Abstract
Roundabout 1 (Robo1) is the cognate receptor for secreted axon guidance molecule, Slits, which function to direct cellular migration during neuronal development and angiogenesis. The Slit2–Robo1 signaling is modulated by heparan sulfate, a sulfated linear polysaccharide that is abundantly expressed on the cell surface and in the extracellular matrix. Biochemical studies have further shown that heparan sulfate binds to both Slit2 and Robo1 facilitating the ligand–receptor interaction. The structural requirements for heparan sulfate interaction with Robo1 remain unknown. In this report, surface plasmon resonance (SPR) spectroscopy was used to examine the interaction between Robo1 and heparin and other GAGs and determined that heparin binds to Robo1 with an affinity of ~650 nM. SPR solution competition studies with chemically modified heparins further determined that although all sulfo groups on heparin are important for the Robo1–heparin interaction, the N-sulfo and 6-O-sulfo groups are essential for the Robo1–heparin binding. Examination of differently sized heparin oligosaccharides and different GAGs also demonstrated that Robo1 prefers to bind full-length heparin chains and that GAGs with higher sulfation levels show increased Robo1 binding affinities.
Keywords: Heparin, Robo1, Binding, Surface plasmon resonance
1. Introduction
Mammals have four roundabout receptors (Robo1–4) [1]. Robo1–3 have been established to be the cognate receptors for secreted axon guidance molecules (Slit1–3) [2,3], whereas the binding of Slit to Robo4 has not been universally accepted. Binding of Slit2 and Slit3 to Robo4 has been shown using supernatant from myc-tagged Slit2 or Slit3 transfected cells or purified recombinant Slits added to Robo4 expressing cells or in ELISA assays [4–6]. A recent study by Koch et al. [4] found that Robo4 bound another axon guidance molecule, UNC5B. Therefore, these studies suggest that the biological functions of Robo4 might be initiated by interacting with both Slit and UNC5B. Robo1–3 are mainly expressed in nervous systems, while Robo4 is specifically expressed in vascular endothelial cells and hematopoietic stem cells [2]. Slit–Robo interaction is essential for development of the nervous system. For example, binding of Slit2 to Robo1 triggers cytoskeletal rearrangements within the axon growth cone, resulting in axon repulsion. This fundamental function of the Slit–Robo interaction is conserved between invertebrates and vertebrates [7]. Recent studies have further revealed that the Slit–Robo interaction also directs axon branching [8] and neuron migration [9]. In addition, Slit–Robo interaction also regulates non-neuron-related functions, such as muscle precursor cell migration [10], leukocyte trafficking [11], development of lung [12], kidney [13], heart, and diaphragm [14,15], inflammation [16,17], tumor metastasis [18,19], angiogenesis [6,20–22] and hematopoietic stem cell trafficking [3]. These studies establish that Slit–Robo signaling plays fundamental roles in both physiological and pathological processes.
Heparan sulfate (HS) belongs to glycosaminoglycan (GAG) family of anionic and often highly sulfated, complex, polydisperse linear polysaccharides. HS is abundantly expressed on the cell surface and in the extracellular matrix where it interacts with a large number of protein ligands [23–25]. HS–protein interactions have been demonstrated to critically modulate the biological activity of the proteins through various mechanisms, including immobilization of proteins in the matrix, regulation of enzyme activity, binding of ligands to their receptors, and protection of proteins against degradation [25]. As a result, diverse patho-physiological processes are modulated by the interaction between heparin/HS and proteins including: blood coagulation, cell growth and differentiation, host defense, viral infection, lipid transport and metabolism, cell-to-cell and cell-to-matrix signaling, inflammation and cancer [23–27]. An understanding of HS–protein interactions at the molecular level is of fundamental importance to biology and might aid in the development of highly specific glycan-based therapeutic agents [24,28].
Compelling evidence has demonstrated that HS critically regulates Slit–Robo function during nervous system development [29–35]. Slit1 and Slit2 bind the HS proteoglycan glypican-1 through its GAG chain [36]. Additionally, Drosophila Robo forms a complex with the GAG chain of syndecan [33] and also binds to heparin [30,37]. These observations suggest that HS may interact with Slit and Robo simultaneously, resulting in the formation of ternary Slit/Robo/HS signaling complexes on cell surface thereby facilitating the Slit–Robo signaling, reminiscent of the well-accepted FGF/FGFR/HS and VEGF/VEGFR/HS models [38–42]. In previous studies we used heparin, a highly sulfated form of HS that is abundantly available, as a model of HS to determine the structural features required for HS to interact with Slit2 and Slit3, and demonstrated Slit shows a heparin/HS-specific interaction with high affinity and depends on the size, the degree of sulfation, the presence of N- and 6-O-sulfo groups and carboxyl moiety of the polysaccharide [43,44]. In the present study, we carried out surface plasmon resonance (SPR) analysis to determine the HS structure that is important for HS interaction with Robo1.
2. Materials and methods
2.1. Materials
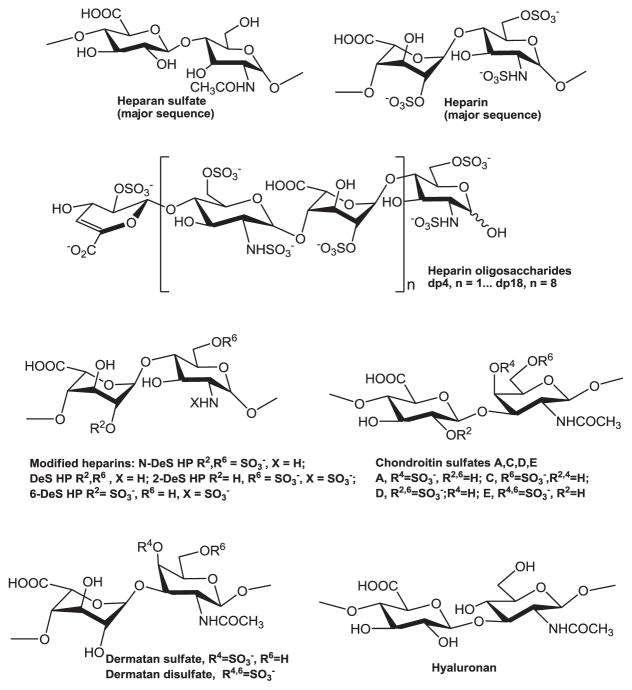
The GAGs used in this study include porcine intestinal heparin (16 kDa), low molecular weight (LMW) heparin (Enoxaparin, 4.8 kDa) and porcine intestinal HS (11.7 kDa) (Celsus Laboratories, Cincinnati, OH); porcine rib cartilage chondroitin sulfate A (20 kDa) (Sigma, St. Louis, MO), porcine intestine dermatan sulfate (also known as chondroitin sulfate B, 30 kDa, Sigma), dermatan disulfate (4,6 disulfo DS, 33 kDa, Celsus) prepared through the chemical 6-O-sulfonation of dermatan sulfate [45], shark cartilage chondroitin sulfate C (20 kDa, Sigma), whale cartilage chondroitin sulfate D (20 kDa, Seikagaku, Tokyo, Japan), squid cartilage chondroitin sulfate E (20 kDa, Seikagaku), and Streptococcus zooepidemicus hyaluronic acid, sodium salt (100 kDa, Sigma). Fully desulfonated heparin (14 kDa), N-desulfonated heparin (14 kDa) and 2-O-desulfonated IdoA heparin (13 kDa) were all prepared based on literature procedures [46]. A 6-O-desulfonated heparin (13 kDa) was prepared as described previously [47,48]. Heparin oligosaccharides, including disaccharide (dp2), tetrasaccharide (dp4), hexasaccharide (dp6), octasaccharide (dp8), decasaccharide (dp10), dodecasaccharide (dp12), tetradecasaccharide (dp14), hexadecasaccharide (dp16) and octadecasaccharide (dp18), were prepared from controlled partial heparin lyase 1 treatment of bovine lung heparin (Sigma) followed by size fractionation. Chemical structures of these GAGs are shown in Fig. 1. Sucrose octasulfate (SOS) was from Toronto Research Chemicals, Toronto, Canada. Sensor SA chips were from GE Health-care (Uppsala, Sweden). SPR measurements were performed on a BIAcore 3000 (GE Healthcare) operated using BIAcore 3000 control and BIAevaluation software (version 4.0.1).
Fig. 1.
Chemical structures of heparin, heparin-derived oligosaccharides and other GAGs.
2.2. Protein expression and purification
Human Robo1 (Ig domains 1 and 2) was expressed as a soluble secreted fusion protein in HEK293 by transient transfection of HEK293 suspension cultures. The coding region of the human roundabout homolog 1 precursor (NP_002932, Uniprot Q9Y6N7) Ig domains 1 and 2 (residues 58–266) was chemically synthesized by GeneArt AG (Regensburg, Germany) with an additional NH2-terminal fusion of the 7 amino acid recognition sequence of the tobacco etch virus (TEV) protease and a termination codon at the 3′ end of the coding region. The synthetic DNA contained a flanking 5′ EcoRI site and a 3′ HindIII site and was subcloned into a similarly digested, chemically synthesized [5] CMV promoter with intron, post-regulatory element, termination, and terminal repeat sequences as previously described [49]. The final fusion protein sequence was comprised of a 25 amino acid NH2-terminal signal sequence from the Trypanosoma cruzi lysosomal α-mannosidase [50] followed by an 8xHis tag, 17 amino acid AviTag [51], “super-folder” GFP [52], the TEV protease recognition site, and the Ig1-2 domains of Robo1. This expression vector was designated Robo1-pGEn2 and the recombinant product termed Robo1-GFP.
Suspension culture HEK293f cells (Life Technologies, Grand Island, NY) were transfected and recombinant products were purified essentially as described [49] by chromatography on Ni-NTA superflow (Qiagen, Valencia, CA) followed by elution in 25 mM HEPES, 300 mM NaCl, 300 mM imidazole, pH 7.0. Fractions containing fluorescence were pooled and concentrated to ~1 mg/ml using an ultrafiltration pressure cell membrane (Millipore, Billerica, MA) with a 10 kDa molecular weight cutoff. For removal of the GFP fusion tags, the concentrated Robo1-GFP preparation was treated with recombinant His-tagged GFP-TEV protease (1:20 ratio) at 4 °C overnight followed by 15-fold dilution in 25 mM HEPES, 300 mM NaCl, pH 7.0. The sample was then passed through a Ni-NTA column to remove the cleaved fusion tag and His-tagged GFP-TEV and further purified by Superdex 75 chromatography. The final Robo1 preparation was concentrated by ultrafiltration and buffer exchanged into 100 mM ammonium bicarbonate, pH 7.4 to a final concentration of 0.6 mg/ml.
2.3. Preparation of heparin biochip
Biotinylated heparin was prepared by reaction of sulfo-N-hydroxysuccinimide long-chain biotin (Piece, Rockford, IL) with free amino groups of unsubstituted glucosamine residues in the polysaccharide chain following a published procedure [53]. The biotinylated heparin was immobilized to streptavidin (SA) chip based on the manufacturer’s protocol. In brief, 20-μL solution of the heparin–biotin conjugate (0.1 mg/mL) in HBS-EP running buffer was injected over flow cell 2 (FC2) of the SA chip at a flow rate of 10 μL/min. The successful immobilization of heparin was confirmed by the observation of a ~250 resonance unit (RU) increase in the sensorchip. The control flow cell (FC1) was prepared by 1 min injection with saturated biotin.
2.4. Measurement of interaction between heparin and Robo1 using BIAcore
Protein samples were diluted in HBS-EP buffer (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, 0.005% surfactant P20, pH 7.4). Different dilutions of protein samples were injected at a flow rate of 30 μL/min. At the end of the sample injection, the same buffer was flowed over the sensor surface to facilitate dissociation. After a 3 min dissociation time, the sensor surface was regenerated by injecting with 30 μL of 2 M NaCl to get fully regenerated surface. The response was monitored as a function of time (sensorgram) at 25 °C. The sensorgrams of various Robo1 concentrations were globally fitted to either a 1:1 Langmuir model or a two-state binding (conformational change) model as described by the following equation:
where the equilibrium constants of each binding step are K1 = ka1/kd1 and K2 = ka2/kd2, and the overall equilibrium binding constant (or association constant) is calculated as KA = K1(1 + K2) and the dissociation constant KD = 1/KA. In this model, Robo1 (A) binds to heparin (B) to form an initial complex (AB) and then undergoes subsequent binding or conformational change to form a more stable complex (ABx) [56].
2.5. Variable contact time of the interaction of Robo1 with heparin
Robo1 (500 nM) in HBS-EP running buffer was injected at different injection times (30, 60, 90 and 120 s) over flow cell 1 (control) and flow cell 2 of the SA sensor chip. A kinetic injection mode was used, flowing the protein for (30, 60, 90 and 120 s) and monitoring the dissociation for 3 min. The response was monitored as a function of time, and after each analyte injection the surface was regenerated with 1 min injection of 2 M NaCl. The conformational change that occurs in the complex was examined by comparison of the dissociation phase of the normalized sensorgrams.
2.6. Solution competition study between heparin on chip surface and heparin-derived oligosaccharides in solution using SPR
Robo1 protein (500 nM) mixed with 1000 nM of heparin oligosaccharides, including dp2–dp18, in HBS-EP buffer were injected over heparin chip at a flow rate of 30 μL/min. After each run, the dissociation and the regeneration were performed as described above. For each set of competition experiments, a control experiment (only protein without any added heparin or oligosaccharides) was performed to make sure the surface was completely regenerated and that the results obtained between runs were comparable.
2.7. Solution competition study between heparin on chip surface and GAGs, chemical modified heparin in solution using SPR
For testing of inhibition of other GAGs and chemical modified heparins to the Robo1–heparin interaction, Robo1 at 500 nM was pre-mixed with 1000 nM of GAG or chemical modified heparin and injected over the heparin chip at a flow-rate of 30 μL/min. After each run, a dissociation period and regeneration protocol was performed as described above. Different concentrations of chemical modified heparin (from 2.5, 5, 10 and 20 μM) were also tested in this solution competition SPR analysis.
3. Results
3.1. Kinetics measurement of Robo1–heparin interactions
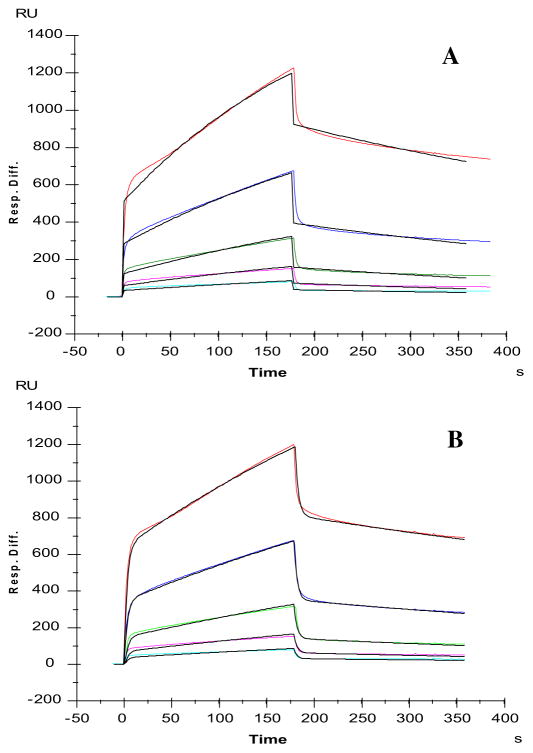
Previous biochemical and genetic experiments have shown that HS (which serves as an essential co-receptor) is required for Slit– Robo signaling and have also shown that Robo1 is a heparin binding-protein [37,55]. However, the HS structure that is required for HS to interact with Robo1 remains unknown. To address this issue, we used heparin as a molecular model of HS and carried out SPR analysis to characterize GAG binding to Robo1. Heparin was immobilized on SA sensor chips and sensorgrams of Robo1–heparin interaction were obtained by injecting different concentrations of Robo1 (Fig. 2). The sensorgrams of various Robo1 concentrations were globally fitted to either the 1:1 Langmuir model or the two-state (conformational change) binding model. The binding kinetics are presented in Table 1. These sensorgrams fit to a Langmuir 1:1 binding model (chi-squared value: 399) resulting that Robo1 binds to heparin with a dissociation constant KD of ~650 nM (Fig. 2A). A much better fitting profile (chi-squared value: 72) was obtained when fitting with the two-state (conformational change) binding model (Fig. 2B), suggesting the binding of heparin to Robo1 may induce a conformation change in Robo1. Global fitting of the sensorgrams provided on and off rate constants of the binding and the conformational change equilibrium from which an apparent affinity constant KD of 1.4 μM was calculated.
Fig. 2.
SPR sensorgrams of Robo1–heparin interaction. Concentrations of Robo1 (from top to bottom): 1000, 500, 250, 125 and 63 nM, respectively. A: Fitting with a Langmuir 1:1 binding model; B: Fitting with a two stages binding (conformation change) model. The black curves are the fitting curves using models from BIAevaluate 4.0.1.
Table 1.
Summary of kinetic data on Robo1–heparin interaction using different fitting models.a
| Fitting with a Langmuir 1:1 binding model | ka = 3.3 × 103 (±1.3 × 103) (1/MS) | kd = 2.0 × 10−3 (±0.4 × 10−3) (1/S) | Apparent affinity constant KD = 6.5 × 10−7 (±1.3 × 10−7) M |
| Fitting with a 2-stage binding model | ka1 = 4.0 × 104 (±1.5 × 104) (1/MS) ka2 = 7.6 × 10−3 (±0.8 × 10−3) (1/S) | kd1 = 0.35 (±0.05) (1/S) kd2 = 1.3 × 10−3 (±0.1 × 10−3) (1/S) | Apparent affinity constant KD = 1.4 × 10−6 M |
The data with (±) in parentheses are the standard deviations (SD) from triplicate binding experiments. ka is association rate constant, kd is disassociation rate constant; ka1 is the first stage (binding) association rate constant, kd1 is the first stage (binding) disassociation rate constant, ka2 is second stage (conformation change) rate constant, kd2 is second stage (conformation change) reverse rate constant.
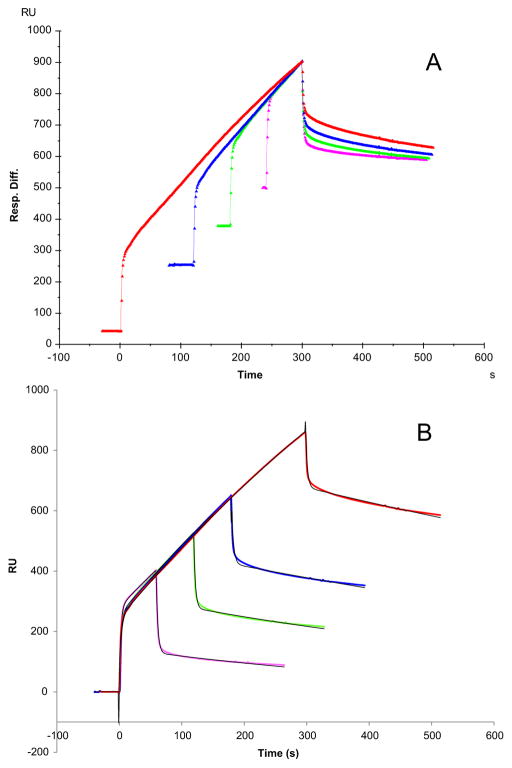
3.2. Variable contact time of the interaction of Robo1 with heparin
Additional information of the conformational change can be achieved with an SPR experiment that varied the contact time between ligand and analyte [56]. Longer association times allow the conformational change to take place to a greater extent, generating a more stable conformation, which is reflected by a slower dissociation rate. To further confirm if the Robo1–heparin binding causes conformational change, different association times of the interaction of Robo1 with heparin were performed for different injection times (30, 60, 90, and 120 s). The resulting sensorgrams were normalized in Fig. 3A, and the dissociation phase of each was compared. The fitted Kd values were 1.7 × 10−3, 1.4 × 10−3, 8.3 × 10−4 and 8.1 × 10−4 (1/s), respectively, for 30, 60, 90, and 120 s injection times (Fig. 3B), indicating that Robo1 and heparin show a slower dissociation rate when longer association times are used. These results are consistent with best fitting profile using the conformational change fitting model (Fig. 2).
Fig. 3.
Sensorgrams of Robo1–heparin interaction with variable contact times. Robo1 concentration was 500 nM. A: normalized sensorgrams by alignment of dissociation phase; B: sensorgrams fitting with a two stages binding (conformation change) model. The black curves are the fitting curves.
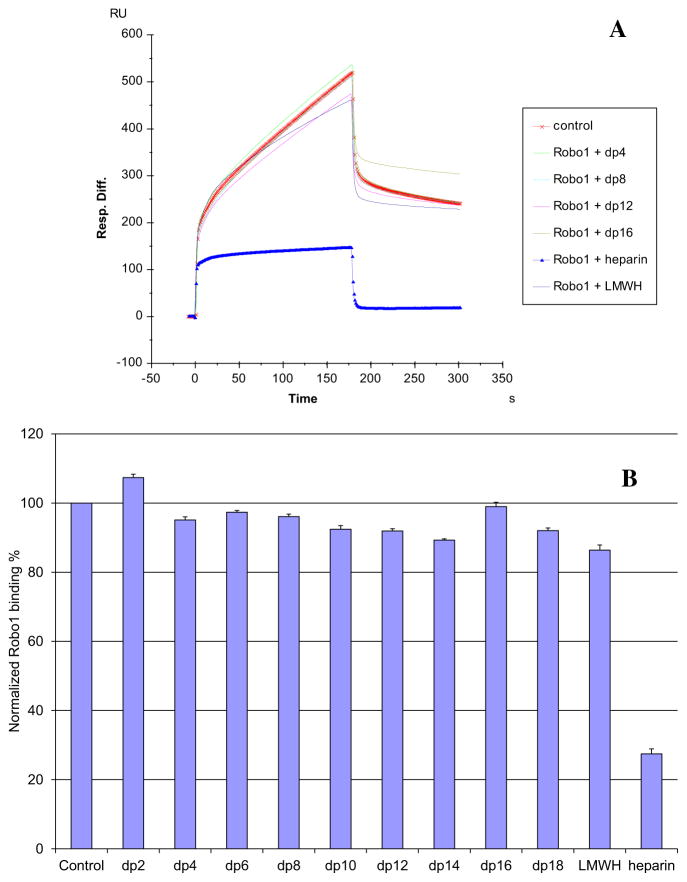
3.3. Competition study on the interaction between immobilized heparin and Robo1 using heparin-derived oligosaccharides and SPR analysis
Solution/surface competition experiments were performed by SPR to examine the effect of heparin size on the heparin–Robo1 interaction. Different sized heparin-derived oligosaccharides, ranging from dp2 to dp18 were examined in the solution phase at a concentration of 1000 nM (Fig. 4). No competition was observed for disaccharide (dp2) and little competition was apparent for oligosaccharides ranging in size from dp4 to dp 18 and for low molecular weight heparin. Only soluble heparin, having an average molecular weight of 12,000, could effectively compete with surface immobilized heparin for Robo1 binding.
Fig. 4.
A: Sensorgrams of competition between heparin oligosaccharides in solution and surface immobilized heparin. Robo1 concentration was 500 nM, and concentrations of heparin oligosaccharides in solution were 1000 nM. B: Bar graphs (based on triplicate experiments with standard deviation) of normalized Robo1 binding preference to surface immobilized heparin by competing with different sized heparin oligosaccharides in solution.
3.4. SPR solution competition study using different chemically modified heparins
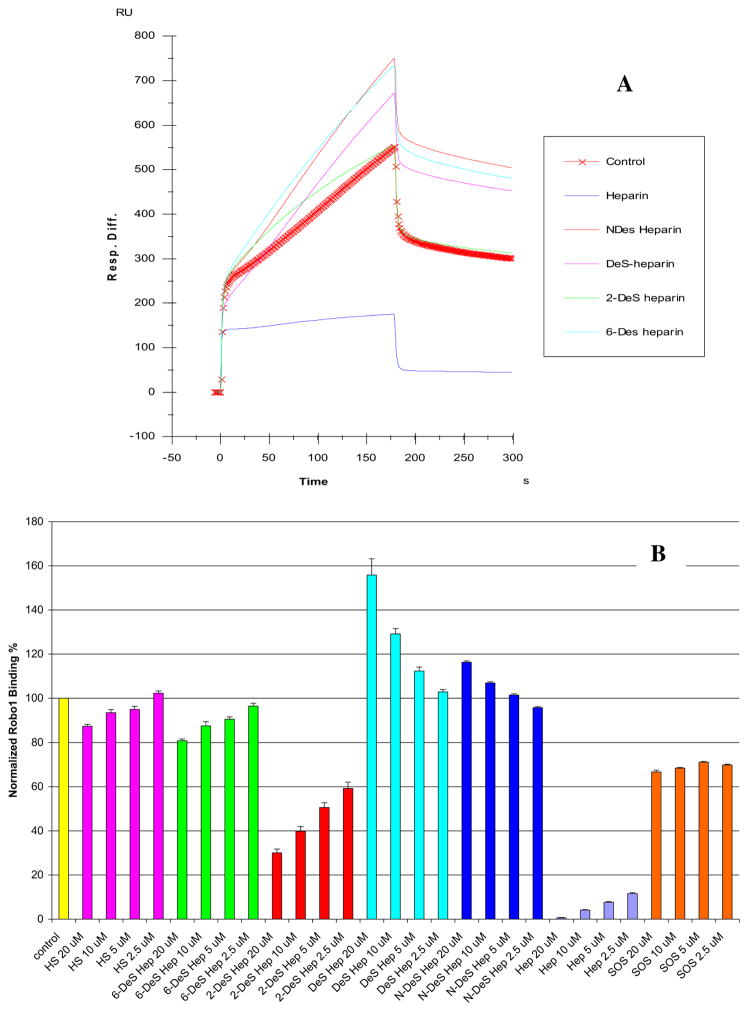
Sensorgrams from the SPR competition experiments using chemically modified heparins (1 μM) are displayed in Fig. 5 (top). The results show that all the four chemically modified heparins (fully desulfonated heparin, N-desulfonated heparin, 2-O-desulfonated heparin and 6-O-desulfonated heparin) showed no inhibitory activities. Next, SPR competition experiments were performed with increasing concentrations (from 2.5, 5, 10 and 20 μM) of chemically modified heparin, heparin, HS, or SOS (as a non GAG control), and results were summarized and shown in Fig. 5 (bottom). The findings include: i) N-desulfonated heparin and fully desulfonated heparin showed no inhibitory activities at all concentrations tested; ii) Heparin showed the strongest concentration-dependent inhibitory activity, HS, 2-O-desulfonated heparin and 6-O-desulfonated heparin showed weak concentration-dependent inhibitory activities; iii) SOS showed also a weak inhibitory activity, which was not concentration-dependent. These results suggest that heparin fine structure plays an important role in its interaction with Robo1. The N-sulfo and the 6-O-sulfo groups (rather than the 2-O-sulfo groups) of heparin appear to be essential for heparin interaction with Robo1.
Fig. 5.
A: Sensorgrams of competition between chemically modified heparins in solution and surface immobilized heparin. Robo1 concentration was 500 nM, and concentrations of chemically modified heparin in solution were 1000 nM. B: Bar graph (based on triplicate solution competition SPR experiments, error bars are standard deviation) of normalized Robo1 binding preference to surface immobilized heparin by competing with HS or different chemically modified heparin in solution.
3.5. SPR solution competition study of different GAGs
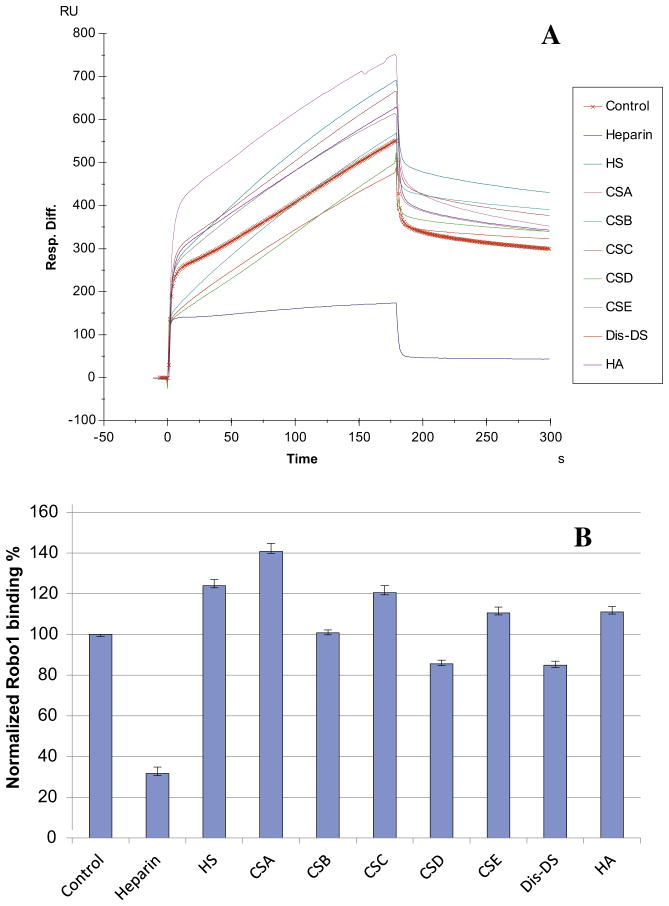
The SPR competition assay was also utilized to determine the binding preference of Robo1 to various GAGs (Fig. 1). The same concentration (1000 nM) of different soluble GAGs was present in the Robo1/immobilized heparin interaction solution. SPR competition sensorgrams and bar graphs of the GAG competition results are displayed in Fig. 6. Only soluble heparin produced a strong inhibition. CSD and Dis-DS showed weak inhibitory activities and the rest of tested GAGs showed no inhibitory activities.
Fig. 6.
(A) Sensorgrams of competition between GAGs in solution and surface immobilized heparin. Robo1 concentration was 500 nM, and concentrations of GAGs in solution were 1000 nM. Other GAGs used: HS, heparan sulfate; CSA, chondroitin sulfate A; CSB, chondroitin sulfate B; CSC, chondroitin sulfate C; CSD, chondroitin sulfate D; CSE, chondroitin sulfate E; Dis-DS, dermatan disulfate; HA, hyaluronic acid. (B) Bar graphs (based on triplicate experiments with standard deviation) of normalized Robo1 binding preference to surface immobilized heparin by competing with different concentrations of GAGs in solution.
4. Discussion
Biochemical and genetic studies have shown that HSPG/GAGs play important roles in regulating key developmental signaling pathways, such as the pathways for Wnt, Hedgehog (Hh), and fibroblast growth factors (FGFs) [22–27]. Previous studies have reported that HS is absolutely required for Slit–Robo signaling [36]. HSPGs serve as essential co-receptors in Slit–Robo signaling by promoting the formation of a ternary Slit–Robo–HS signaling complex [36,54,60]. However, quantitative biophysical binding studies to Robo, exploring a broad range of GAG fine structures, have not been done. In this study we applied SPR analysis to examine the interaction between Robo1 and heparin and other GAGs.
SPR competition experiments were performed with different sized heparin-derived oligosaccharides (from dp2 to dp18). Chain length requirements for heparin protein interactions have been reported for binding to other proteins, including: Shh–heparin, FGF–heparin and IL-7–heparin interactions [57–59]. These chain size requirements are smaller than those observed for Robo1. This result is in partial agreement with Fukuhara et al. [54], who predicted that longer HS/heparin chains (five or more disaccharide units) might be required to span the Slit–Robo interface like a bridge without fully entering into the crevice near the Robo IG1-2 boundary. The large heparin chain length requirement for Robo1 binding appears to be critical for accommodation of the protein as shorter saccharide chains may not suffice in neutralizing the positive charged heparin binding regions of Robo. There is an electrostatic surface of positive charge on Robo near the interdomain regions of Robo molecules [54]. The small difference between dp18 and LMWH is either due to the slightly larger average size of LMWH or the presence of small amounts of larger (>dp20) heparin chains in the LMWH.
SPR competition experiments with different GAGs and chemically modified heparin clearly showed that the inhibitory activity was dependent on the level of GAG sulfation and fine structure. For example, the inhibitory activity of heparin is greatly reduced when it is desulfonated, and the inhibitory activity of dermatan sulfate increases when it is sulfonated to Dis-DS. The inhibitory effects of soluble GAGs on the Robo1/immobilized heparin interaction was greatest for heparin with 2.8 sulfo groups per disaccharide repeating unit, followed by CSD and Dis-DS with 1.5–2 sulfo groups per disaccharide. HS with 1.2 sulfo groups per disaccharide, CSB, CSA and CSC with <1 sulfo group per mole disaccharide showed no inhibitory effects at 1 μM concentration. Surprisingly, CSE (with 1.5–2 sulfo groups per disaccharide) showed no inhibitory effect at a concentration of 1 μM. These results suggest that GAG fine structure plays a role in interaction with Robo1.
In conclusion, our SPR analysis shows that Robo1 is a heparin-binding protein with a high affinity (KD ~ 650 nM) for heparin. The binding of heparin to Robo1 appears to result in a conformation change within Robo1, although this may need to be confirmed using alternative experimental approaches. The SPR solution competition study shows that Robo1 binding to heparin is chain length dependent and Robo1 prefers to bind full chain heparin. In addition, higher sulfation levels of GAGs enhance their binding affinities to Robo1. Low sulfated GAGs (HS, CSA, CSC) having low/no binding affinity to Robo1. While all sulfo groups on heparin are involved in the Robo1–heparin interaction, the N-sulfo and 6-O-sulfo groups appear to play a particularly important role. The Slit– Robo1 interaction has been shown to critically modulate some physiological processes, such as neuronal development, axon migration and angiogenesis [2–4,6–17,20–22] and pathological processes, such as tumor genesis and metastasis [18,19,60]. Our kinetic and structural analysis of HS/heparin in interaction with Robo1 may provide useful information for the rational design of HS/heparin-based agent to promote or block the Slit–Robo1-mediated biological processes. However, to exploit this potential application, further studies will be needed to establish the precise molecular mechanism of this interaction. Site-directed mutagenesis would also be useful for determining the Robo1 residues that are required for HS/heparin-binding and the spatial structural arrangement of Robo1/heparin complex. Meanwhile, study on the structure–activity relationship in the HS/heparin modulation of Slit–Robo1-mediated cell signaling is underway.
Acknowledgments
This work was supported by grants from the National Institutes of Health in the form of GM-38060 (R.J.L.), NIH R01HL093339 (L.W.) and RR005351/GM103390 (L.W. and K.M.)
Abbreviations
- GAG
glycosaminoglycan
- Robo1
roundabout 1
- Slit
secreted axon guidance molecules
- SPR
surface plasmon resonance
- HS
heparan sulfate
- CSA
chondroitin sulfate A
- CSB
chondroitin sulfate B
- CSC
chondroitin sulfate C
- CSD
chondroitin sulfate D
- CSE
chondroitin sulfate E
- Dis-DS
dermatan disulfate
- HA
hyaluronic acid
- SA
streptavidin
- RU
resonance unit
- dp
degree of polymerization
References
- 1.Park KW, Morrison CM, Sorensen LK, Jones CA, Rao Y, Chien CB, Wu JY, Urness LD, Li DY. Robo4 is a vascular-specific receptor that inhibits endothelial migration. Dev Biol. 2003;261:251–267. doi: 10.1016/s0012-1606(03)00258-6. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P, Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature. 2005;436:193–200. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- 3.Smith-Berdan S, Nguyen A, Hassanein D, Zimmer M, Ugarte F, Ciriza J, Li D, Garcia-Ojeda ME, Hinck L, Forsberg EC. Robo4 cooperates with CXCR4 to specify hematopoietic stem cell localization to bone marrow niches. Cell Stem Cell. 2011;8:72–83. doi: 10.1016/j.stem.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koch AW, Mathivet T, Larrivee B, Tong RK, Kowalski J, Pibouin-Fragner L, Bouvree K, Stawicki S, Nicholes K, Rathore N, Scales SJ, Luis E, del Toro R, Freitas C, Breant C, Michaud A, Corvol P, Thomas JL, Wu Y, Peale F, Watts RJ, Tessier-Lavigne M, Bagri A, Eichmann A. Robo4 maintains vessel integrity and inhibits angiogenesis by interacting with UNC5B. Dev Cell. 2011;20:33–46. doi: 10.1016/j.devcel.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Suchting S, Heal P, Tahtis K, Stewart LM, Bicknell R. Soluble Robo4 receptor inhibits in vivo angiogenesis and endothelial cell migration. FASEB J. 2005;19:121–123. doi: 10.1096/fj.04-1991fje. [DOI] [PubMed] [Google Scholar]
- 6.Zhang B, Dietrich UM, Geng JG, Bicknell R, Esko JD, Wang L. Repulsive axon guidance molecule Slit3 is a novel angiogenic factor. Blood. 2009;114:4300–4309. doi: 10.1182/blood-2008-12-193326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- 8.Wang KH, Brose K, Arnott D, Kidd T, Goodman CS, Henzel W, Tessier-Lavigne M. Biochemical purification of a mammalian slit protein as a positive regulator of sensory axon elongation and branching. Cell. 1999;96:771–784. doi: 10.1016/s0092-8674(00)80588-7. [DOI] [PubMed] [Google Scholar]
- 9.Wu W, Wong K, Chen J, Jiang Z, Dupuis S, Wu JY, Rao Y. Directional guidance of neuronal migration in the olfactory system by the protein Slit. Nature. 1999;400:331–336. doi: 10.1038/22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kramer SG, Kidd T, Simpson JH, Goodman CS. Switching repulsion to attraction: changing responses to slit during transition in mesoderm migration. Science. 2001;292:737–740. doi: 10.1126/science.1058766. [DOI] [PubMed] [Google Scholar]
- 11.Wu JY, Feng L, Park HT, Havlioglu N, Wen L, Tang H, Bacon KB, Jiang Z, Zhang X, Rao Y. The neuronal repellent Slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature. 2001;410:948–952. doi: 10.1038/35073616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xian J, Clark KJ, Fordham R, Pannell R, Rabbitts TH, Rabbitts PH. Inadequate lung development and bronchial hyperplasia in mice with a targeted deletion in the Dutt1/Robo1 gene. Proc Natl Acad Sci U S A. 2001;98:15062–15066. doi: 10.1073/pnas.251407098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grieshammer U, Le M, Plump AS, Wang F, Tessier-Lavigne M, Martin GR. SLIT2-mediated ROBO2 signaling restricts kidney induction to a single site. Dev Cell. 2004;6:709–717. doi: 10.1016/s1534-5807(04)00108-x. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Zhang L, Wang D, Shen H, Jiang M, Mei P, Hayden PS, Sedor JR, Hu H. Congenital diaphragmatic hernia, kidney agenesis and cardiac defects associated with Slit3-deficiency in mice. Mech Dev. 2003;120:1059–1070. doi: 10.1016/s0925-4773(03)00161-8. [DOI] [PubMed] [Google Scholar]
- 15.Yuan W, Rao Y, Babiuk RP, Greer JJ, Wu JY, Ornitz DM. A genetic model for a central (septum transversum) congenital diaphragmatic hernia in mice lacking Slit3. Proc Natl Acad Sci U S A. 2003;100:5217–5222. doi: 10.1073/pnas.0730709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan H, Zu G, Xie Y, Tang H, Johnson M, Xu X, Kevil C, Xiong WC, Elmets C, Rao Y, Wu JY, Xu H. Neuronal repellent Slit2 inhibits dendritic cell migration and the development of immune responses. J Immunol. 2003;171:6519–6526. doi: 10.4049/jimmunol.171.12.6519. [DOI] [PubMed] [Google Scholar]
- 17.Kanellis J, Garcia GE, Li P, Parra G, Wilson CB, Rao Y, Han S, Smith CW, Johnson RJ, Wu JY, Feng L. Modulation of inflammation by slit protein in vivo in experimental crescentic glomerulonephritis. Am J Pathol. 2004;165:341–352. doi: 10.1016/S0002-9440(10)63301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasad A, Fernandis AZ, Rao Y, Ganju RK. Slit protein-mediated inhibition of CXCR4-induced chemotactic and chemoinvasive signaling pathways in breast cancer cells. J Biol Chem. 2004;279:9115–9124. doi: 10.1074/jbc.M308083200. [DOI] [PubMed] [Google Scholar]
- 19.Schmid BC, Rezniczek GA, Fabjani G, Yoneda T, Leodolter S, Zeillinger R. The neuronal guidance cue Slit2 induces targeted migration and may play a role in brain metastasis of breast cancer cells, Breast Cancer Res. Treat. 2007 doi: 10.1007/s10549-007-9504-0. [DOI] [PubMed] [Google Scholar]
- 20.Wang LJ, Zhao Y, Han B, Ma YG, Zhang J, Yang DM, Mao JW, Tang FT, Li WD, Yang Y, Wang R, Geng JG. Targeting slit-roundabout signaling inhibits tumor angiogenesis in chemical-induced squamous cell carcinogenesis. Cancer Sci. 2008;99:510–517. doi: 10.1111/j.1349-7006.2007.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones CA, London NR, Chen H, Park KW, Sauvaget D, Stockton RA, Wythe JD, Suh W, Larrieu-Lahargue F, Mukouyama YS, Lindblom P, Seth P, Frias A, Nishiya N, Ginsberg MH, Gerhardt H, Zhang K, Li DY. Robo4 stabilizes the vascular network by inhibiting pathologic angiogenesis and endothelial hyperpermeability. Nat Med. 2008;14:448–453. doi: 10.1038/nm1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bedell VM, Yeo SY, Park KW, Chung J, Seth P, Shivalingappa V, Zhao J, Obara T, Sukhatme VP, Drummond IA, Li DY, Ramchandran R. Round-about4 is essential for angiogenesis in vivo. Proc Natl Acad Sci U S A. 2005;102:6373–6378. doi: 10.1073/pnas.0408318102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capila I, Linhardt RJ. Heparin–protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 24.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 25.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 26.Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 27.Powell AK, Yates EA, Fernig DG, Turnbull JE. Interactions of heparin/heparan sulfate with proteins: appraisal of structural factors and experimental approaches. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- 28.Sasisekharan R, Raman R, Prabhakar V. Glycomics approach to structure– function relationships of glycosaminoglycans. Annu Rev Biomed Eng. 2006;8:181–231. doi: 10.1146/annurev.bioeng.8.061505.095745. [DOI] [PubMed] [Google Scholar]
- 29.Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302:1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- 30.Hohenester E, Hussain S, Howitt JA. Interaction of the guidance molecule slit with cellular receptors. Biochem Soc Trans. 2006;34:418–421. doi: 10.1042/BST0340418. [DOI] [PubMed] [Google Scholar]
- 31.Rhiner C, Gysi S, Frohli E, Hengartner MO, Hajnal A. Syndecan regulates cell migration and axon guidance in C. elegans. Development. 2005;132:4621–4633. doi: 10.1242/dev.02042. [DOI] [PubMed] [Google Scholar]
- 32.Steigemann P, Molitor A, Fellert S, Jackle H, Vorbruggen G. Heparan sulfate proteoglycan syndecan promotes axonal and myotube guidance by slit/robo signaling. Curr Biol. 2004;14:225–230. doi: 10.1016/j.cub.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 33.Johnson KG, Ghose A, Epstein E, Lincecum J, O’Connor MB, Van Vactor D. Axonal heparan sulfate proteoglycans regulate the distribution and efficiency of the repellent slit during midline axon guidance. Curr Biol. 2004;14:499–504. doi: 10.1016/j.cub.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Hagino S, Iseki K, Mori T, Zhang Y, Hikake T, Yokoya S, Takeuchi M, Hasimoto H, Kikuchi S, Wanaka A. Slit and glypican-1 mRNAs are coex-pressed in the reactive astrocytes of the injured adult brain. Glia. 2003;42:130–138. doi: 10.1002/glia.10207. [DOI] [PubMed] [Google Scholar]
- 35.Hu H. Cell-surface heparan sulfate is involved in the repulsive guidance activities of Slit2 protein. Nat Neurosci. 2001;4:695–701. doi: 10.1038/89482. [DOI] [PubMed] [Google Scholar]
- 36.Ronca F, Andersen JS, Paech V, Margolis RU. Characterization of Slit protein interactions with glypican-1. J Biol Chem. 2001;276:29141–29147. doi: 10.1074/jbc.M100240200. [DOI] [PubMed] [Google Scholar]
- 37.Hussain SA, Piper M, Fukuhara N, Strochlic L, Cho G, Howitt JA, Ahmed Y, Powell AK, Turnbull JE, Holt CE, Hohenester E. A molecular mechanism for the heparan sulfate dependence of slit–robo signaling. J Biol Chem. 2006;281:39693–39698. doi: 10.1074/jbc.M609384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cebe-Suarez S, Zehnder-Fjallman A, Ballmer-Hofer K. The role of VEGF receptors in angiogenesis; complex partnerships. Cell Mol Life Sci. 2006;63:601–615. doi: 10.1007/s00018-005-5426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tessler S, Rockwell P, Hicklin D, Cohen T, Levi BZ, Witte L, Lemischka IR, Neufeld G. Heparin modulates the interaction of VEGF165 with soluble and cell associated flk-1 receptors. J Biol Chem. 1994;269:12456–12461. [PubMed] [Google Scholar]
- 40.Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M. Crystal structure of a ternary FGF– FGFR–heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 41.Gitay-Goren H, Soker S, Vlodavsky I, Neufeld G. The binding of vascular endothelial growth factor to its receptors is dependent on cell surface-associated heparin-like molecules. J Biol Chem. 1992;267:6093–6098. [PubMed] [Google Scholar]
- 42.Rapraeger AC. Heparan sulfate-growth factor interactions. Methods Cell Biol. 2002;69:83–109. doi: 10.1016/s0091-679x(02)69009-0. [DOI] [PubMed] [Google Scholar]
- 43.Zhang F, Ronca F, Linhardt RJ, Margolis RU. Structural determinants of heparan sulfate interactions with slit proteins. Biochem Biophys Res Commun. 2004;317:352–357. doi: 10.1016/j.bbrc.2004.03.059. [DOI] [PubMed] [Google Scholar]
- 44.Condac E, Strachan H, Gutierrez-Sanchez G, Brainard B, Giese C, Heiss C, Johnson D, Azadi P, Bergmann C, Orlando R, Esmon CT, Harenberg J, Moremen K, Wang L. The C-terminal fragment of axon guidance molecule Slit3 binds heparin and neutralizes heparin’s anticoagulant activity. Glycobiology. 2012;22:1183–1192. doi: 10.1093/glycob/cws087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brister SJ, Buchanan MR, Griffin CC, Van Gorp CL, Linhardt RJ. U.S. Patent #5,922,690. Dermatandisulfate, an inhibitor of thrombin and complement activation. 1999
- 46.Yates EA, Santini F, Guerrini M, Naggi A, Torri G, Casu B. 1H and 13C NMR spectral assignments of the major sequences of twelve systematically modified heparin derivatives. Carbohydr Res. 1996;294:15–27. doi: 10.1016/s0008-6215(96)90611-4. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Brown JR, Varki A, Esko JD. Heparin’s anti-inflammatory effects require glucosamine 6-O-sulfation and are mediated by blockade of L- and P-selectins. J Clin Invest. 2002;110:127–136. doi: 10.1172/JCI14996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang S, Condac E, Qiu H, Jiang J, Gutierrez-Sanchez G, Bergmann C, Handel T, Wang L. Heparin-induced leukocytosis requires 6-O-sulfation and is caused by blockade of selectin- and CXCL12 protein-mediated leukocyte trafficking in mice. J Biol Chem. 2012;287:5542–5553. doi: 10.1074/jbc.M111.314716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barb AW, Meng L, Gao Z, Johnson RW, Moremen KW, Prestegard JH. NMR characterization of immunoglobulin G Fc glycan motion on enzymatic silylation. Biochemistry. 2012;51:4618–4626. doi: 10.1021/bi300319q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandersall-Nairn AS, Merkle RK, O’Brien K, Oeltmann TN, Moremen KW. Cloning, expression, purification, and characterization of the acid alpha-mannosidase from Trypanosoma cruzi. Glycobiology. 1998;8:1183–1194. doi: 10.1093/glycob/8.12.1183. [DOI] [PubMed] [Google Scholar]
- 51.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holo-enzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 53.Hernaiz M, Liu J, Rosenberg RD, Linhardt RJ. Enzymatic modification of heparan sulfate on a biochip promotes its interaction with antithrombin III. Biochem Biophys Res Commun. 2000;276:292–297. doi: 10.1006/bbrc.2000.3453. [DOI] [PubMed] [Google Scholar]
- 54.Futamura M, Dhanasekaran P, Handa T, Phillips MC, Lund-Katz S, Saito H. Two-step mechanism of binding of apolipoprotein E to heparin: implications for the kinetics of apolipoprotein E–heparan sulfate proteoglycan complex formation on cell surfaces. J Biol Chem. 2005;280:5414–5422. doi: 10.1074/jbc.M411719200. [DOI] [PubMed] [Google Scholar]
- 55.Fukuhara N, Howitt JA, Hussain SA, Hohenester E. Structural and functional analysis of slit and heparin binding to immunoglobulin-like domains 1 and 2 of Drosophila Robo. J Biol Chem. 2008;283:16226–16234. doi: 10.1074/jbc.M800688200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munoz E, Xu D, Kemp M, Zhang F, Liu J, Linhardt RJ. Affinity, kinetic, and structural study of the interaction of 3-O-sulfotransferase isoform 1 with heparan sulfate. Biochemistry. 2006;45:5122–5128. doi: 10.1021/bi052403n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang F, McLellan JS, Ayala AM, Leahy DJ, Linhardt RJ. Kinetic and structural studies on interactions between heparin or heparan sulfate and proteins of the hedgehog signaling pathway. Biochemistry. 2007;46:3933–3941. doi: 10.1021/bi6025424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang F, Zhang Z, Lin X, Beenken A, Eliseenkova AV, Mohammadi M, Linhardt RJ. Compositional analysis of heparin/heparan sulfate interacting with fibroblast growth factor. fibroblast growth factor receptor complexes. Biochemistry. 2009;48:8379–8386. doi: 10.1021/bi9006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang F, Liang X, Pu D, George KI, Holland PJ, Walsh ST, Linhardt RJ. Biophysical characterization of glycosaminoglycan-IL-7interactions using SPR. Biochimie. 2012;94:242–249. doi: 10.1016/j.biochi.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang B, Xiao Y, Ding BB, Zhang N, Yuan X, Gui L, Qian KX, Duan S, Chen Z, Rao Y, Geng JG. Induction of tumor angiogenesis by Slit–Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell. 2003;4:19–29. doi: 10.1016/s1535-6108(03)00164-8. [DOI] [PubMed] [Google Scholar]