkbeiner S. A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc Natl Acad Sci U S A. 2010;107:16982–16987. doi: 10.1073/pnas.1004498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xie C, Greggio E, Parisiadou L, Shim H, Sun L, Chandran J, Lin X, Lai C, Yang WJ, Moore DJ, Dawson TM, Dawson VL, Chiosis G, Cookson MR, Cai H. The chaperone activity of heat shock protein 90 is critical for maintaining the stability of leucine-rich repeat kinase 2. J Neurosci. 2008;28:3384–3391. doi: 10.1523/JNEUROSCI.0185-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yan MH, Fujioka H, Liu J, Wilson-Delfosse A, Chen SG, Perry G, Casadesus G, Zhu X. LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum Mol Genet. 2012;21:1931–1944. doi: 10.1093/hmg/dds003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- Xi J, Liu Y, Liu H, Chen H, Emborg ME, Zhang SC. Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem Cells. 2012;30:1655–1663. doi: 10.1002/stem.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Müller-Myhsok B, Dickson DW, Meitinger T, Strom TM, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]