Abstract
Background
Variation of gene expression can lead to phenotypic variation and have therefore been assumed to contribute the diversity of wine yeast (Saccharomyces cerevisiae) properties. However, the molecular bases of this variation of gene expression are unknown. We addressed these questions by carrying out an integrated genetical-genomic study in fermentation conditions. We report here quantitative trait loci (QTL) mapping based on expression profiling in a segregating population generated by a cross between a derivative of the popular wine strain EC1118 and the laboratory strain S288c.
Results
Most of the fermentation traits studied appeared to be under multi-allelic control. We mapped five phenotypic QTLs and 1465 expression QTLs. Several expression QTLs overlapped in hotspots. Among the linkages unraveled here, several were associated with metabolic processes essential for wine fermentation such as glucose sensing or nitrogen and vitamin metabolism. Variations affecting the regulation of drug detoxification and export (TPO1, PDR12 or QDR2) were linked to variation in four genes encoding transcription factors (PDR8, WAR1, YRR1 and HAP1). We demonstrated that the allelic variation of WAR1 and TPO1 affected sorbic and octanoic acid resistance, respectively. Moreover, analysis of the transcription factors phylogeny suggests they evolved with a specific adaptation of the strains to wine fermentation conditions. Unexpectedly, we found that the variation of fermentation rates was associated with a partial disomy of chromosome 16. This disomy resulted from the well known 8–16 translocation.
Conclusions
This large data set made it possible to decipher the effects of genetic variation on gene expression during fermentation and certain wine fermentation properties. Our findings shed a new light on the adaptation mechanisms required by yeast to cope with the multiple stresses generated by wine fermentation. In this context, the detoxification and export systems appear to be of particular importance, probably due to nitrogen starvation. Furthermore, we show that the well characterized 8–16 translocation located in SSU1, which is associated with sulfite resistance, can lead to a partial chromosomic amplification in the progeny of strains that carry it, greatly improving fermentation kinetics. This amplification has been detected among other wine yeasts.
Keywords: Wine yeast, Fermentation, QTL, Transcriptome, Partial disomy, Detoxification
Background
Since the development of wine-making, wine yeasts have undergone a specific pattern of evolution and have become highly effective in the fermentation of grape juice [1]. They are able to withstand various stresses, such as low pH or high levels of ethanol. These yeasts are also resistant to inhibitors, such as sulfites [2] and medium-chain fatty acids [3], and are able to continue fermentation for long periods of nutrients starvation. In addition, commercial wine yeasts make a specific contribution to the aroma bouquet of the wine. Knowledge of the genomic bases of these specific features is a prerequisite for understanding the mechanisms underlying adaptation [1,4,5]. Such knowledge would also provide a basis for the improvement of industrial wine yeast strains [6]. Various studies have enhanced our understanding of genomic structural variation in wine yeast strains. Genome sequencing has led to the identification of genomic rearrangements and mutations specific to wine yeasts (reviewed in [7-9]). One of the best examples is the translocation between chromosomes 8 and 16 described by Pérez-Ortín et al.[10] leading to overexpression of the sulfite exporter gene SSU1 and to the resistance of higher levels of sulfite. Dunn et al.[11,12] have shown, using comparative genomic hybridization (aCGH), that copy-number variations for several groups of genes (involved in drug responses and ion sensors) are also characteristic of wine strains. Based on an analysis of the wine yeast EC1118 sequence, Novo et al.[13] showed that three loci widely conserved in wine strains had been gained by horizontal transfer events from non Saccharomyces yeast [14]. Comparison of sequenced genomes has suggested that nucleotide polymorphisms are the major source of phenotypic diversity [15,16]. However the relationships between such genetic variations and phenotypic diversity remain unclear, particularly in the context of alcoholic fermentation.
Quantitative trait locus (QTL)-based approaches are widely used in yeast, to link allelic variations to phenotypic diversity [17-20]. This approach has been applied to wine strains to identify the origin of fermentation traits. Marullo et al. [21] used this approach to show that mutations of the asparaginase gene ASP1 underlie differences in acetic acid production between two wine strains. QTL-based approaches have proved efficient for identifying the traits associated with a single mutation, but most fermentation traits are under complex polygenic control which is much more difficult to resolve. Using new approaches based on the analysis of very large populations of segregants; Extreme QTL mapping (X-QTL); Ehrenreich et al.[22,23] demonstrated the feasibility of identifying the genetic determinants of complex phenotypes, such as drug resistance.
Trait variation in may also results from changes in gene expression of the underlying regulatory networks. Brem et al.[24] have demonstrated that regulatory variations could be linked to genomic alterations by QTL approaches. Searches for expression QTLs (eQTLs) are now widely used in yeast to decipher regulatory networks variations [25-29], reviewed in [30,31].
Such approaches can be used to elucidate the origin of regulatory network variations and their impact on fermentation traits, such as the fermentation duration, the nitrogen consumption and the metabolite production. We previously generated a segregating population by crossing the lab strain S288c and a spore derivative of the wine yeast EC1118, for eQTL linkage analysis [4]. One of the main results of this previous study was the detection of a large impact of the ABZ1 (encoding para-amino benzoate (PABA) synthase) allele on nitrogen assimilation, resulting in large variations in the fermentation rate [4]. Furthermore, a recent QTL study using this lineage implicated ABZ1 allele variation in aromatic compounds production [32] probably due to the involvement of this gene in amino-acid metabolisms.
We report here of the results of a new search for fermentation traits QTL and eQTL with an enlarged segregant population in the late stages of alcoholic fermentation, more relevant conditions for addressing the stress response. The use of this approach provided us with a broad view of expression variation in alcoholic fermentation and allowed us to identify the genetic origin of variation for several regulatory networks involved in key processes, such as detoxification and sulfate assimilation. An unexpected result of this study was the finding that fermentation rate was controlled by a partial disomy of chromosome 16, revealing a new role in wine yeasts for a well known translocation.
Results
Phenotypics and transcriptomic analysis of the 59A×S288c lineage
We phenotyped 44 segregants obtained from a cross between the laboratory strain S288c and the wine yeast derivative 59A (as described in Methods). Fermentations were performed in a synthetic medium simulating a grape must (SM425) and containing para-amino benzoate (PABA) to counteract the effect of the ABZ1 allele [4]. Our analysis were performed in more stringent conditions than that of Ambroset et al.[4]: the amounts of ergosterol and oleic acid used were half those generally used (the final concentrations were 7.5 mg/l for ergosterol and 2.5 μl/l for oleic acid) and fermentations were performed at 24°C. We estimated the kinetic properties regarding the lag time and the fermentation rate at 2 stages of the fermentation: Rmax (maximal fermentation rate) and R70 (fermentation rate at 70% of fermentation). The fermentation kinetics of parental strains and of several segregants are shown in Figure 1. Population size and metabolite quantities were determined at the end of the fermentation.
Figure 1.
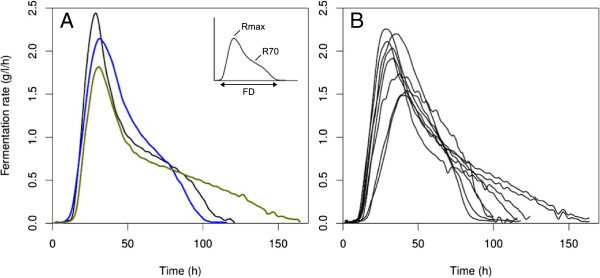
Fermentation profiles for parental strains and for segregants. (A) Fermentation profile for the two parental strains and for 59A×S288c: 59A in blue, S288c in green, 59A×S288c in black. (B) Fermentation profile for 8 segregants. Sugar was completely consumed for all segregants.
We first checked that phenotypes affected by ABZ1 allele, such as Rmax, were corrected by the addition of PABA and independent of the allelic form (data not shown). Most of the phenotypes had a high heritability (80% to 97%), indicating that genetic variations had a major impact on overall variations. The dry weight was the only phenotype displaying a low heritability (>50%) and was disregarded. R70 values followed a bimodal distribution, suggesting that the phenotype is mainly controlled by one locus. The other phenotypes, such as Rmax and the amounts of metabolites followed continuous distributions, indicating a probable polygenic control (Additional file 1).
Transcriptome profiles were obtained at 70% of fermentation progress (66 g/l CO2 released), corresponding to late stationary phase, 20 to 40 hours after the end of the growth phase (depending on the segregants). At this point in the fermentation, the yeast is subject to nutrient starvation and ethanol stress (8% v/v). We compared RNA abundance between segregants by Agilent mono-color labeling and hybridization on oligonuclotides microarrays as described in Methods. We identified 1610 genes differentially expressed between 59A and S288c with a log2 of fold-change (LogFC) of more than 0.7 in either direction (adjPv<0.01).
We assessed the correlation between the fermentation rate (R70) and gene expression and found large sets of genes positively correlated (347 genes involved in cell trafficking and nitrogen reutilization; Funspec [33]) and negatively correlated (351 genes involved in oxidative stress and respiration; Funspec).
Clustering analysis revealed a partial disomy of chromosome 16 affecting genes expression
A clustering analysis of the whole gene expression dataset revealed significant associations (Pearson correlation coefficient ≥ 0,84) for 887 genes (Additional file 2). Most of the gene clusters obtained were enriched in functional classes, such as mating, mitochondrial translation, ribosomal proteins, ergosterol synthesis and sulfur assimilation consistent with the known coordination of expression of these genes. Some clusters also corresponded to genes correlated with phenotypes (R70, Rmax). Other clusters included physically associated genes (ASP cluster, regions A B and C [13], telomeric genes).
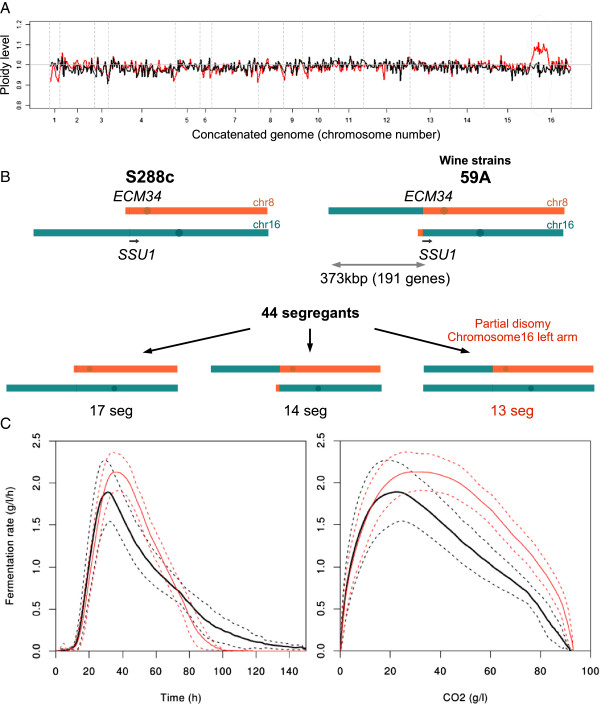
We identified a cluster of 37 genes displaying a higher expression in 13 segregants. These genes were physically linked and all were located in the first 373 kbp at the start of chromosome 16 (Additional file 2, cluster “chr16 left-arm genes”). A careful mining of the CGH data (from the Affymetrix chip signals) revealed that these 13 segregants carried a 373 kbp duplicated region on the left arm of chromosome 16 (Figure 2A). No other chromosomal aberration was observed. This partial disomy resulted from the translocation of an arm of chromosome 16 onto the chromosome 8 originating from the wine yeast strain, in addition to the presence of a standard chromosome 16 (Figure 2B). This 8–16 translocation is known to be responsible for higher level of sulfite resistance and was found in many wine yeasts [10]. We examined the impact of this partial disomy on transcriptome profiles by comparing the expression data for the 13 disomic segregants with those for the other 31 segregants. As expected, the genes of the duplicated region were more strongly expressed in segregants carrying the disomic region (155 of the 191 genes in the area concerned, adjPv<0.05), consistent with the doubling of gene copy number. We also observed differences in expression for 1305 other genes (adjPv<0.01) not located in this area. The upregulated genes include a high proportion of genes involved in methionine biosynthesis (17 genes, Pv=10-9 -FunSpec [33]) whereas the down-regulated genes included a high proportion of genes involve in the oxidative stress response (20 genes, Pv=10-6 -FunSpec) and aerobic respiration (37 genes, Pv=10-14 -FunSpec). The pattern of functional enrichment of the deregulated genes suggests that disomic strains are less affected by stresses of alcoholic fermentation conditions. Consistent with this hypothesis, we observed that the partial disomy had a major impact on fermentation kinetics (Figure 2C). Partial disomy was associated with significantly higher fermentation rates, a shorter duration of fermentation (t-test Pv<10-6) and an absence of sluggish fermentation profiles. Levels of acetic acid production were also lower for the disomic segregants (data not shown, t-test Pv<0.01), supporting our hypothesis of lower levels of stress in these clones.
Figure 2.
The 8–16 translocation and impact on fermentation. (A) CGH profile for the segregant 7a (in red) which has the partial disomy, in comparison to S288C CGH profile (in black). (B) Parental and daughter genotype: Crossing 59A which has the translocation with S288c gives four types of segregants: without translocation (S288c like), with the translocation (59A like), with two chr16 left arm (partial disomy) or without chr16 left arm but this combination is not viable, due to the loss of essentials genes (e.g. NSL1, NAB3, RPL5). (C) Average fermentation profile for the normal segregants (black) and disomic segregants (red). The doted lines represent standard error. Kinetics are represented in function of time (left) or of CO2 release (right).
Phenotypic and expression QTL linkages
We generated a new high-density genetic map (1.81 markers/10 kbp Additional file 3) from the Affymetrix S98 microarrays genotyping data for the 44 segregants. Linkage analysis was performed by the interval mapping method, for QTLs and eQTLs.
Three phenotypic QTLs were identified with a false discovery rate (FDR) threshold of 0.05: one QTL explaining flocculation on chromosome 1 encompassing FLO1, another explaining clumpiness and encompassing AMN1 and a third, explaining fermentation rate (R70), on the left arm of the chromosome 16 (140 kbp), overlapping with the duplicated region and clearly triggered by the partial disomy. Limiting the population to the 31 segregants with no partial duplication of chromosome 16 eliminated the detection of the R70 QTL on chromosome 16. It also led to the detection of three QTLs for metabolites levels, mapping to chromosome 2 (208 kbp) for pyruvic acid and chromosome 14 (460 kbp) for succinic acid and glycerol.
Genome wide linkage analysis of gene expression led to the detection of 1063 eQTLs as summarize in Table 1. Dividing the FDR by two only slightly reduce the number of eQTLs, demonstrating the robustness of the data. A high amount (56%) of local-eQTLs (or cis-eQTLs, see Methods) were detected with higher average LOD than trans-eQTLs (Table 1). Some of the genes differentially expressed between the parents displaying cis-eQTLs corresponded to genes absent from one of the parental strains. As expected, genes from the three new regions acquired by horizontal transfer in EC1118 [13] and conserved in 59A formed three clusters (Additional file 2), each controlled in cis-regulations. We did not consider these genes, nor the 111 genes missing from EC1118 [13] for subsequent analyses, reducing the number of eQTLs to 967.
Table 1.
Summary of eQTLs detected
| LODscore threshold | 4 | 4.4 | 4 (without absent genes) |
|---|---|---|---|
| trans-eQTL |
468 (5.31*) |
315 |
443 |
| cis-eQTL |
595 (8.78*) |
508 |
524 |
| Total |
1063 |
823 |
967 |
| FDR | 0.10 | 0.05 | 0.10 |
* Average LODscore.
We found that 113 of the 1460 genes affected by the partial disomy displayed eQTLs, and 48 of these eQTLs mapped to the deleted or duplicated regions on chromosomes 8 and 16. Given that the partial disomy could affect the eQTL data, we adapted the analysis (analysis 1) to take this effect into account. Two new eQTL linkage analyses were performed as described in Figure 3A: (a) a search with a population limited to 31 non disomic segregants (analysis 2) (b) a filtering of the data for the 13 disomic segregants, eliminating the markers in the duplicated region and ignoring the 1460 genes affected by the partial disomy (analysis 3).
Figure 3.
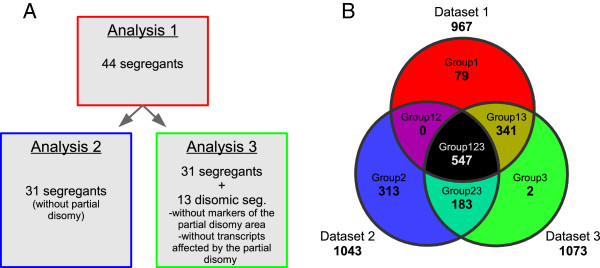
Analyses to by-pass the effect of partial disomy on transcriptome. (A) Principle of the analyses: Analysis 2 was the safest way to eliminate all the partial disomy effect and would have been applicable with a larger population. However, using a 31 segregants population reduced significantly our ability to detect small QTLs. Analysis 3 bypassed the majority of disomy effects while keeping a good detectability for the other traits. However, if the disomy has a partial effect on transcripts expression, this analysis kept this effect as a noise for eQTL detection. (B) Venn diagram for the comparison of the 3 analyses. The color used here will be used in Figure 4.
These two new data sets were compared with the first one on a Venn diagram Figure 3B and in Additional file 4. Most of the eQTLs (547) were common to all the three data sets (group 123 on Figure 3B) and corresponded to eQTLs with high LOD-score (cis-eQTLs). In analysis 2, restricted to the 31 non disomic segregants, the detection power was lower: at lod4, the FDR was 0.13. Data set 3 overlapped strongly with data set 1, with 888 of 967 eQTLs common to these two data sets. This suggests that the partial disomy slightly decreased our ability to detect true eQTLs, but had only a limited effect on the robustness of the data. The eQTL analysis was performed for the union of the three data sets (1465 eQTLs), taking the “Venn” group for each eQTL into account. We, therefore, concluded that we could overcome the effect of the partial disomy.
Variations in expression networks are triggered by few loci resulting in the formation of hotspots
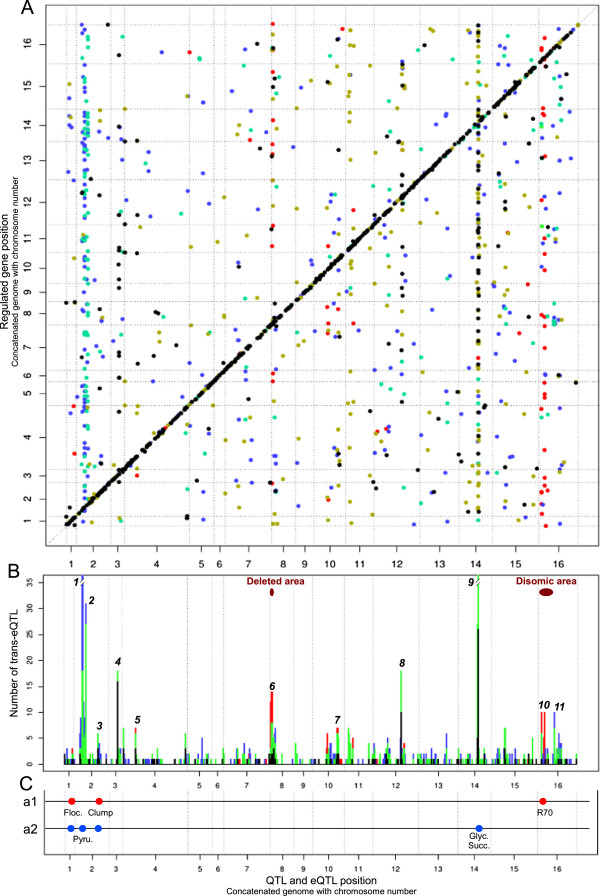
The locations of the phenotypic QTLs and eQTLs are summarized in Figure 4 and Table 2. Many eQTLs overlapped in several hotspots and most were common to the three analyses. However, the disomy was responsible for two hotspots on chromosomes 8 and 16 (hotspot 6 and hotspot 10, in red Figure 4B). Furthermore, 113 of the eQTLs discovered in the two new analyses are overlapping in two hotspots on chromosome 2, at 208 kbp and 283 kbp (hotspots 1 and 2 in blue/green Figure 4A-4B). Due to the genetic similarities between EC1118 and RM11-1a, some phenotypic QTLs and eQTLs are identical to those previously identified for the cross between BY4716 and RM11-1a (BxR population: hotspot 3, 4, 8 and 9, [25,34,35]).
Figure 4.
QTL and eQTL localization. (A) Localization of the 1465 eQTLs in function of the corresponding transcript position. Colors correspond to the group in the Venn diagram Figure 3B. Spot in the first diagonal correspond to cis-eQTLs. (B) Trans-eQTL density and formation of hotspot. Descriptions of each hotspot are in Table 1. Black peaks correspond to the trans eQTL in common of the 3 analyses, Green peaks correspond to trans-eQTL detected at least in analysis 3. Blue peaks correspond trans-eQTL specific of analysis 2. Red peaks correspond trans-eQTL specific of analysis 1. (C) Phenotypic QTLs detected with 44 segregants (a1, red) and with 31 segregants (a2, blue).
Table 2.
Hotspot description
| Hotspot | Localization a | Number of eQTL b | Functional enrichment | Phenotypic QTL | Knowon or candidate gene regulator | Functional validation | Comments |
|---|---|---|---|---|---|---|---|
| 1 |
chr2:207 |
66 |
No functional enrichment |
Pyruvate |
SCT1 |
Not realized |
Hidden by the disomy, Sct1p is involved in lipid desaturation and diverges in the two strains by the size of a glutamic acid repeated sequence |
| 2 |
chr2:277 |
47 |
No functional enrichment |
- |
Unknow |
- |
Hidden by the disomy |
| 3 |
chr2:560 |
7 |
Daughter cell separation |
Clumpness, OD, cell pop. |
AMN1 |
Yvert et al. 2003 [34] |
Impacting on cellular population estimation |
| 4 |
chr3:198 |
20 |
Mating, pheromone |
- |
MAT |
Brem et al. 2005 [35] |
|
| 5 |
chr4:309 |
9 |
Thiamine synthesis |
- |
THI3 |
Not realized |
THI3 bears two non-synonymous SNPs and a frame-shift in its coding sequence |
| 6 |
chr8:20 |
27 |
Helicase |
- |
- |
- |
Disomy consequence |
| 7 |
chr10:590 |
18 |
Mitochondrial protein |
- |
GRR1 |
Not realized |
GRR1 has a strong cis-eQTL (LOD = 13.4) and several alteration of its coding sequence |
| 8 |
chr12:667 |
33 |
Fatty acid metabolism, detoxification |
- |
HAP1/PDR8 |
Yvert et al. 2003 [34] |
Genes involved in the response to oxygen (nine targets of HAP1, Additional file 5) |
| 9 |
chr14:456 |
101 |
Mitochondrial protein |
Succinate, Glycerol |
MKT1 |
Smith and Kruglyak 2008 [25] |
Perturbation of the mitochondrial function affect the production of metabolites |
| 10 |
chr16 left-arm |
50 |
No functional enrichment |
R70 |
- |
- |
Disomy consequence |
| 11 | chr16:370 | 17 | chr8 translocated gene | - | SSU1 translocation | Not realized | Translocation consequence |
aLocalization of the center of hotspot given in chromosome:kilo base pair.
bNumber of total eQTL from the union of the 3 datasets.
However, we also found some linkages not previously identified. Hotspot 5 on chromosome 4 was found to be enriched in genes involved in thiamine biosynthesis pathway and is probably controlled by variations of the THI3 regulator gene located in the hotspot area. Hotspot 7 on chromosome 10 brings together 18 genes with various functions (mitochondria, glucose sensing: SNF3, YDL199c, GRR1) and GRR1 seams to be the most relevant candidate gene for the control of this hotspot. It is indeed involved in glucose repression and amino acid sensing [36].
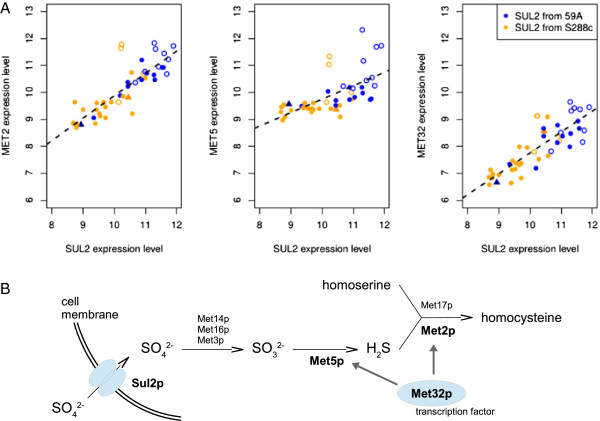
We also found variations in several networks involving fewer genes but potentially involved in the wine-making process. The high-affinity sulfate permease gene, SUL2, has a cis-eQTL (LOD 4.5) overlapping with trans-eQTLs for three other genes of the sulfur pathway MET2, MET5 and MET32. SUL2 has two SNPs in its promoter (but none in the consensus sequences for transcription factors binding sites, Yeastract [37]) and five non-synonymous SNPs in its coding sequence. Changes in sulfate transport efficiency are clearly responsible for the higher expression level of the three other genes involved in sulfur amino-acid metabolism (Figure 5).
Figure 5.
Control of genes involved in sulfuric amino-acid synthesis by SUL2 expression. (A) Correlation between MET2, MET5 or MET32 expression and SUL2 expression. Orange spots correspond to the segregants with SUL2 from S288c. Blue spots correspond to the segregants with SUL2 from 59A. Open circles correspond to disomic segregants. Doted line corresponds to the linear regression. (B) Sulfate assimilation via SUL2 and the four genes MET2, MET5, MET32 and MET3. Genes in bold character display a linkage on SUL2.
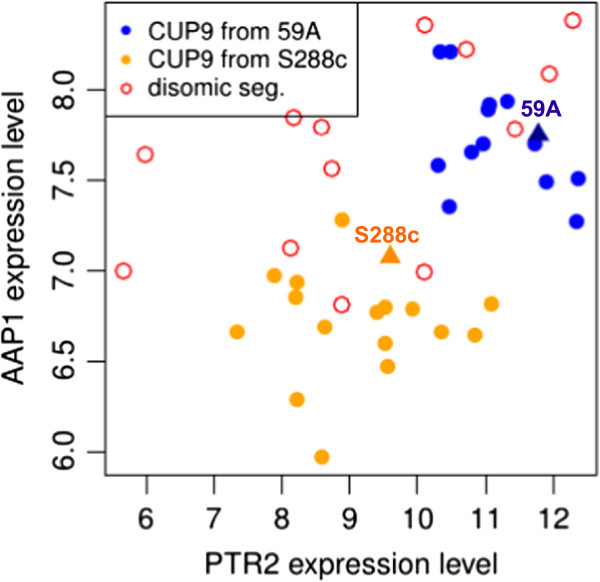
Similarly, we observed a perturbation of the nitrogen uptake system that might have a significant impact on wine yeast performance [38]. The membrane peptide transporter gene, PTR2, and the arginine/alanine aminopeptidase gene, AAP1, both have eQTLs mapping to CUP9 (chromosome 16) which encodes the known transcriptional repressor of PTR2. CUP9 has two non-synonymous SNPs in its coding sequence and the 59A allele clearly increases the expression of PTR2 and, probably, also that of AAP1 (Figure 6). Byrd et al.[39] described the activation of PTR2 by ubiquitin-dependent Cup9p degradation in the presence of peptide [40]. However, in the 59A strain, PTR2 has a frame-shift in its sequence and is, therefore, probably non-functional [41]. The involvement of CUP9 in the control of AAP1 is consistent with a broader role of this regulator in peptide metabolism. The synthetic must used here did not contain peptides, but these molecules may have been generated by endogenous nitrogen metabolism. The mutation in 59A may act by modulating the repressive activity or degradation of Cup9p.
Figure 6.
Correlation between PTR2 and AAP1 expression.CUP9 allelic origin is likely to control both genes. Red open circles correspond to disomic segregants, CUP9 being localized in the left arm of chromosome 16. Orange spots correspond the segregants with CUP9 from S288c. Blue spots correspond the segregants with CUP9 from 59A.
Finally, a flocculation QTL was found on chromosome 1, mapping to FLO1 as previously described [24] (Figure 4C). Flocculation phenotype was found linked to FLO1 expression level (Additional file 6). Surprisingly, multiple QTL searches on the basis of flocculation phenotype did yield a locus containing FLO8. However, a second QTL was found on chromosome 1, in a 45 kbp region (Additional file 6). This region contains the gene OAF1 involved in fatty-acid and peroxisome biogenesis. It is possible that OAF1 mutations triggered variation of cell surface hydrophobicity that could impact flocculation [42].
Variations in detoxification mechanisms are triggered by mutations in transcription factor genes
In this linkage analysis, we detected changes in the detoxification network. Several membrane transporters involved in drug export displayed eQTLs (Table 3). Some of these genes were under local regulation (cis-eQTL): SVS1, required for resistance to vanadate, the two polyamine exporter, TPO1 and TPO2, and the ion transporters ALR2 and SSU1. The cis-eQTL of SSU1 is due to the 8–16 translocation. Other detoxification system genes (e.g. SNG1, PDR12, QDR2) displayed distant control. Several eQTLs associated variations in the expression of these transporters with variations of zinc-finger transcription factor genes: HAP1, PDR8, YRR1 and WAR1.
Table 3.
Genes involved in detoxification having an eQTL
| Name | eQTL | |||||
|---|---|---|---|---|---|---|
|
Standard |
Systematic |
Localization
a
|
Lod score |
Regulation
b
|
Venn group |
SGD Function |
|
Ions Detoxification |
|
|
|
|
|
|
|
MNR2 |
YKL064W |
chr2:151 |
4,49 |
unknown |
23 |
Putative magnesium transporter; has similarity to Alr1p and Alr2p which mediate influx of Mg2+ and other divalent cations |
|
ALR2 |
YFL050C |
chr6:30 |
5,88 |
Self |
123 |
Probable Mg(2+) transporter; overexpression confers increased tolerance to Al(3+) and Ga(3+) ions; plays a role in regulating Ty1 transposition |
|
SSU1 |
YPL092W |
chr16:371 |
5,92 |
Self |
123 |
Plasma membrane sulfite pump involved in sulfite metabolism and required for efficient sulfite efflux; major facilitator superfamily protein |
|
Drug Transport |
|
|
|
|
|
|
|
TPO2 |
YGR138C |
chr7:761 |
17,28 |
Self |
123 |
Polyamine transport protein specific for spermine; localizes to the plasma membrane; transcription of TPO2 is regulated by Haa1p; member of the major facilitator superfamily |
|
SNG1 |
YGR197C |
chr15:639 |
5,24 |
YRR1 |
1 |
Protein involved in nitrosoguanidine (MNNG) resistance; expression is regulated by transcription factors involved in multidrug resistance |
|
ERC1 |
YHR032W |
chr2:624 |
4,09 |
unknown |
13 |
Member of the multi-drug and toxin extrusion (MATE) family of the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily |
|
PDR11 |
YIL013C |
chr12:659 |
4,78 |
HAP1/PDR8 |
123 |
ATP-binding cassette (ABC) transporter multidrug transporter involved in MDR; mediates sterol uptake when sterol biosynthesis is compromisedregulated by Pdr1p |
|
QDR2 |
YIL121W |
chr12:680 |
10,31 |
HAP1/PDR8 |
123 |
Multidrug transporter of the major facilitator superfamily required for resistance to quinidine barban cisplatin and bleomycin; may have a role in potassium uptake |
|
GEX2 |
YKR106W |
chr11:658 |
13,66 |
Self |
123 |
Proton:glutathione antiporter localized to the vacuolar and plasma membranes; almost identical to paralog Gex1p; potential role in resistance to oxidative stress |
|
TPO1 |
YLL028W |
chr12:86 |
9,89 |
Self |
123 |
Polyamine transporter that recognizes spermine putrescine and spermidine; catalyzes uptake of polyamines at alkaline pH and excretion at acidic pH |
|
TPO4 |
YOR273C |
chr12:659 |
4,61 |
HAP1/PDR8 |
13 |
Polyamine transport protein recognizes spermine putrescine and spermidine; localizes to the plasma membrane; member of the major facilitator superfamily |
|
PDR12 |
YPL058C |
chr13:116 |
7,50 |
WAR1 |
123 |
Plasma membrane ATP-binding cassette (ABC) transporter weak-acid-inducible multidrug transporter required for weak organic acid resistance; regulated by War1p |
|
Transcription factor |
|
|
|
|
|
|
|
HAP1 |
YLR256W |
chr12:673 |
6,22 |
Self |
123 |
Zinc finger transcription factor involved in the complex regulation of gene expression in response to levels of heme and oxygen |
|
PDR8 |
YLR266C |
chr16:138 |
5,00 |
unknown |
2 |
Transcription factor; targets include ATP-binding cassette (ABC) transporters, major facilitator superfamily transporters, and other genes involved in the PDR phenomenon |
|
WAR1 |
YML076C |
chr12:673 |
6,57 |
HAP1/PDR8 |
123 |
Homodimeric Zn2Cys6 zinc finger transcription factor; binds to a weak acid response element to induce transcription of PDR12 and FUN34 |
|
YRR1 |
YOR162C |
chr15:639 |
5,03 |
Self |
13 |
Zn2-Cys6 zinc-finger transcription factor that activates genes involved in multidrug resistance; paralog of Yrm1p acting on an overlapping set of target genes |
|
FZF1 |
YGL254W |
chr7:21 |
4,02 |
Self |
123 |
Transcription factor involved in sulfite metabolism, identified regulatory target is SSU1, overexpression suppresses sulfite-sensitivity |
|
Other/Unknown |
|
|
|
|
|
|
|
SVS1 |
YPL163C |
chr16:251 |
9,55 |
Self |
123 |
Cell wall and vacuolar protein required for wild-type resistance to vanadate |
|
YKR104W |
YKR104W |
chr11:651 |
4,63 |
Self |
13 |
Putative transporter of the multidrug resistance-associated protein (MRP) subfamily; contains a stop codon in S288C |
|
YLR046C |
YLR046C |
chr12:238 |
5,72 |
Self |
123 |
Putative membrane protein transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in multidrug resistance |
|
YJL216C |
YJL216C |
chr10:42 |
11,36 |
Self |
123 |
Protein of unknown function similar to alpha-D-glucosidases; transcriptionally activated by both Pdr8p and Yrm1p along with other genes involved in the PDR phenomenon |
| YLR179C | YLR179C | chr12:689 | 4,08 | HAP1/PDR8 | 23 | Protein of unknown function transcription is activated by paralogous proteins Yrm1p and Yrr1p along with proteins involved in multidrug resistance; not essential |
aLocalization of the center of hotspot given in chromosome:kilo base pair.
b“Self” mean the eQTL is closer than 40 kpb to the gene controlled.
The TPO1 gene, which displays a cis-eQTL, has three SNPs in its promoter area and three non synonymous SNPs in its coding sequence. The S288c allele of TPO1 is more strongly expressed than the wine yeast allele. As TPO1 is involved in octanoic acid resistance [3], we investigated the effect of variations of TPO1 expression on octanoic acid resistance. We measured population growth in the synthetic must medium (pH3.3) supplemented with octanoic acid (0.2 mM). Octanoic acid resistance dependent principally on the TPO1 allele (t-test Pv<0.005, Figure 7) but was also weakly correlated with TPO1 expression level (data not shown). Our data therefore suggest that the form of TPO1 encoded by the S288c allele is more effective, conferring a higher octanoic acid resistance.
Figure 7.

TPO1 allele controls is own expression and octanoic acid resistance. (A) Box plot showing TPO1 allele effect over TPO1 expression level in the population. (B) Box plot comparing segregants resistance to octanoic acid by TPO1 allele (t-test Pv<0.005). Values for parental strains are indicated.
PDR11, TPO4 and QDR2 all had an eQTL in hotspot 8 mapping to HAP1 (Table 3). Another gene, PDR8, encoding a pleiotropic drug resistance transcription factor lies in the same locus, 25 kbp away from HAP1, and may control some of these genes (Additional file 5). Consistent with this hypothesis, Steyer et al.[32], recently showed that high nerolidol production were associated with the S288c allele of PDR8. They demonstrated that QDR2 regulation by PDR8 accounted the variation of nerolidol release into the medium. We assessed the impact of PDR8 allelic variation on gene expression by performing an allelic switch for PDR8 and constructed 59A strain expressing the PDR8 allele from S288c (59A PDR8-S288c). We compared the transcriptome of this strain with that of wild type 59A in fermentation conditions (the same conditions as for the global analysis). We detected variation of expression for very few genes (adjPv<0.01, Table 4). QDR2 was the gene most strongly upregulated by allelic replacement, with a LogFC of 2.24, consistent with the results of Steyer et al.[32]. Among the other targets of PDR8, we only found YLR179c controlled by the allelic form of PDR8. Both QDR2 and YLR179c had an eQTL mapping to PDR8. In addition to a slight down regulation of PDR8 itself, we identified three other genes (YLR149c, GIT1 and QDR1) as upregulated in the constructed strain and potentially playing a role in detoxification. These genes have not previously been described as PDR8-controlled. We did not detect the targets of PDR8 reported by Hikkel et al.[43]. This was probably due to the specific conditions of alcoholic stress and nutrient starvation used in our study. Furthermore, we performed an allelic switch with a smaller impact than the over-expression of PDR8 used in other studies. Finally, only QDR2 and YLR179c were controlled by PDR8 in hotspot 8, suggesting that expressions of the other genes are triggered by HAP1.
Table 4.
Gene expression modified by PDR8 allelic switch
| ORF | Name | eQTL | LogFC | AdjPv | SGD function |
|---|---|---|---|---|---|
|
YIL121W |
QDR2 |
hotspot8 |
2,234 |
<10e-05 |
Multidrug transporter of the major facilitator superfamily required for resistance to quinidine barban cisplatin and bleomycin |
|
YLR179C |
YLR179C |
hotspot8 |
0,897 |
0,0026 |
Protein of unknown function transcription is activated by paralogous proteins Yrm1p and Yrr1p along with proteins involved in multidrug resistance |
|
YLR149C |
YLR149C |
- |
1,349 |
<10e-05 |
Putative protein of unknown function; overexpression causes a cell cycle delay or arrest; null mutation results in a decrease in plasma membrane electron transport |
|
YCR098C |
GIT1 |
- |
0,548 |
0,005 |
Plasma membrane permease, mediates uptake of glycerophosphoinositol and glycerophosphocholine as sources of the nutrients inositol and phosphate |
|
YIL120W |
QDR1 |
- |
0,472 |
0,0046 |
Multidrug transporter of the major facilitator superfamily, required for resistance to quinidine, ketoconazole, fluconazole, and barban |
| YLR266C | PDR8 | chr16:138 | −0,493 | 0,0032 | Transcription factor; targets include ATP-binding cassette (ABC) transporters, major facilitator superfamily transporters, and other genes involved in the pleiotropic drug resistance (PDR) phenomenon |
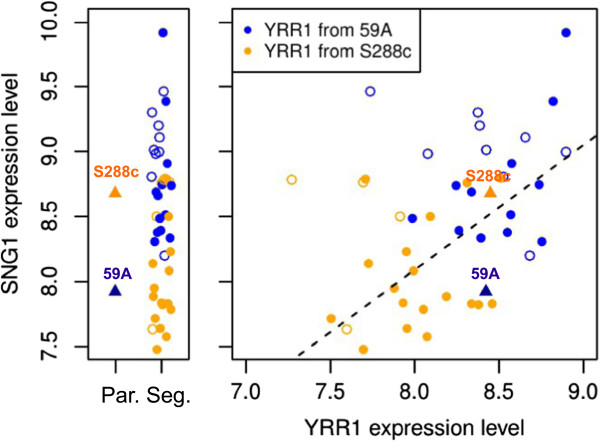
The gene paralog of PDR8, YRR1, displays a cis-eQTL on chromosome 15 and has four SNPs in its promoter region and six non-synonymous SNPs in its coding sequence. SNG1, one of the targets of YRR1 responsible for nitrosoguanidine resistance [44], has an eQTL mapping to the position of YRR1. Surprisingly, we found no linkages of this locus to other YRR1 target genes. Moreover, the inverse patterns of parental behavior and locus segregation for SNG1 expression indicates that other loci are involved in SNG1 control (Figure 8).
Figure 8.
YRR1 control over SNG1 expression. Orange spots correspond the segregants with YRR1 from S288c, blue spots correspond the segregants with YRR1 from 59A. Open circles correspond to disomic segregants, There is a permutation between parent and the allelic effect in the segregants population, involving polygenic control.
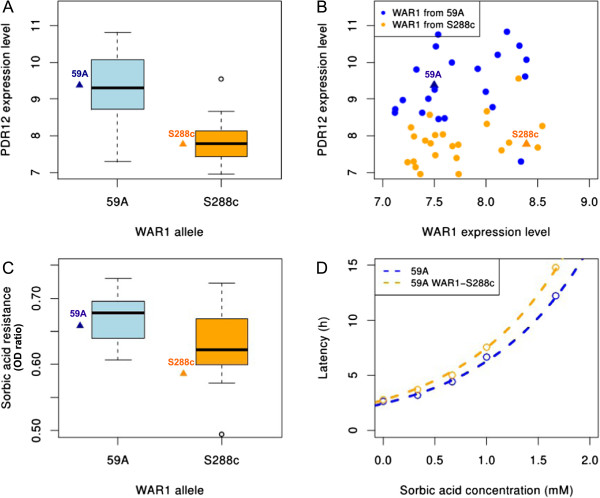
The PDR12 gene, encoding a plasma membrane ABC transporter responsible for organic acid efflux, had an eQTL mapping to its known transcription factor WAR1 (chromosome 13), an activator of weak acid stress response. The WAR1 allele of 59A has five non synonymous SNPs in its coding sequence and was associated with higher levels of PDR12 expression level (Figure 9A). In addition, WAR1 was itself one of the genes for which expression was affected by HAP1 variation (hotspot 8). The level of WAR1 expression had no significant impact on PDR12 transcript abundance. Instead PDR12 expression levels were influenced by the type of WAR1 allele (Figure 9B). PDR12 was the only known WAR1-dependent gene with an eQTL mapping at WAR1 (even with a LODscore below the threshold) [45]. As PDR12 has been implicated in sorbic acid resistance [46], we investigated whether variation of the expression of this gene modulated resistance to this acid. We assessed the growth of segregants in the synthetic medium (pH3.3) supplemented with sorbic acid (0.5 mM, see Methods). Sorbic acid resistance appeared to be partly controlled by the WAR1 allele (Figure 9C, t-test Pv<0.005) consistent with the effect of WAR1 allele on PDR12 expression. We then performed an allelic switch for WAR1 and constructed a 59A strain expressing the WAR1 allele from S288c (59A WAR1-S288c). The growth profiles of the strains were monitored in the synthetic must medium supplemented with various amount of sorbic acid. The lag phase was longer for 59A WAR1-S288c than for 59A wild-type (Figure 9D). This phenotypic difference was subtler than that observed with a knockout strain, but these results nevertheless confirm that sorbic acid resistance is linked to PDR12 expression and to WAR1 genetic variation. Variations of detoxification networks may thus be linked to variations of several transcription factor genes. Two of these linkages were validated by allelic switch experiments. Moreover, we demonstrated that the levels of resistance to octanoic acid and sorbic acid were clearly modulated by these genetic variations.
Figure 9.
Control of sorbic acid resistance by WAR1 allele. (A) Box plot showing WAR1 allele effect over PDR12 expression level in the population. Values for parental strains are indicated. (B) Confrontation of WAR1 expression level and PTR12 expression level show no correlation. Orange spots correspond the segregants with WAR1 from S288c. Blue spots correspond the segregants with WAR1 from 59A. (C) Box plot comparing segregants resistance to sorbic acid by they WAR1 allele (t-test Pv<0.005). Values for parental strains are indicated. (D) Lag phase duration in function of the sorbic acid concentration for 59A wildtype and for the allelic replacement of WAR1.
As polymorphism may result from specific adaptation, we tried to infer the phylogeny of War1p and Yrr1p from nucleotidic sequence available for different strains in SGD. Interestingly; the two allelic versions (wine and laboratory) of these two genes diverge from their S. paradoxus orthologue, and are clearly divergent but apparently in a different manner. For WAR1, S288C and 59A alleles are located on different branches, whereas for YRR1, the phylogeny indicates two clusters one containing palm wine and Asian alleles and the second cluster containing 59A wine, rum and bread alleles, as well as S288C alleles. However in this case S288C and 59A are also located on different branches of this cluster (Additional file 7, MEGA 5 [47]). In order to detect if these two genes have evolved neutrally, we performed a McDonald Kreitman test calculating the neutrality index (NI) [48]. This test was applied to a set of 15 WAR1 alleles from strains isolated from various substrates and NI was 2.28 (Pv = 0.02), indicating a significant excess of non-neutral mutations. As the cluster of Asian alleles might inflate this test, we removed the sequence and performed the test again, which indicate a similar result (NI = 2.56 and Pv = 0.03). We performed the similar analysis on YRR1 alleles. We obtained NI = 2.51 (Pv = 0.001) for the whole set of alleles and NI = 3.93 (Pv = 0.002) after removal of the Asian and palm wine alleles. This suggests that WAR1 and YRR1 do not evolve neutrally: these two genes are either subject to the accumulation of slightly deleterious mutations that are eliminated by negative selection during speciation, or alternatively they present substantial diversity that might be associated with balanced selection resulting from specific adaptation of the strain to wine fermentation conditions.
Discussion
In this study, we developed a genetical genomic approach to get insight into variation of gene regulatory networks during alcoholic fermentation and to assess the impact of this variation on wine yeast properties. We mapped 1465 eQTLs with 601 local eQTLs and 864 distant eQTLs (several distributed in 11 hotspots). As heritability exceeded 80% for 80% of the transcripts, genetic variations clearly had a major impact on global mRNA levels. The differences between the transcripts differentially expressed between the two parental strains and those displaying eQTLs confirmed that multigenetic controls are underlying most variations in transcripts abundance. These observations are consistent with the results obtained for other genetical genomic approaches in various organisms [30]. We observed several overlaps between phenotypic QTLs (glycerol, pyruvate) and eQTLs, consistent with the involvement of regulatory networks variation in yeast trait variations. Some of these linkages have been described before (clumpiness, floculation, mating type), whereas others are probably specific to our conditions or strains. We show that various mechanisms contribute to variations of expression and highlight the role of transcription factors for toxic compound export and the contribution of large chromosomal variations, a partial chromosomal duplication in this case, to the control of fermentation rate.
The transcriptome analysis was performed during the stationary phase in starvation and alcohol stress. Therefore our approach was adapted to describe regulatory change associated to this condition. However, variations in regulatory network involved in nitrogen consumption and cell growth were not considered here. For example, expressions of only few genes were found correlated to Rmax.
Genetic control of fermentation rate
Ambroset et al.[4] showed that fermentation rate (Rmax) depended on the PABA biosynthesis capacity and ABZ1 allele. By bypassing the effect of ABZ1, we identified another genetic control of the fermentation rate: the improvement of fermentation kinetics by partial disomy of the left arm of chromosome 16. In the system studied here, this partial disomy was present only in the offspring, due to cross carried out. This link cannot therefore be considered a “real” QTL, but it nevertheless highlights the role of gene copy number variations in the control of a key phenotype [11].
The mechanisms underlying the improvement of fermentation rate associated with this disomy are unclear, however, and will not be easy to decipher, given the size of the chromosomal region and the large number of genes it contains (190 genes). However, candidate genes may be identified on the basis of their patterns of expression, because the fermentation phenotype is thought to be triggered by a change in expression level. One or several of the 155 genes overexpressed in the translocated area may improve tolerance to starvation or, potentially, to alcohol. We investigated the properties of the set of genes in this region of chromosome 16 and evaluated their correlation with fermentation rate (R70). We observed that levels of expression of SAM3 and SAM4, involved in S-adenosyl methionine (SAdM) pool control, were correlated with R70. Interestingly, the expression levels of other genes, not located in the duplicated region, involved in SAdM synthesis (SAM1, SAM2) and involved in methyl transfers (CHO2, NNT1, SET7) were also correlated with R70. We suspect that SAM3 and SAM4 improve of the fermentation rate of the disomic segregants by increasing the availability of SAdM. However, the addition of SAdM (final concentration: 0.1 μM) to the fermentation medium did not improve the fermentation rate of the 59A strain (data not shown). We previously showed that the methyl donor synthesis and methionine biosynthesis pathways had a strong impact on fermentation rate [4], highlighting the key role of these metabolic pathways in alcoholic fermentation. Additional experiments are required to determine the mechanisms underlying this control of fermentation rate.
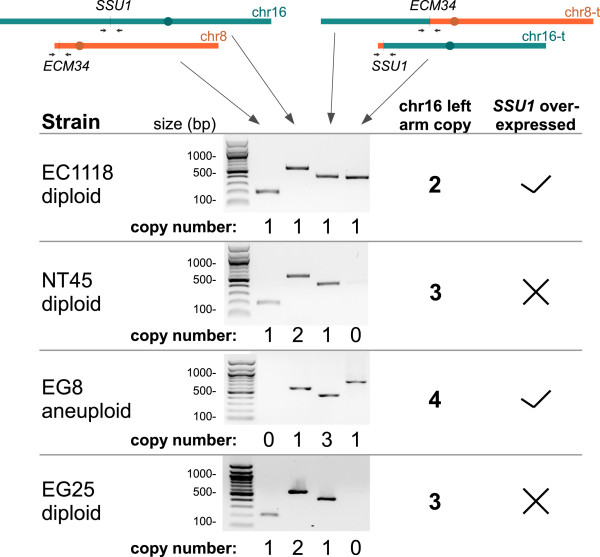
As this chromosomal amplification increased the fermentation capacity of wine yeasts, we checked for its presence in wild and industrial strain. Dunn et al.[12] reported a aCGH analysis of 83 S. cerevisiae strains including 69 wine strains. We used their data to search for a partial trisomy (for diploid strains) of the left arm of chromosome 16. We detected such partial trisomy in the wine yeast strain NT45. Moreover, in their characterization of the S. cerevisiae/S. kudriavzevii triploid hybrid EG8, Erny et al.[49] detected a major amplification (four copies) of the left arm of the S. cerevisiae chromosome 16. The diploid S. cerevisiae strain EG25, which is genetically related to the S. cerevisiae moiety of the EG8 genome (with no S. kudriavzevii DNA), also displays trisomy of this region (Legras JL, personal communication). We determined the form of the chromosomes 16 and 8 (normal or translocated) present, by PCR for these three strains (Figure 10). The NT45 and EG25 strains carry three chromosomes 16 left arms, originating from two normal chromosomes 16 and one copy of chromosome 8 carrying the translocation from chromosome 16. The hybrid aneuploid strain EG8 carries four left arms of chromosome 16: one from a normal chromosome 16 and three from the three chromosomes 8, all carrying the translocation. This findings confirm that amplification of the left arm of chromosome 16 occurs in wild/industrial strains. The situation described here suggests a spectacular evolutionary event occurring in two independent steps: a translocation selected due to its effect on sulfite resistance followed by amplification of the chromosomal region concerned, improving fermentation properties.
Figure 10.
Confirmation by PCR of chromosome left arm amplification. Confirmation over three strain: NT45, EG8 and EG25 selected by aCGH data. EG8 displays a chromosome 8 trisomy. SSU1 over-expression is carried by chromosome 16 translocated.
We evaluated the contribution of the disomy effect to eQTL linkage, by modifying the input dataset. We found that a large set of new eQTLs had been masked by the effect of this disomic. Most of these eQTLs (113) formed 2 hotspots on chromosome 2. Several of the genes controlled by these hotspots were found correlated with R70. Thus, these loci may partially control fermentation rate, despite the lack of detection of a QTL for this phenotype.
Importance of variations in detoxification systems
The analysis of functional enrichment for all eQTLs (in FunSpec [33]) highlighted a set of 22 eQTLs linking genetic variation to expression level of genes involved in detoxification. Our data show the importance of variations in transcription factor genes and their impact on detoxification systems: PDR8, WAR1 and YRR1, in addition to HAP1 (as summarized in Figure 11).
Figure 11.
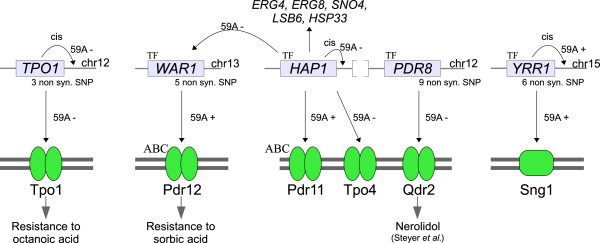
Summary of the variations in the detoxification regulatory network. Small arrows represent an allelic control on expressions. ABC transporter and transcription factor (TF) are indicated.
During fermentations, yeast produces toxic compounds that may lead to stuck fermentations [50]. This is the case for the medium-chain fatty acids generated as by-products of lipid biosynthesis. PDR12 has been shown to be involved in weak organic acid [46], and Legras et al.[3] have shown that PDR12, together with TPO1, is also a key transporter involved in octanoic acid and decanoic acid resistance. We were able to link genetic variations of TPO1 to its expression and to octanoic acid resistance. PDR12 has an eQTL in the gene encoding its known transcription factor, WAR1, that we could link to variations of sorbic acid resistance, but not to octanoic acid resistance, suggesting that the allelic effect of TPO1 outweighs the control of medium-chain fatty acid resistance.
For the same population, Steyer et al. [32] shown that the allele of PDR8 gene controls the export of nerolidol via QDR2. We confirm here that PDR8 allelic variation controls the expression of QDR2 and YLR179c, the function of which remain unknown. We observed that several drug exporters (TPO1, TPO2, TPO4 and QDR2) were less strongly expressed by the wine form, consistent with the deletions in wine yeasts reported by Dunn et al. (PDR3, SNQ1, QDR1[11]). The exact physiological role of several of these transporters remains unclear. Qdr2p was initially identified as a drug efflux system but its natural targets remain a matter of debate [51,52]. It was subsequently suggested that Qdr2p might export amino acids in starvation conditions [53]. Such a function would be consistent with the lower levels of expression of this gene in wine yeast, given the major effect of nitrogen homeostasis on wine yeasts fermentation capacity.
In addition to the toxic compounds produced by the yeast during fermentation, natural grape musts contain various phenolic compounds [54], which act as key wine preservatives and contribute to aging. Stilbene compounds are clearly toxic to fungi, such as S. cerevisiae and Aspergillus flavus[55]. Pan et al.[56] studied the transcriptomic response to pterostilbene (an analog of resveratrol) and show that YRR1 and SNG1 were among the upregulated genes in response to the drug. We identified eQTLs for these two genes, which were upregulated by the wine allele of YRR1 transcription factor encoding gene.
As the polymorphism observed here may result from specific adaptation to exposure to these compounds, we tried to infer to phylogeny of War1p and Yrr1p and evaluate if these genes evolve neutrally. Interestingly the McDonald Kreitman test [48] revealed a non neutral evolution for these two genes (NI calculated was greater than two): this indicates very likely that the substantial diversity observed for WAR1 and YRR1 results from balanced selection due to specific adaptation of the strain to wine fermentation conditions. This is consistent with the analysis of Steyer et al.[32], who reported also a non neutral evolution of PDR8 according to the results of a McDonald test.
Unlike Yvert et al.[34], who reported that distant QTLs were not often due to variations of transcription factor genes, we found that most of the variation of detoxification gene expression was linked to transcription factors. Surprisingly, variations in these three genes (PDR8, YRR1, WAR1) encoding zinc-finger transcription factors modified the expression of only one or two of their known targets. This may be partly due to the different environment in which PDR8 targets have been characterized. We performed our expression analysis in stationary phase under a stressful condition whereas cell growths in rich medium were used for the Chip on Chip assay of Hikkel et al.[43]. In addition, Jothi et al.[57] have shown that these transcription factors belong to the lower layer of the hierarchy of the yeast transcription factors network, with direct effects on a limited number of targets. This may make them more tolerant to variation than genes higher up in this hierarchy.
Regulatory network variations involved in wine fermentation
In this study, we investigated, for the first time, the expression profiles of the genes present in the three regions introgressed into EC1118 by horizontal transfer [13]. As we worked with a second parental strain devoid of these three regions, we could not identify real eQTLs for these genes. We searched for links, using the populations of segregant possessing each region (half of segregants). On chromosome 6, at 145kpb, an eQTL was identified that trigger the expression of the two genes encoding polypeptide transporters (FOT) describes by Damon et al.[41]. The new fructose transporter gene, FSY1, [58] also had an eQTL on chromosome 13, at 446 kbp (data not shown). No clear candidate gene was identified in these loci, but our findings could lead to revealing the role of these new genes in fermentation. No phenotypic QTL or trans-eQTL were found in these three regions. More detailed analysis, with a larger set of segregants, should be carried out to assess the variations of expression associated with these regions.
The phenotypic data indicated that most of the traits considered were under multigenic control. This may reflect the large genetic distance between our two parental strains, but Marullo et al.[59] aslo found, in a study using two commercial wine yeasts and a population of 51 segregants, a preponderance of multigenic traits, such as ethanol tolerance and kinetic properties. We were nevertheless able to associate several phenotypic QTLs with eQTLs. Pyruvic acid levels, which were strongly correlated with R70, were linked to a major hotspot on chromosome 2. A candidate gene, SCT1, was identified. This gene is involved in lipid desaturation, suggesting that it may act by modulating the plasma membrane stability under alcoholic stress.
However, within the eQTLs set, several new linkages are probably involved in the management of the multi-stress conditions in wine fermentation. Changes were detected for genes involved in glucose sensing (GRR1 cis-eQTL that may control SNF3 expression) or thiamine synthesis (affected by THI3). Functional validations of these results are required, together with further investigations to determine the role of such genetic variation in the adaptation of strains to the wine making process. We also showed that the sulfur amino-acid pathway (MET32, MET2, MET5) was partially enhanced by SUL2 mutation, probably through effects on sulfate uptake. Such activation has not been described before and may reflect the conditions of nitrogen starvation in this study.
Conclusion
In this study, we developed an approach to deciphering the variation of transcriptional networks in yeast, during an industrial wine-making process resulting in multiple stresses. The results obtained were consistent with those of other studies. However, the wine fermentation conditions used here led to the identification of specific variations of regulatory networks not revealed previously in classical laboratory environment. We show that expression variations affect networks of genes involved in various pathways and cellular functions, through diverse mechanisms involving both local and distal regulations and copy number changes driven by chromosomal rearrangements. We focus here on variations of detoxification systems and identified and we confirm the key role of variations in transcription factor genes in modulating the expression of these systems. These results also raise questions about the role of these export systems in fermentation conditions, in which cells are starved and stressed.
The eQTL linkage analysis was generally highly consistent, but most of the phenotype changes were not linked genetic variations and were considered to be under multi-locus control. We found that fermentation rate depended on a gene copy numbers effect of a duplication of the left arm of chromosome 16. This partial disomy was also found in natural yeasts and appears to be a potential new target for yeast optimization for the wine making process.
Methods
Strains, growth conditions and fermentation medium
The two parental strains used for the segregating population are S288c (MATα; SUC2, gal2) strain and 59A (MATa; ho), which is a haploid derivative of EC1118 (HO/ho), an industrial wine strain [4]. Mutations between the two parental strains are accessible in the GenYeasTrait database previously described (http://genome.jouy.inra.fr/genyeastrait/[4]).
We increased the population of 30 F1 segregants described in [4] with 14 randomly selected segregants. Thereby we disposed of two complete tetrads in the analyzed population, and the other segregants were selected from 24 different asci of two or three viable spores. The phenotyping was done on fermentation in 0,9l of synthetic SM425 medium (with 425 mg/l of assimilate nitrogen pH3.3 and previously describe as SM300 [60]) which mimics natural grape must. This medium was supplemented by 1 mg/l of p-aminobenzoic acid (PABA, sigma A-9878), 14.5 mg/l of iron (III) chloride (FeCl3, sigma F-7134), 10 μl/l of antifoam silicone (rhodorsil Prolabo 27429.297). The sugar source was glucose (10%) and fructose (10%) and the quantity of anaerobic factor was reduced by ½. After a reactivation of strain on YPED (48 h) and a preculture in shake flasks containing 30 ml of SM425 (12 h), 900 ml of medium were inoculated at 106 cell/ml. Fermenters were equipped with airlock to maintain anaerobiosis. Fermentation were performed at 24°C and preculture at 28°C.
Fermentation kinetics and phenotypic measures
Each parental fermentation and phenotypic measures were realized in four replicates and segregants fermentation in two replicates. The fermentation kinetics were measured by the loss of weight due to CO2 release. The weight was record every 20 min and the data were smoothed by polynomial method. The final population size was estimated at 45 g/l of CO2 release and 66 gCO2/l using electronic particle counter (Beckman Coulter) and optic density at 600 nm (OD600, spectrophotometer SECOMAN-UVLine9400). Dry weights were measured at 88 gCO2/l by filtration using nitrocellulose filters (pore size 0.45 μm; Millipore) and drying at 100°C (48 h).
The metabolic compounds amounts in the medium was measured after the end of the fermentation by high-pressure liquid chromatography (HPLC) using ion-exclusion column (HPX-87H BIO-RAD) with 8 mM H2SO4 mobile phase. Glucose, ethanol, glycerol and succinate were detected by refractometry. Acetic acid and pyruvic acid were detected by UV absorption (as described in [61]). Measures repeatability between replicate was controlled and the phenotype heritability H 2 was calculated as follow (describe in [35]):
where Var env is the pooled variance among parental measurement and Var seg is the variance among phenotype values for the segregants. Phenotypes with low H 2 are not statistically valid and were discarded.
Resistance phenotyping was performed in the same SM425 medium (pH3.3) than fermentation with a completion of the right amount of toxic compound stock solution and ethanol for a final rate of 1.7% (vol/vol). Octanoic acid stock solution: 69.3 mM in ethanol. Sorbic acid stock solution: 33.7 mM in water. The growth was taken with OD600 with and without toxic compound, and resistance indicator was measured by latency differences, ratio of specific growth rate and ratio of OD600 at 20 hours (end of exponential stage). We here only considerate the ratio of OD600, showing the lower experimental variation. Three replicates were done for each parental growth.
Molecular biology and strains construction
PDR8 allelic switch in 59A was obtained in three steps: 1) depletion of PDR8 in 59A using hphMX4 cassette for hygromycin resistance (pAG32). Primers sequences for cassette amplification and verification were obtained from Euroscarf. 2) preparation of a replacement cassette containing PDR8-loxP-kanMX4-loxP by the insertion of loxP-kanMX4-loxP (pUG6 [62]) in the terminator of PDR8 in S288c strain (primers in Additional file 8). 3) allelic switch by depletion of the hphMX4 cassette of 59A PDR8Δ::hph by the PDR8-loxP-kanMX4-loxP replacement cassette from S288c and selection on YEPD containing G418 (200 μg/l). The loss of hphMX4 cassette was controlled by PCR and the absence of growth on hygromycin (200 μg/l).
WAR1 allelic switch in 59A was obtained in two steps: 1) depletion of WAR1 in 59A using loxP-kanMX4-loxP (pUG6). Primers sequences for cassette amplification and verification were obtained form Euroscarf. 2) allelic switch by depletion of the kanMX cassette by WAR1 sequence form S288c DNA (primers in Additional file 8) and selection on YEPD containing 300mg/l sorbic acid, pH4.2. The loss of kanMX cassette was controlled by PCR and the absence of growth on YEPD supplemented by G418 (200 μg/l).
The 8–16 translocation was confirmed by PCR, using the same primers than Pérez-Ortín et al.[10].
Transcriptomic profiling
The transcriptome profiling was performed only one time for each segregants and in three technical replicates for each two biological replicates of the parental strains. At 66 gCO2/l release (70% of fermentation progress), 109 cells were sampled, pelleted, washed with DEPC-treated water and freezed in methanol at −80°C. This condition is corresponding to late stationary phase; cells are not in transition situation which could have increase experimental variation on transcriptome. Total RNA extractions were performed with Trizol reagent (Gibco BRL, Life Technologies), purified by isopropanol precipitation then with RNeasy kit (Qiagen). Cy3-labeled cRNA was synthesized with the One color RNA Spike-In kit (Agilent Technologies) and purified with RNeasy kit (Qiagen). Quality and quantity of RNA were controlled at each step by spectrometry (NanoDrop 1000, Thermo Scientific).
Agilent gene expression microarrays 8x15k was used for the micro array hybridization, with one-color method (Agilent Technologies, Santa Clara, CA, USA). Array design is based on ID 016322 completed with the 39 genes from the new region of EC1118 [13] and available on GEO with GPL16012 as accession number. A quantity of 600ng of labeled cRNA were hybridized for 17 h in 65°C in a rotative hybridization oven (Corning) using the Expression Hybridization kit (Agilent Technologies, 5188–5242). Plates were washed with expression wash buffer kit (Agilent Technologies, 5188–5325 5188–5326). The array pictures were analyzed on a GenePix 4000B laser Scanner (Axon Instruments) and with the GenePix PRO7 software.
Data normalization and statistical analyses were performed using R 2.13.1 software and the limma package [63-66]. Normalization was done by the quantile method considering the whole array data set (55 arrays). The normalized LOG2 of the spot-median intensity was used as the quantitative evaluation of gene expression (5.3 corresponding to background signal, 16 to spot saturation). Biological and technical repeatability were estimated higher than 87% and 96% respectively using parental strain replicate by the intra-class coefficient of correlation [67].
Comparative transcriptomic between parental strain and between disomic/normal segregants were performed with a modified t-test using the Benjamini and Hochberg false discovery rate as multiple testing correction of the t-test p-values (adjPv) [68]. The threshold used was the adjPv lower than 0.01, and a filter with a log2 of fold-change (logFC) greater than 0.7 or lower than −0.7 was applied for parental comparison.
Hierarchical clustering was performed using cluster v3.0 (Centered correlation and complete linkage [69]) and displayed with JavaTreeView v1.1.5r2 [70]. As previously described in Yvert et. al.[34], we defined a statistically significant degree of correlation between genes by permutation testing (n=10) and focused further analyses to clusters in which all pairwise correlations are greater than 0.84. At this threshold, fewer than three clusters of two genes and no cluster of more than two genes are expected by chance. Functional analysis of transcriptomic output was realized using Funspec with the Bonferroni correction at p-value cutoff of 0.05 [33]. Complete array data set is available on Gene Expression Omnibus database (global analysis: GSE41025, PDR8 allelic switch analysis: GSE41738).
Genotyping
The genotyping of the 14 new strains was performed with the same method than the first analysis [4]: Genomic DNA was isolated, fragmented, labeled, and hybridized on Affymetrix YGS98 microarrays, one time for each strain. A new marker map was designed doing the same selection than the precedent analysis described in Ambroset et al. 2011 [4] and inspired from Brem et al. 2002 [24]. This resulted in a map of 2140 markers.
To study genome duplication, comparative genomic hybridization (CGH-like) analysis was performed using Affymetrix normalized logarithm of PM signal of all the probes. A smoothing taking the average over 200 probes was done.
We completed the 2140 markers map with 46 markers localized in area of low markers density. These markers were based on single nucleotide polymorphism detection using Illumina veracode technology. This resulted in a good genotyping cover with an average density of 1.81 markers each 10 kbp.
Linkage analysis
The linkage analysis was performed using R/qtl package [71] on the three datasets corresponding to the three analyses (Analysis1, Analysis2 and Analysis3). The normal model with Haley-Knott regression method was used resulting in logarithm of odds (LOD) score for each marker and pseudo-marker every 2.5 cM (7.5 kbp) (interval mapping method). An interval estimate of the location of each QTL or eQTL was obtained as the 1-LOD support interval: the region in which the LOD score is within 1 unit of the peak LOD score. For phenotypic QTLs, individual LOD score threshold for a false discovery rate of 0.05 was determined doing 1000 permutations. For eQTLs, the permutation was performed 20 times, and the average number of transcripts showing linkage at a specific LOD score threshold was used to calculate FDR.
The eQTLs closer than 40 kbp to the localization of the gene were considerate as “local” eQTLs or cis-eQTLs. This could be due to a mutation in the gene promoter (real cis-eQTL), a mutation in the protein that control its own expression as a feedback, a mutation in an other gene close that control its expression (trans-eQTL co-localized). They also can be artifacts due to mutations in sequence that reduce the mRNA affinity with the micro-array probe.
Phylogeny and neutrality tests
To infer the evolutionary history of WAR1 and YRR1, we collected their sequences from genomes available at SGD (http://www.yeastgenome.org/). All uncompleted or frameshift-containing sequences where discarded from this set. The phylogenies were inferred with MEGA [47] by the Maximum Likelihood method based on the Kimura 2-parameter model [72]. The trees with the highest log likelihood are shown. The trees are drawn to scale, with branch lengths proportional to the number of substitutions per site.
The Neutrality Index correspond to the comparison of the ratio of non synonymous to synonymous polymorphism (intra species) to the ratio of non synonymous to synonymous divergence with the nearest species. An NI lower than one reflects a paucity of non synonymous polymorphism relative to non synonymous divergence, and is indicative of positive selection; an NI greater than one indicates negative selection of deleterious alleles driving divergence between species or balancing selection. The significance of the NI test [48] was calculated using the http://bioinf3.uab.cat/mkt/MKT.asp website.
Availability of supporting data
Sequence of 59A strain is available in http://genome.jouy.inra.fr/genyeastrait/. Transcriptomic row data are available in Gene Expression Omnibus database (global analysis: GSE41025, PDR8 allelic switch analysis: GSE41738). Other supporting data are included in the additional files.
Abbreviations
QTL: Quantitative trait locus; X-QTL: Extreme QTL; eQTL: Expression QTL; cis-eQTL: eQTL closer than 40 kbp to the localization of the gene (“local” eQTL); trans-eQTL: eQTL farer than 40 kbp to the localization of the gene (“distant” eQTL); aCGH: Comparative genomic hybridization in array; logFC: Log2 of Fold-Change; FDR: False discovery rate; LOD: Logarithm of odds; SAdM: S-adenosyl methionine; PABA: Para-amino benzoate; Rmax: Maximal fermentation rate; R70: Fermentation rate at 70% of fermentation progress; OD600: Optic density at 600 nm; Pv: P-value; adjPv: Adjusted P-value; kbp: Kilo base-pair.
Competing interests
The authors declare that they have no competing interests.
Author's contributions
CB participated in the design of the study, performed the data acquisition and analysis, and drafted the manuscript. CA participated in the design of the study, and realized the 59AxS288c lineage. IS performed the statistical analyses, the transcriptomic data processing and the QTL and eQTL linkages. JLL performed the phylogenic analysis and provided guidance over the data analysis and the manuscript preparation. BB conceived and coordinated the study, and provided guidance over the manuscript preparation. All authors read and approved the final manuscript.
Supplementary Material
Example of phenotypic value distribution among the population. Parental values are indicated in the top with open circle and black circle for 59A and S288c respectively. Heritablity (H2) is indicated. R70 exhibits a bimodal distribution while other phenotypes have a continuous distribution.
Clustering analysis.
Markers map. 2140 markers are from Affymetrix genotyping, 46 markers were based on single nucleotide polymorphism detection using Illumina veracode technology to cover area of low Affymetrix markers density. The global density is 1.81 markers each 10 kbp.
Comparison of partial disomy correction analyses.
Hotspot 8 genes triggered by HAP1 or/and PDR8.
Flocculation phenotype triggered by FLO1 expression.
Phylogenic trees of the protein sequence of the two transcription factor involved in drug detoxification network variation. Trees were drawn by MEGA5 software with the maximum likelihood method from the genome sequences available [SGD]. Parental strains are indicated by blue and yellow spot for 59A and S288c respectively.
Primer used.
Contributor Information
Christian Brion, Email: christian@brion.fr.
Chloé Ambroset, Email: chlo.ambroset@yahoo.fr.
Isabelle Sanchez, Email: sanchezi@supagro.inra.fr.
Jean-Luc Legras, Email: jean-luc.legras@supagro.inra.fr.
Bruno Blondin, Email: blondin@supagro.inra.fr.
Acknowledgment
C. Brion was supported by a grant form French Ministry for Higher Education and Research. C. Ambroset was supported by a fellowship from the INRA Département Microbiologie et Chaîne Alimentaire (MICA) and the region Languedoc-Roussillon. This work was partly supported by Agence Nationale de la Recherche grant Genyeastrait ANR-07-BLAN-0205.
References
- Camarasa C, Sanchez I, Brial P, Bigey F, Dequin S. Phenotypic landscape of Saccharomyces cerevisiae during wine fermentation: evidence for origin-dependent metabolic traits. PLoS ONE. 2011;6:e25147. doi: 10.1371/journal.pone.0025147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divol B, du Toit M, Duckitt E. Surviving in the presence of sulphur dioxide: strategies developed by wine yeasts. Appl Microbiol Biotechnol. 2012;95:601–613. doi: 10.1007/s00253-012-4186-x. [DOI] [PubMed] [Google Scholar]
- Legras JL, Erny C, Jeune CL, Lollier M, Adolphe Y, Demuyter C, Delobel P, Blondin B, Karst F. Activation of two different resistance mechanisms in Saccharomyces cerevisiae upon exposure to octanoic and decanoic acids. Appl Environ Microbiol. 2010;76:7526–7535. doi: 10.1128/AEM.01280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambroset C, Petit M, Brion C, Sanchez I, Delobel P, Guérin C, Chiapello H, Nicolas P, Bigey F, Dequin S, Blondin B. Deciphering the molecular basis of wine yeast fermentation traits using a combined genetic and genomic approach. G3: Genes, Genomes, Genetics. 2011;1:263–281. doi: 10.1534/g3.111.000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer J, Zörgö E, Cubillos FA, Zia A, Gjuvsland A, Simpson JT, Forsmark A, Durbin R, Omholt SW, Louis EJ, Liti G, Moses A, Blomberg A. Trait variation in yeast is defined by population history. PLoS Genet. 2011;7:e1002111. doi: 10.1371/journal.pgen.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marullo P, Mansour C, Dufour M, Albertin W, Sicard D, Bely M, Dubourdieu D. Genetic improvement of thermo-tolerance in wine Saccharomyces cerevisiae strains by a backcross approach. FEMS Yeast Res. 2009;9:1148–1160. doi: 10.1111/j.1567-1364.2009.00550.x. [DOI] [PubMed] [Google Scholar]
- Dequin S, Casaregola S. The genomes of fermentative Saccharomyces. C R Biol. 2011;334:687–693. doi: 10.1016/j.crvi.2011.05.019. [DOI] [PubMed] [Google Scholar]
- Sicard D, Legras J-L. Bread, beer and wine: yeast domestication in the Saccharomyces sensu stricto complex. C R Biol. 2011;334:229–236. doi: 10.1016/j.crvi.2010.12.016. [DOI] [PubMed] [Google Scholar]
- Bisson LF. Geographic origin and diversity of wine strains of saccharomyces. Am J Enol Vitic. 2012;63:165–176. doi: 10.5344/ajev.2012.11083. [DOI] [Google Scholar]
- Pérez-Ortín JE, Querol A, Puig S, Barrio E. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 2002;12:1533–1539. doi: 10.1101/gr.436602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn B, Levine RP, Sherlock G. Microarray karyotyping of commercial wine yeast strains reveals shared, as well as unique, genomic signatures. BMC Genomics. 2005;6:53. doi: 10.1186/1471-2164-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn B, Richter C, Kvitek DJ, Pugh T, Sherlock G. Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 2012;22:908–924. doi: 10.1101/gr.130310.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo M, Bigey F, Beyne E, Galeote V, Gavory F, Mallet S, Cambon B, Legras J-L, Wincker P, Casaregola S, Dequin S. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc Natl Acad Sci USA. 2009;106:16333–16338. doi: 10.1073/pnas.0904673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeote V, Bigey F, Beyne E, Novo M, Legras J-L, Casaregola S, Dequin S. Amplification of a Zygosaccharomyces bailii DNA segment in wine yeast genomes by extrachromosomal circular DNA formation. PLoS One. 2011;6:e17872. doi: 10.1371/journal.pone.0017872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O’Kelly MJT, Van A O, Barton DBH, Bailes E, Nguyen AN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of S. cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH, Davis RW. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 2002;416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- Hu XH, Wang MH, Tan T, Li JR, Yang H, Leach L, Zhang RM, Luo ZW. Genetic dissection of ethanol tolerance in the budding yeast Saccharomyces cerevisiae. Genetics. 2007;175:1479–1487. doi: 10.1534/genetics.106.065292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos FA, Billi E, Zörgö E, Parts L, Fargier P, Omholt S, Blomberg A, Warringer J, Louis EJ, Liti G. Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol Ecol. 2011;20:1401–1413. doi: 10.1111/j.1365-294X.2011.05005.x. [DOI] [PubMed] [Google Scholar]
- Salinas F, Cubillos FA, Soto D, Garcia V, Bergström A, Warringer J, Ganga MA, Louis EJ, Liti G, Martinez C. The genetic basis of natural variation in oenological traits in Saccharomyces cerevisiae. PLoS ONE. 2012;7:e49640. doi: 10.1371/journal.pone.0049640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marullo P, Aigle M, Bely M, Masneuf-Pomarède I, Durrens P, Dubourdieu D, Yvert G. Single QTL mapping and nucleotide-level resolution of a physiologic trait in wine Saccharomyces cerevisiae strains. FEMS Yeast Res. 2007;7:941–952. doi: 10.1111/j.1567-1364.2007.00252.x. [DOI] [PubMed] [Google Scholar]
- Ehrenreich IM, Gerke JP, Kruglyak L. Genetic dissection of complex traits in yeast: insights from studies of gene expression and other phenotypes in the BYxRM cross. Cold Spring Harb Symp Quant Biol. 2009;74:145–153. doi: 10.1101/sqb.2009.74.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Torabi N, Jia Y, Kent J, Martis S, Shapiro JA, Gresham D, Caudy AA, Kruglyak L. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature. 2010;464:1039–1042. doi: 10.1038/nature08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- Smith EN, Kruglyak L. Gene-environment interaction in yeast gene expression. PLoS Biol. 2008;6:e83. doi: 10.1371/journal.pbio.0060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB, Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc Natl Acad Sci USA. 2005;102:1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Landry CR, Hartl DL, Cavalieri D. Cascading transcriptional effects of a naturally occurring frameshift mutation in Saccharomyces cerevisiae. Mol Ecol. 2008;17:2985–2997. doi: 10.1111/j.1365-294X.2008.03765.x. [DOI] [PubMed] [Google Scholar]
- Zhu J, Sova P, Xu Q, Dombek KM, Xu EY, Vu H, Tu Z, Brem RB, Bumgarner RE, Schadt EE. Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation. PLoS Biol. 2012;10:e1001301. doi: 10.1371/journal.pbio.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-I, Dudley AM, Drubin D, Silver PA, Krogan NJ, Pe’er D, Koller D. Learning a prior on regulatory potential from eQTL data. PLoS Genet. 2009;5:e1000358. doi: 10.1371/journal.pgen.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RBH, Chan EKF, Cowley MJ, Little PFR. The influence of genetic variation on gene expression. Genome Res. 2007;17:1707–1716. doi: 10.1101/gr.6981507. [DOI] [PubMed] [Google Scholar]
- Kliebenstein D. Quantitative genomics: analyzing intraspecific variation using global gene expression polymorphisms or eQTLs. Annu Rev Plant Biol. 2009;60:93–114. doi: 10.1146/annurev.arplant.043008.092114. [DOI] [PubMed] [Google Scholar]
- Steyer D, Ambroset C, Brion C, Claudel P, Delobel P, Sanchez I, Erny C, Blondin B, Karst F, Legras J-L. QTL mapping of the production of wine aroma compounds by yeast. BMC Genomics. 2012;13:573. doi: 10.1186/1471-2164-13-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Grigull J, Mohammad N, Hughes TR. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinforma. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- Brem RB, Storey JD, Whittle J, Kruglyak L. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature. 2005;436:701–703. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqui I, Vissers S, Bernard F, de Craene J-O, Boles E, Urrestarazu A, André B. Amino acid signaling in saccharomyces cerevisiae: a permease-like sensor of external amino acids and f-box protein grr1p are required for transcriptional induction of the agp1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol. 1999;19:989–1001. doi: 10.1128/mcb.19.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdulrehman D, Monteiro PT, Teixeira MC, Mira NP, Lourenco AB, dos Santos SC, Cabrito TR, Francisco AP, Madeira SC, Aires RS, Oliveira AL, Sa-Correia I, Freitas AT. YEASTRACT: providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic Acids Res. 2010;39:D136–D140. doi: 10.1093/nar/gkq964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Moreno R, Morales P, Gonzalez R, Mas A, Beltran G. Biomass production and alcoholic fermentation performance of Saccharomyces cerevisiae as a function of nitrogen source. FEMS Yeast Res. 2012;12:477–485. doi: 10.1111/j.1567-1364.2012.00802.x. [DOI] [PubMed] [Google Scholar]
- Byrd C, Turner GC, Varshavsky A. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GC, Du F, Varshavsky A. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature. 2000;405:579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- Damon C, Vallon L, Zimmermann S, Haider MZ, Galeote V, Dequin S, Luis P, Fraissinet-Tachet L, Marmeisse R. A novel fungal family of oligopeptide transporters identified by functional metatranscriptomics of soil eukaryotes. ISME J. 2011;5:1871–1880. doi: 10.1038/ismej.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straver MH, Aar PCVD, Smit G, Kijne JW. Determinants of flocculence of brewer’s yeast during fermentation in wort. Yeast. 1993;9:527–532. doi: 10.1002/yea.320090509. [DOI] [PubMed] [Google Scholar]
- Hikkel I, Lucau-Danila A, Delaveau T, Marc P, Devaux F, Jacq C. A general strategy to uncover transcription factor properties identifies a new regulator of drug resistance in yeast. J Biol Chem. 2003;278:11427–11432. doi: 10.1074/jbc.M208549200. [DOI] [PubMed] [Google Scholar]
- Grey M, Pich CT, Haase E, Brendel M. SNG1–a new gene involved in nitrosoguanidine resistance in Saccharomyces cerevisiae. Mutat Res. 1995;346:207–214. doi: 10.1016/0165-7992(95)90037-3. [DOI] [PubMed] [Google Scholar]
- Schuller C. Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in saccharomyces cerevisiae. Mol Biol Cell. 2003;15:706–720. doi: 10.1091/mbc.E03-05-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P, Mahé Y, Thompson S, Pandjaitan R, Holyoak C, Egner R, Mühlbauer M, Coote P, Kuchler K. The pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 1998;17:4257–4265. doi: 10.1093/emboj/17.15.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Erny C, Raoult P, Alais A, Butterlin G, Delobel P, Matei-Radoi F, Casaregola S, Legras JL. Ecological success of a group of Saccharomyces cerevisiae/Saccharomyces kudriavzevii hybrids in the northern european wine-making environment. Appl Environ Microbiol. 2012;78:3256–3265. doi: 10.1128/AEM.06752-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre H, Charpentier C. Biochemical aspects of stuck and sluggish fermentation in grape must. J Ind Microbiol Biotechnol. 1998;20:20–27. doi: 10.1038/sj.jim.2900442. [DOI] [Google Scholar]
- Vargas RC, Tenreiro S, Teixeira MC, Fernandes AR, Sá-Correia I. Saccharomyces cerevisiae multidrug transporter Qdr2p (Yil121wp): localization and function as a quinidine resistance determinant. Antimicrob Agents Chemother. 2004;48:2531–2537. doi: 10.1128/AAC.48.7.2531-2537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos G, Cabedo M, Rull B, Yenush L, Serrano R, Mulet JM. Role of the yeast multidrug transporter Qdr2 in cation homeostasis and the oxidative stress response. FEMS Yeast Res. 2012;13(1):97–106. doi: 10.1111/1567-1364.12013. [DOI] [PubMed] [Google Scholar]
- Vargas RC, García-Salcedo R, Tenreiro S, Teixeira MC, Fernandes AR, Ramos J, Sá-Correia I. Saccharomyces cerevisiae multidrug resistance transporter Qdr2 is implicated in potassium uptake, providing a physiological advantage to quinidine-stressed Cells. Eukaryot Cell. 2007;6:134–142. doi: 10.1128/EC.00290-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AL. Wine Phenolics. Ann N Y Acad Sci. 2002;957:21–36. doi: 10.1111/j.1749-6632.2002.tb02903.x. [DOI] [PubMed] [Google Scholar]
- Kim JH, Campbell BC, Mahoney NE, Chan KL, Molyneux RJ. Identification of phenolics for control of Aspergillus flavus using Saccharomyces cerevisiae in a model target-gene bioassay. J Agric Food Chem. 2004;52:7814–7821. doi: 10.1021/jf0487093. [DOI] [PubMed] [Google Scholar]
- Pan Z, Agarwal AK, Xu T, Feng Q, Baerson SR, Duke SO, Rimando AM. Identification of molecular pathways affected by pterostilbene, a natural dimethylether analog of resveratrol. BMC Med Genomics. 2008;1:7. doi: 10.1186/1755-8794-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Balaji S, Wuster A, Grochow JA, Gsponer J, Przytycka TM, Aravind L, Babu MM. Genomic analysis reveals a tight link between transcription factor dynamics and regulatory network architecture. Mol Syst Biol. 2009;5:294. doi: 10.1038/msb.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeote V, Novo M, Salema-Oom M, Brion C, Valério E, Gonçalves P, Dequin S. FSY1, a horizontally transferred gene in the Saccharomyces cerevisiae EC1118 wine yeast strain, encodes a high-affinity fructose/H+ symporter. Microbiology (Reading, Engl.) 2010;156:3754–3761. doi: 10.1099/mic.0.041673-0. [DOI] [PubMed] [Google Scholar]
- Marullo P, Bely M, Masneuf-Pomarède I, Pons M, Aigle M, Dubourdieu D. Breeding strategies for combining fermentative qualities and reducing off-flavor production in a wine yeast model. FEMS Yeast Res. 2006;6:268–279. doi: 10.1111/j.1567-1364.2006.00034.x. [DOI] [PubMed] [Google Scholar]
- Bely M, Sablayrolles J-M, Barre P. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in oenological conditions. J Ferment Bioeng. 1990;70:246–252. doi: 10.1016/0922-338X(90)90057-4. [DOI] [Google Scholar]
- Calull M, Marcé RM, Borrull F. Determination of carboxylic acids, sugars, glycerol and ethanol in wine and grape must by ion-exchange high-performance liquid chromatography with refractive index detection. J Chromatogr, A. 1992;590:215–222. doi: 10.1016/0021-9673(92)85384-6. [DOI] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. URL http://www.R-project.org. [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Smyth G. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editor. New York: Springer; 2005. limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- Quan H, Shih WJ. Assessing reproducibility by the within-subject coefficient of variation with random effects models. Biometrics. 1996;52:1195–1203. doi: 10.2307/2532835. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- De Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Example of phenotypic value distribution among the population. Parental values are indicated in the top with open circle and black circle for 59A and S288c respectively. Heritablity (H2) is indicated. R70 exhibits a bimodal distribution while other phenotypes have a continuous distribution.
Clustering analysis.
Markers map. 2140 markers are from Affymetrix genotyping, 46 markers were based on single nucleotide polymorphism detection using Illumina veracode technology to cover area of low Affymetrix markers density. The global density is 1.81 markers each 10 kbp.
Comparison of partial disomy correction analyses.
Hotspot 8 genes triggered by HAP1 or/and PDR8.
Flocculation phenotype triggered by FLO1 expression.
Phylogenic trees of the protein sequence of the two transcription factor involved in drug detoxification network variation. Trees were drawn by MEGA5 software with the maximum likelihood method from the genome sequences available [SGD]. Parental strains are indicated by blue and yellow spot for 59A and S288c respectively.
Primer used.