SUMMARY
Direct lineage reprogramming is a promising approach for human disease modeling and regenerative medicine with poorly understood mechanisms. Here we reveal a hierarchical mechanism in the direct conversion of fibroblasts into induced neuronal (iN) cells mediated by the transcription factors Ascl1, Brn2, and Myt1l. Ascl1 acts as an “on target” pioneer factor by immediately occupying most cognate genomic sites in fibroblasts. In contrast, Brn2 and Myt1l do not access fibroblast chromatin productively on their own; instead Ascl1 recruits Brn2 to Ascl1 sites genome-wide. A unique trivalent chromatin signature in the host cells predicts the permissiveness for Ascl1 pioneering activity among different cell types. Finally, we identified Zfp238 as a key Ascl1 target gene that can partially substitute for Ascl1 during iN cell reprogramming. Thus, precise match between pioneer factor and the chromatin context at key target genes is determinative for trans-differentiation to neurons and likely other cell types.
INTRODUCTION
The lineage identity of differentiated somatic cells is considered to be very stable due to rigid chromatin configurations, inheritable DNA modifications and re-enforcing transcription factor networks (Vierbuchen and Wernig, 2012). However, various experimental conditions including nuclear transfer into oocytes, cell fusion, and overexpression of transcription factors have been shown to overcome these epigenetic barriers and induce cell fate reprogramming to both pluripotency and unrelated somatic cell fates (Graf and Enver, 2009; Jaenisch and Young, 2008; Vierbuchen and Wernig, 2011). We recently discovered that three neuronal transcription factors Ascl1, Brn2, and Myt1l (BAM factors) are sufficient to convert mesodermal fibroblasts or endodermal hepatocytes into fully functional neuronal cells, termed induced neuronal (iN) cells (Marro et al., 2011; Vierbuchen et al., 2010). The generation of human iN cells is much less efficient and requires additional factors such as NeuroD1 or microRNAs (Ambasudhan et al., 2011; Pang et al., 2011; Qiang et al., 2011; Yoo et al., 2011).
Our previous findings suggested that of the three factors, Ascl1 is the central driver of reprogramming since only Ascl1 is sufficient to induce immature iN cells in mouse embryonic fibroblasts (MEFs). In contrast, neither Brn2 nor Myt1l alone achieve any morphological changes in MEFs. However, when combined with Ascl1, Brn2 and Myt1l greatly improved the conversion efficiency and both were required for the induction of fully reprogrammed iN cells. Ascl1 is a well-studied pro-neural gene of the basic helix-loop-helix (bHLH) family of transcription factors that specifically bind DNA sequences containing an E-box motif (Bertrand et al., 2002). It is prominently expressed in a subset of central and peripheral neural progenitors (Guillemot et al., 1993; Lo et al., 1991). Overexpression of Ascl1 and related factors in the developing spinal cord induces rapid neuronal differentiation (Ma et al., 1999; Nakada et al., 2004). Accordingly, Ascl1-mutant mice show severe defects in neurogenesis (Guillemot et al., 1993). Ascl1 regulates and is regulated by the Notch pathway, which mediates lateral inhibition (Bertrand et al., 2002; Guillemot et al., 1993).
The other two iN cell factors are less well characterized. Brn2 (also known as Pou3f2) belongs to the Pou-Homeodomain family of transcription factors. Brn2 is expressed in ventricular zone progenitor cells throughout the neuraxis and downregulated upon differentiation, except in cortical development where it remains expressed in layer II/III and V pyramidal neurons (Dominguez et al., 2012). Combined deletion of Brn1 and Brn2 in mice resulted in severe proliferation defects of cortical progenitor cells and migration defects of upper-layer neurons ultimately leading to a disorganized and thinned cortex (Sugitani et al., 2002). Very little is known about Myt1l. It contains multiple zinc finger domains of the Cys-Cys-His-Cys (C2HC) type that are thought to interact with DNA (Kim and Hudson, 1992). Myt1l is expressed throughout the central and peripheral nervous system in early postmitotic neurons (Cahoy et al., 2008; Weiner and Chun, 1997). Interestingly, the Xenopus ortholog of Myt1 family transcription factors X-Myt1 is required for proper neuronal differentiation and synergizes with proneural bHLH transcription factors to promote ectopic neurogenesis in non-neural ectoderm (Bellefroid et al., 1996).
These observations argue that the three iN cell reprogramming factors promote neuronal differentiation in the context of a neural progenitor cell. It is, however, completely unclear how these factors can exert their proper function in distantly related cell types such as fibroblasts. It has to be assumed that both the chromatin configuration at neuronal genes and the expression of transcriptional co-regulators are not favorable for neuronal induction because fibroblasts normally never give rise to neurons and the reprogramming factors were not evolutionarily selected to induce transdifferentiation.
Here, we used an integrative genomic approach to characterize the molecular mechanism governing the early phase of MEF-to-iN cell reprogramming.
RESULTS
BAM factors induce rapid and global transcriptional changes in fibroblasts
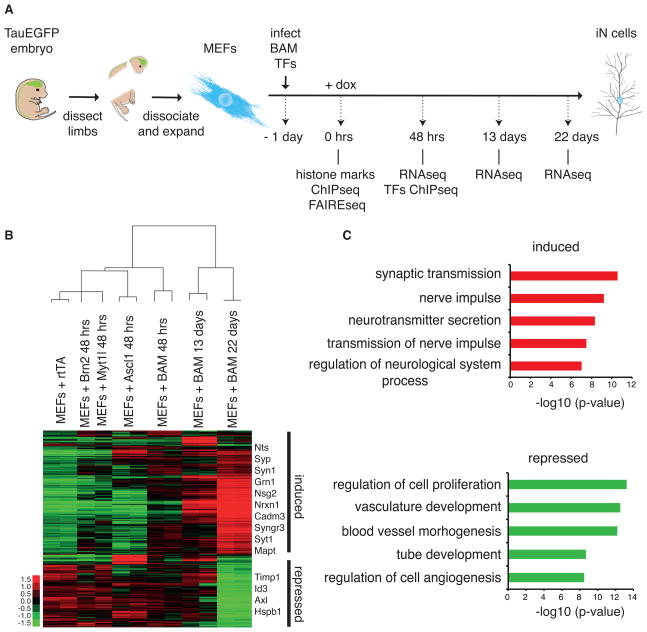
To begin to elucidate the molecular mechanism underlying iN cell reprogramming, we have determined the genome-wide transcriptional changes by RNA sequencing at various time points of reprogramming (Figure 1A). In addition to control cells infected with rtTA virus and treated with doxycycline (dox), we have profiled MEFs 0 and 48 hours after the induction of the single or combined BAM factors with dox, as well as TauEGFP-FACS-sorted early and late iN cells, day 13 and 22 after dox, respectively. We observed that 2,522 genes significantly changed expression over this time course (fold change >2, p-value <0.05) (Figure 1B and S2A–B). Genes that increased expression were enriched for gene ontology (GO) terms associated with neuronal activity (Figure 1C).
Figure 1. Global transcriptional responses to the BAM reprogramming factors.
(A) Schematic representation of the overall experimental design of this study
(B) Hierarchical clustering and heatmap of genome-wide expression analysis during the iN cell reprogramming process by RNA-seq across indicated time points (n=2 biological replicates except n=1 for MEF+Brn2 and MEF+Myt1l). The day 13 and day 22 samples were FACS-purified for TauEGFP-positive cells. Shown are those 2,522 genes that changed expression at least two-fold at any time point. This emphasizes the global changes between MEFs and mature iN cells. Compare to Figure 7A showing a subset of the same data emphasizing the short term transcriptional effect. Fold change is represented in logarithmic scale normalized to the mean expression value of a gene across all samples. Right: names of selected genes significantly induced and repressed upon reprogramming.
(C) Top 5 most significant Gene ontology (GO) terms enriched in the group of induced and repressed genes.
See also Figure S1.
We observed that many more genes were up-regulated than down-regulated at 48 hours when MEFs were infected with Ascl1 or all three factors, whereas Brn2 and Myt1l had subtle transcriptional effects when expressed alone (Figure 1B and 7A). Principal component analysis (PCA) showed that Ascl1 uniquely underlies the top two principle components of gene expression in MEFs transduced with the BAM factors (Figure S1C). These data suggest that Ascl1 functions mainly as a transcriptional activator as had been proposed before (Castro and Guillemot, 2011) and that Ascl1 may be the strongest driver of the neuronal transcriptional program.
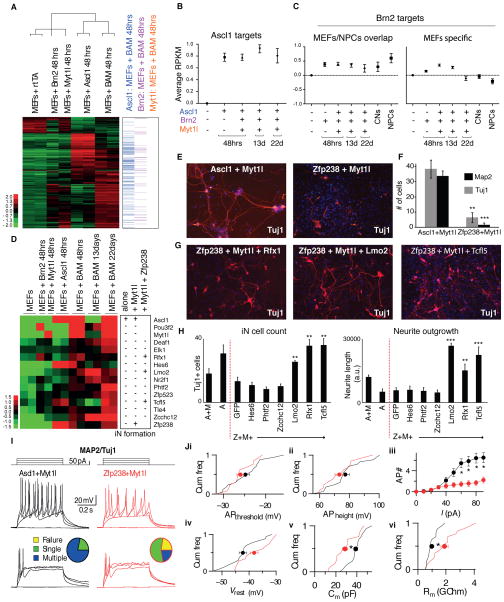
Figure 7. Zfp238 is a critical Ascl1 target and iN cell reprogramming mediator.
(A) Left: Heat map representing RNA-seq data in MEFs 48h after infection with the indicated factors. Shown are 790 genes that changed expression at least two-fold in any given condition. Fold change of genes expression is represented in logarithmic scale. Of the 790 genes, 607 are up-regulated in MEFs+Ascl1 vs. MEFs and 183 are down-regulated. 143 of the 790 genes classify as Ascl1 targets in MEFs+BAM of which 133 are up- and 10 are down-regulated. Right: Ascl1, Brn2 or Myt1l targets within these 790 genes. Genes were identified as targets based on binding peaks within −10 to +2 kb from the TSS of genes. Data are represented as mean +/− SEM.
(B) Quantification of average changes in gene expression of Ascl1 target genes in MEFs 48h after BAM induction, which were identified in (A) and are depicted across various time points of reprogramming. (13d, and 22d samples were TauEGFP-sorted). Y-axis represents the average expression (normalized reads in log2 scale) for the identified RefSeq genes. Data are represented as mean +/− SEM.
(C) Left: Quantification of average change in gene expression of Brn2 target genes based on overlapping peaks between MEFs and NPCs, depicted across time points (as in (B) and neuronal cell populations. Right: Quantification of average change in gene expression of Brn2 target genes based on MEF-specific peaks, depicted across time points and neuronal cell populations. Y-axis represents the average expression (normalized reads in log2 scale) for the identified RefSeq genes.
(D) Left: Expression level heat map (log2 based) of the BAM factors and twelve selected transcription factors induced during reprogramming. Right: schematic summarizing screen strategy and results for iN cell formation upon expression of the candidate factors alone, in combination with Myt1l, or in combination of Zfp238 and Myt1l.
(E) Immunofluorescent detection of Tuj1 (red) and DAPI (blue) of iN cells derived from MEFs after infection with Ascl1+Myt1l or Zfp238+Myt1l.
(F) Quantification of reprogramming efficiency by measuring number of cells with neuronal morphology and positive for MAP2 and Tuj1. Shown are averages of average numbers of labeled cells per 10x visual field in at least 10 randomly picked fields (Error bars = SEM, n = 3 biological replicates, t-test ** p < 0.01, *** p < 0.001).
(G) Immunofluorescent detection of Tuj1 (red) and DAPI (blue) iN cells derived from MEFs after infection with Zfp238+Myt1l and Rfx1 (left), Lmo2 (middle), or Tcfl5 (right).
(H) Average number of Tuj1+ cells and total neurite length of MEFs expressing Zfp238+Myt1L (Z+M) plus the indicated factor. Also shown are MEFs infected with Ascl1 (A) and Ascl1+Myt1l (A+M). A t-test was performed compared to the GFP control (Error bars = SEM, n=5 visual fields, **P<0.01, ***P<0.001).
(I–J). Electrophysiological characterization of iN cells derived from MEFs by overexpressing either Ascl1 (black) or Zfp238 (red) in combination with Myt1l.
(I) Cells patched in current-clamp mode. Action potential (AP)-generation pattern induced by current pulses of step-wise increasing amplitudes (top panel: stimulation protocol). Example traces from cells generating multiple (middle panels) or single (bottom panels) APs in either condition. Pie-charts indicate fraction of cells firing single (green), multiple (blue) or no (yellow) action potentials (n = 18 cells / 3 batches).
(J) Cumulative plots (Cum freq) representing intrinsic properties (lines) of iN cells capable of firing action potentials in Ascl1+Myt1l (n = 18 cells / 3 batches) and Zfp+Myt1l (n = 14 cells / 3 batches) conditions with their corresponding averages ± SE (filled circles with error bars). No significant differences found in AP threshold (APthreshold, p > 0.1) (i) or AP height (APheight, p > 0.2) (ii). (iii) However, the number of APs fired (AP #) plotted with respect to current pulses (I) of increasing amplitude was significantly different between the 2 conditions (asterisks, p < 0.02). (iv) No significant differences found in resting membrane-potential (Vrest, p > 0.05). Cumulative plots with average values ± SE show significant difference (asterisks) in capacitance (P > 0.005) (Cm,v) and membrane resistance (p > 0.002) (Rm, vi) measured at holding potential −70 mV.
See also Figure S7.
Genome-wide maps of BAM factors occupancy reveal Ascl1 dominant targeting capacity
Based on the extensive changes in transcription observed 48 hours after initiation of reprogramming, we performed Chromatin Immunoprecipitation followed by high throughput sequencing (ChIP-seq) for all three BAM factors in MEFs 48 hours after induction of all three factors. We readily identified 5,902 significant Ascl1-occupied loci upon filtering high quality peaks based on at least 20 fold enrichment and less than 5% FDR (Figure 2A). In contrast, Brn2 and Myt1l ChIP-seq required substantial technical optimization (see materials and methods, Figures S2B and S2C). Under optimized conditions Brn2 ChIP-seq revealed 2,354 peaks with at least 5-fold enrichment and less than 5% FDR. Using the same criteria, Myt1l ChIP-seq showed only minimal occupancy, despite extensive cross-linking and sonication optimization and use of different epitope tagging strategies. After optimization, we identified 475 peaks with at least 2-fold enrichment and less than 5% FDR. Most of these putative Myt1l isolated sites colocalized with histone modifications associated with accessible chromatin (Figure S2C). We confirmed that overexpressed Myt1l is present in fibroblast nuclei by immunofluorescence and in the chromatin fraction by Western blot. However, the relative abundance of Myt1l on chromatin was less than Ascl1 (Figure S2A).
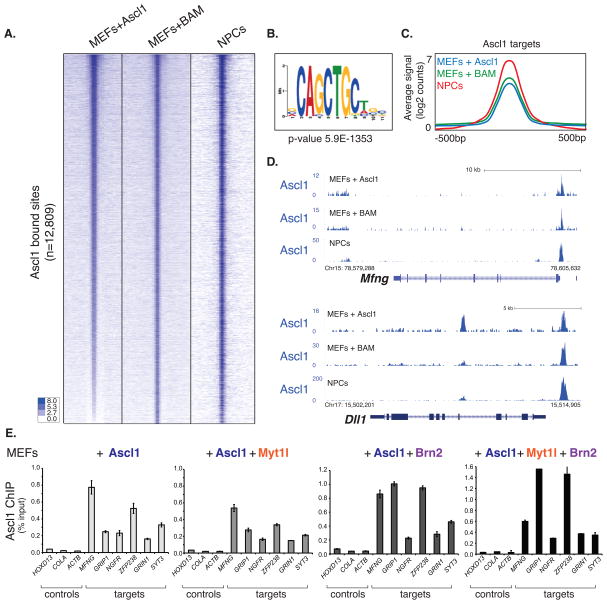
Figure 2. Genome-wide maps of BAM factors occupancy reveal Ascl1 dominant targeting capacity.
(A) Heat maps of normalized tag densities in log2 scale representing genome-wide occupancy profile for Ascl1, Brn2, and Myt1l in MEFs 48h after induction of the BAM factors. For each bound site, the signal is displayed within a 4 kb window centered around the peak summit. Peaks are sorted based on intensity. The signal at the corresponding genomic regions of the transcription factor binding is displayed across the other two data sets. Note little enrichment of Brn2 and Myt1l at Ascl1 bound sites; however, strong enrichment for Ascl1 in Brn2 sites and some in Myt1l.
(B) Left: Average signal intensity of Ascl1, Brn2, Myt1l ChIP-seq peaks at respective target sites represented as the average of log2 normalized reads. Right: Ascl1, Brn2, and Myt1l high confidence peaks genomic classification from ChIPseq in BAM-infected MEFs. Starting at the top (darkest shade) and moving in clockwise orientation with color gradient.
(C) BAM factors occupancy profile at the Zfp238 locus revealing strong binding of Ascl1 but not Brn2 or Myt1l. The y-axis represents the total number of mapped reads. The genomic scale is in kilobases (kb).
See also Figure S2.
Ascl1 binds similar genomic sites in fibroblasts and neural progenitor cells independent of Brn2 and Myt1l
Given the apparent dominant role of Ascl1 we next asked whether the other two reprogramming factors are required for Ascl1 to access its genomic targets in fibroblasts. We determined the Ascl1 binding sites by ChIP-seq in MEFs 48 hours after infection with Ascl1 alone. Surprisingly, the pattern of binding was virtually identical to Ascl1 binding in the context of all three transcription factors, as the correlation was almost identical to that observed between two biological replicates of Ascl1 ChIP-Seq in BAM infected MEFs (~0.8) (Figure 3A and S3B). We confirmed these findings by performing Ascl1 ChIP-qPCR with cells expressing Ascl1 alone, in combination with Brn2 or Myt1l, and with cells expressing all three factors (Figure 3E). Thus, exogenous Ascl1 can access its binding sites in fibroblasts without assistance from Brn2 or Myt1l.
Figure 3. Ascl1 binds similar genomic sites in fibroblasts and neural progenitor cells independent of Brn2 and Myt1l.
(A) Heat maps of normalized tag densities representing genome-wide occupancy profile for Ascl1 across cell samples: MEFs infected with Ascl1, 48h post induction (MEFs+Ascl1), MEFs infected with the BAM factors, 48h post induction (MEFs+BAM), and NPCs infected with Ascl1, 18h post induction (NPCs). For each bound site, the signal is displayed within a 4 kb window centered around the peaks.
(B) The E-box motif is significantly enriched at Ascl1 target sites across data sets (p-value 5.9e-1353).
(C) Average signal intensity of Ascl1 high confidence peaks across the three cell types as defined in (A), represented as the average of normalized reads in log2 scale.
(D) Representative tracks comparing Ascl1 occupancy at two neuronal target genes across the three cell types.
(E) Ascl1 ChIP-qPCR at selected neuronal target genes in MEFs 48h after induction of Ascl1 alone or in combination with the indicated transcription factors. Target gene binding by Ascl1 is not greatly influenced by presence of Brn2 or Myt1l. Error bars = standard deviation of the ΔCT.
See also Figure S3.
To better characterize the nature of the Ascl1 target sites in MEFs we sought to determine its targets in a close to physiological context. During normal development Ascl1 is expressed in a subset of neural precursor cells (NPCs) of the ventricular zone of the neural tube and is believed to instruct neuronal differentiation (Guillemot et al., 1993; Lo et al., 1991). When overexpressed in the neural tube or cultured NPCs Ascl1 induces a rapid exit from the cell cycle and neuronal differentiation (Nakada et al., 2004). We therefore determined the Ascl1 binding sites by ChIP-seq in NPCs with or without overexpressing Ascl1 for 18h. Unexpectedly, the comparison of those results to genome-wide binding site analysis of Ascl1 in MEFs revealed a highly similar occupancy with comparable affinity (Figures 3A, 3C, and S3A–C).
As has been observed in genome wide localization studies for Ascl1 and other bHLH transcription factors, the E-box motif CANNTG was highly enriched across its binding sites based on the MEME algorithm (Figure 3B) (Bailey et al., 2009). The CAGCTG motif was by far the most frequently bound E-box motif as seen for myogenic factor MyoD but surprisingly less so for the other pro-neural bHLH factor NeuroD2 (Fong et al., 2012). We observed the largest fraction of binding sites in the body of genes (often intronic sequences) followed by intergenic and promoter regions (Figure 2B). Many of the genes that are bound by Ascl1 are members of the Notch pathway like Hes6, Dll1, and Mfng, or are otherwise associated with neural development such as NeuroD4 (Figure 3D).
Brn2 is mislocalized during the early stages of the reprogramming
To better characterize the genomic sites bound by Brn2 in BAM-infected MEFs, we also determined the Brn2 target sites in cultured NPCs by ChIP-seq (Figure 4A). In contrast to Ascl1 targets, we found little overlap between Brn2 binding in fibroblasts and NPCs (Figure 4B and 4C). A POU-like motif was significantly enriched at Brn2 target sites in NPCs, but not in Brn2-bound sites in BAM-infected MEFs. Instead, an E-box motif was significantly enriched in a large fraction of these sites (Figure 4A). Direct comparison of Brn2 and Ascl1 targets in BAM-infected MEFs confirmed a high degree of overlap (Figure 2A). This finding raises the possibility that Ascl1 actively recruits Brn2 to some of its targets. To test this hypothesis, we performed Brn2 ChIP-seq in MEFs infected with Brn2 alone. Surprisingly, there was hardly any overlap with either Brn2 binding in NPCs or BAM-infected MEFs (Figure 4A–C). About 600 (i.e. 22.2%) of the statistically significant Brn2 targets in BAM-infected MEFs are also significant Ascl1 targets (Figure 4D). Only 29 of these 600 sites (equivalent to 0.7% of all Brn2 targets in MEFs+Brn2) remain Brn2-bound when Brn2 is expressed alone (Figure 4D and S4A). These results show that Ascl1 recruits Brn2 to many of its targets. We have confirmed the co-localization of Brn2 and Ascl1 on the fibroblast chromatin by reciprocal co-immunoprecipitation experiments. Chromatin immunoprecipitated with antibodies against Brn2 contained a substantial amount of Ascl1 in MEFs infected with both factors (Figure 4E middle panel). Consistent with the ChIP-seq data, chromatin pulled down with Ascl1 antibodies contained relatively less Brn2 (Figure 4E right panel). To define the window of Brn2 action during iN reprogramming, we generated Ascl1-inducible MEFs and infected them with Brn2 one day before, or 2 or 5 days after addition of dox. The neuronal complexity, measured as total neurite length, increased about 6-fold when Brn2 was added at days −1 and 2 and about 2-fold when added at day 5 (Figure 4F and S4B). In summary, these data suggest that the vast majority of Brn2-bound sites in MEFs are non-physiological and in part Brn2 gains access to chromatin sites made accessible by Ascl1. Furthermore, it suggests that Brn2 is primarily needed for later stages of reprogramming by contributing to iN maturation even up to 5 days after initiation of iN reprogramming.
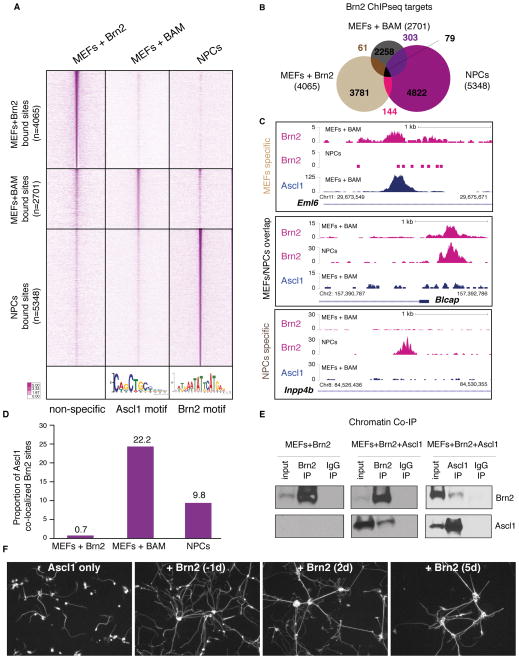
Figure 4. Brn2 is mislocalized during the early stages of the reprogramming.
(A) Heat maps of normalized tag densities representing genome-wide occupancy profile for Brn2 ChIP-seq in MEFs+Brn2 alone, MEFs+BAM, and NPCs. For each bound site, the signal is displayed within a 4 kb window centered around the summits of binding sites. Bottom panel illustrates the most significant motif enriched at Brn2 binding sites in MEFs+BAM (left, an Ascl1 motif, p=3.3e−524) and in NPCs (right, a Brn2 motif, p=3.9e−472). No significant motif was enriched in the MEFs+Brn2 data set.
(B) Venn diagram representing Brn2 high confidence peak overlap between targets in MEFs+Brn2, MEFs+BAM, and NPCs.
(C) Representative tracks illustrating Brn2 occupancy profile in MEFs+BAM and NPCs, and Ascl1 occupancy in MEFs+BAM. Top, middle, and bottom panel depict examples corresponding to 67.5%, 5.5%, and 27% of the cases observed for Brn2 occupancy between data sets respectively.
(D) 22.2% of Brn2 sites in MEFs+BAM co-localize with Ascl1, while only 9.9% of Brn2 sites in NPCs co-localize with Ascl1, which is a significant difference (p < 0.0001 t-test). Only 0.7% of Brn2 peaks in MEFs+Brn2 data set overlap with Ascl1 target sites in MEFs+BAM. Peak overlap was determined by at least 1 base pair overlap between high confidence peaks.
(E) Chromatin co-immunoprecipitation (co-IP) of Brn2 in MEFs infected with Brn2 (left) or Brn2+Ascl1 (middle). Chromatin co-IP of Ascl1 in MEFs infected with Brn2+Ascl1 cells (right). IPs were probed with antibodies against Brn2 (upper row) or Ascl1 (bottom row). Both Ascl1 and Brn2 protein is detectable when chromatin is precipitated with Brn2 or Ascl1 antibodies, respectively demonstrating co-occupancy of the 2 factors on chromatin.
(F) Brn2 contributes to neuronal maturation. Representative Tuj1 staining 11 days after dox-induction in the VA1 cell line (containing dox-inducible Ascl1) infected with Brn2 at different stages. From left to right shows: 1) Negative control (infected with GFP to control for possible virus toxicity effects), 2) positive control infected with Brn2 one day before dox induction, 3–4) infected with Brn2 2 days and 5 days respectively after dox induction. Brn2 contributes to iN maturation even up to 5 days after initiation of iN reprogramming.
See also Figure S4.
Unlike Brn2, Ascl1 acts as a pioneer factor in fibroblasts
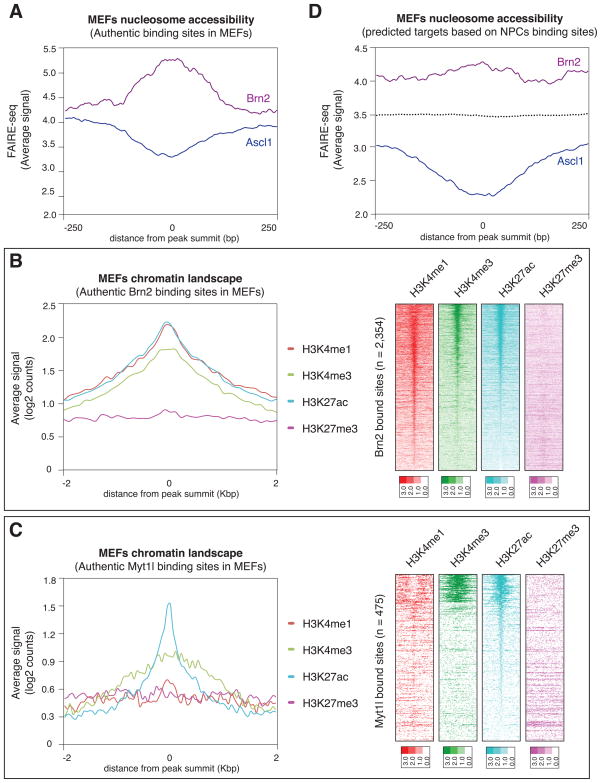
The surprising discovery that Ascl1 binds similar target sites in BAM-infected MEFs as in NPCs raised the question whether those genomic Ascl1 target sites are in an open (nucleosome-free) or closed (nucleosome-bound) state in fibroblasts before infection. Based on the crystal structure of MyoD, bHLH transcription factors are predicted to only be able to bind nucleosome-free DNA (Ma et al., 1994). To measure the non-nucleosomal fractions of the MEF genome, we performed Formaldehyde assisted isolation of regulatory elements followed by high throughput sequencing (FAIRE-seq) in uninfected MEFs (Giresi et al., 2007). Strikingly, the FAIRE analysis showed that Ascl1 target sites in MEFs were on average substantially depleted of nucleosome-free DNA (Figure 5A). These results demonstrate that Ascl1 preferentially binds previously closed chromatin in MEFs as early as within 48 hours and thus fulfills the formal criteria of a pioneer transcription factor (Zaret and Carroll, 2011).
Figure 5. Ascl1, but not Brn2 and Myt1l, acts as a pioneer factor in embryonic fibroblasts.
(A). Average FAIRE-seq signal of uninfected MEFs at Ascl1 and Brn2 target sites in MEFs infected with the BAM factors. For each target site, the signal is displayed +/− 250bp from the peak summit. While Brn2 targets are mostly nucleosome-free, the Ascl1 targets are predominantly nucleosome-bound in MEFs.
(B) Left: Average enrichment of individual histone marks in MEFs based on Brn2 target sites in MEFs after infection with the BAM factors. Right: Heat maps of normalized tag densities representing MEFs chromatin marks at the same Brn2 targets sites. The signal is displayed within an 8 kb window centered around the binding sites.
(C) Left: Average enrichment of individual histone marks in MEFs based on Myt1l target sites in MEFs infected with BAM. Right: Heat maps of tag densities representing indicated histone marks at Myt1l targets sites. The signal is displayed within an 8 kb window centered around the binding sites.
(D) Average FAIRE-seq signal of uninfected MEFs at the genomic sites where Ascl1 (blue) or Brn2 (purple) is bound in NPCs. Dotted line represents the FAIRE-seq signal at random sites.
See also Figure S5.
To gain insight into the differential binding behaviors of Ascl1 and Brn2, we plotted the FAIRE signal from uninfected MEFs at the genomic sites bound by Brn2 in BAM-infected MEFs. We found that, in contrast to Ascl1, Brn2 targets sites in MEFs are predominantly nucleosome-free (Figure 5A). We next sought to characterize the histone marks associated with Brn2 targeting in fibroblasts. We generated H3K27ac ChIP-seq data in MEFs and utilized previously published ChIP-seq data sets for the H3K4me1, H3K4me3, and H3K27me3 histone marks. Consistent with our observation that Brn2 mostly binds accessible chromatin, Brn2 bound sites in fibroblasts are enriched for “active” marks, such as H3K4me1, H3K4me3 and H3K27ac (Figure 5B). The chromatin configurations of the Myt1l targets are also associated with open and active chromatin as demonstrated by the significant enrichment for the “active” marks H3K4me3 and H3K27ac at the 475 high-confidence bound sites (Figures 5C, 2A and 2B).
Given that, unlike Ascl1, Brn2 does not act as a pioneer factor, we then wondered whether the proper Brn2 targets in NPCs are perhaps closed in MEFs, which would explain why Brn2 is barred from binding there. Surprisingly, we found that, similar to the observed Brn2 bound sites in MEFs, the Brn2 target sites in NPCs are quite open in MEFs although to a slightly lesser degree than in the factual binding sites in MEFs (Figure 5D). As expected, the Ascl1 targets in NPCs are in a preferentially closed state in MEFs (Figure 5D and S5A). Thus, nucleosome occupancy is neither predictive for Ascl1 nor for Brn2 binding in MEFs.
A “trivalent” chromatin state predicts accessibility of Ascl1 and permissiveness for iN cell reprogramming among different cell types
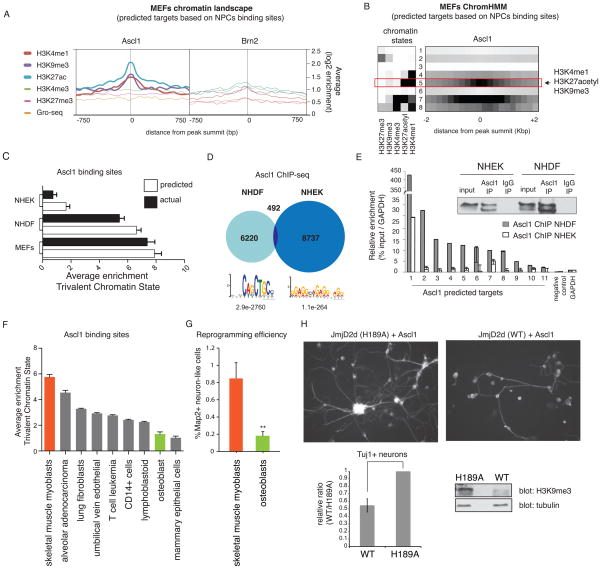
Pioneer factors can access nucleosomal DNA, yet only a very small proportion of all binding sites present in the genome are actually bound by any given transcription factor. Furthermore, transcription factors (including the well-characterized FoxA pioneer factors) bind different sets of target sites depending on the cellular context (Zaret and Carroll, 2011). Thus, additional parameters must exist that determine selective binding. We sought to explore whether a specific signature of pre-existing histone marks in MEFs is associated with Ascl1 target sites. Average enrichments values of the individual marks showed strong enrichment of H3K4me1, H3K27acetyl, and H3K9me3 at Ascl1 target sites based on binding in NPCs. In contrast, none of those histone marks were significantly enriched in MEFs at the sites where Brn2 is bound in NPCs (Figure 6A).
Figure 6. A trivalent chromatin state is characteristic and predictive of Ascl1 chromatin accessibility between different cell types.
(A) Average enrichment of individual histone marks in uninfected MEFs at the sites where Ascl1 (left) or Brn2 (right) is bound in NPCs. For each binding site, the signal is displayed +/− 750bp from peak summit.
(B) MEFs chromatin states based on ChromHMM analysis. Heat map of chromatin states enrichment within −2/+2 kb from Ascl1 binding site in NPCs. Arrow highlights most differentially enriched state between Ascl1 and Brn2 binding sites specified as state #5 (trivalent chromatin state = H3K4me1, H3K27acetyl, and H3K9me3).
(C) Average enrichment of trivalent chromatin state at predicted Ascl1 bound sites across cell types based on Ascl1 binding in NPCs (white columns). In black columns are represented the average enrichment for trivalent chromatin state at Ascl1 bound sites from Ascl1 ChIP-seq in MEFs, NHDF, and NHEK overexpressing cells. NHDF= normal human dermal fibroblasts; NHEK= normal human epidermal keratinocytes. Error bars = SEM.
(D) Venn diagram representing Ascl1 ChIP-seq peak overlap between targets in NHDF and NHEK cells. Bottom: Motif enriched in most confident thousand peaks.
(E) Left: Ascl1 ChIP-qPCR at predicted Ascl1 target genes in NHDF and NHEK cells overexpressing Ascl1 reveals substantially better binding in fibroblasts than keratinocytes. Right: Validation of equivalent pull-down efficiency of Ascl1 in NHEK and NHDF cells by ChIP followed by immunoblotting against Ascl1. Error bars = standard deviation of the ΔCT
(F) Average enrichment of trivalent chromatin state at predicted Ascl1 bound sites based on Ascl1 binding in NPCs across Encode cell types with chromatin data publicly available. In orange is the highest enrichment scores predicted for Ascl1-mediated reprogramming in human skeletal muscle myoblasts. In green is one of the lowest enrichment scores predicted for Ascl1-mediated reprogramming in human osteoblasts. Error bars = SEM.
(G) iN cell conversion efficiency expressed as ratio of neuronal, MAP2+ cells/ seeded cells in skeletal muscle myoblasts and osteoblasts infected with Ascl1, Brn2, Myt1l and NeuroD1. Measurements were carried out 20 days after induction of transgenes. A t-test was performed to compare reprogramming efficiencies of the two cell types (Error bars = SEM, n = 3 biological replicates, **p < 0.01).
(H) Characterization of Ascl1-mediated reprogramming upon global loss of H3K9me3 by overexpression of wild type (WT) or catalytically mutant (H189A) JmjD2d. Left: Tuj1 staining of MEFs expressing WT or H189A JmjD2d after Ascl1 induction for 11 days. Right: Fraction of Tuj1 positive neurons in WT relative to mutant (H189A) JmjD2d overexpressing MEFs. Bottom: Detection of H3K9me3 levels by immunoblot. Error bars = SEM, n = 3 biological replicates.
See also Figure S6.
We next devised a strategy to locate and quantify the trivalent chromatin state comprised of H3K4me1, H3K27ac, and H3K9me3. We wished to confirm the co-occurrence of these histone modifications in an unbiased manner since in particular, the repressive mark H3K9me3 may not be expected to co-exist with the active enhancer marks H3K4me1 and H3K27ac. We adapted a previously described Hidden Markov Model analysis tool ChromHMM (Ernst and Kellis, 2012). ChromHMM was used to define unique combinations of chromatin marks genome-wide in MEFs based on the five available histone modifications, and we then calculated the enrichment of each of these states in a set of user-specified genomic sites (Ernst and Kellis, 2012). This analysis revealed a specific MEF chromatin state (ChromHMM state 5) that is enriched at Ascl1 binding sites (Figure 6B). ChromHMM state 5 in MEFs represents the combination of high enrichment values for H3K4me1 and H3K27ac and low-to-intermediate enrichment levels of H3K9me3, which were also the three histone marks that showed a significant enrichment when averaged across all Ascl1 binding sites (in the following also referred to as “trivalent chromatin state”) (Figure 6B). Notably, the presence of H3K9me3 has been previously identified as “poised” enhancer mark with cell-type-specific control of enhancer activity (Zentner et al., 2011; Zhu et al., 2012). Among MEFs, NPCs, human fibroblasts and keratinocytes this trivalent state ranges from 0.34–1.7% of the genome. Of all HMM chromatin states, state 5 showed the highest enrichment at Ascl1 targets (Figure S6A–B and S6H–I). Sequential immunoprecipitation of mono-nucleosomes isolated from MEFs demonstrated the co-occurrence of the three chromatin states- H3K27ac, H3K4me1, and H3K9me3 on single nucleosomes, validating the existence of the trivalent state (Figure S6C).
Although the genome-wide Ascl1 occupancy is highly similar in MEFs and NPCs, there is a small fraction of sites bound by Ascl1 in NPCs that are not bound in MEFs. Remarkably, those sites have about two-fold lower enrichment for the trivalent chromatin state (Figure S6D). This finding is yet another correlative evidence that the trivalent chromatin state may favor access of Ascl1 to its target sites.
To address this hypothesis experimentally, we asked whether the presence of a trivalent chromatin state could predict Ascl1 binding in other cell types. We first calculated the enrichment for histone signatures at putative Ascl1 binding sites across distinct cell types, for which iN cell reprogramming efficiency was known, based on Ascl1 targets in NPCs. At Ascl1 targets, our analysis predicts an enrichment for a trivalent chromatin state in MEFs and human dermal fibroblasts (NHDF). Conversely, it predicts low levels in normal human keratinocytes (NHEK) (Figure 6C). Therefore, we tested whether Ascl1 can access its cognate sites in human dermal fibroblast vs. keratinocytes. Indeed, 48 hours after induction, the binding of Ascl1 to its predicted target sites was much higher in fibroblasts than in keratinocytes as determined by ChIP-qPCR, despite equivalent Ascl1 expression and IP efficiencies (Figure 6E). To verify these results in a global manner, we mapped Ascl1’s genome-wide target sites in NHDF and NHEK cells ectopically expressing Ascl1 by ChIP-seq (Figure 6D). As predicted by the trivalent state model, Ascl1 occupancy in NHDF is significantly enriched for the canonical Ascl1 E-box, indicating that it is on-target. In contrast in NHEK cells where Ascl1 sites lack the trivalent chromatin state, Ascl1 occupied almost completely different, off-target sites that lacked the motif. Only about 5.3% of the most confident Ascl1 bound sites in NHEK contained an E-box motif when conducted a de novo motif search. In accordance with these results, we have failed thus far to convert keratinocytes into iN cells using transcription factor combinations containing Ascl1 while fibroblasts and hepatocytes can be reprogrammed efficiently (Marro et al., 2011; Pang et al., 2011; Vierbuchen et al., 2010).
We extended our analysis to make predictions for 9 additional human cell types for which there was publicly available data for the same 5 chromatin marks (Figure 6F). We used ChromHMM software to define chromatin states in each cell type, and then calculated the enrichment for the trivalent state at predicted Ascl1 target sites. We found that human skeletal muscle myoblasts have strong enrichment for the trivalent chromatin state at Ascl1 sites, while human osteoblasts showed low levels. Consistent with our predictions, human skeletal muscle myoblasts are reprogrammed with efficiencies similar to reprogramming human fibroblasts when transduced with Ascl1, Brn2, Myt1l and NeuroD1 (Pang et al., 2011) while only very few iN cells formed from infected osteoblasts (Figure 6G and S6E). The ability to predict reprogramming across cell types of previously unknown potential provides strong support for the importance of the trivalent chromatin state in Ascl1-mediated reprogramming.
Conversely, experimental disruption of the trivalent state correlated with impaired MEF reprogramming to iN cells. We expressed JmjD2d, a histone demethylase that “erases” H3K9me3 or a catalytically inactive point mutant JmjD2d (H189A) (Krishnan et al., 2013; Shi et al., 2007; Whetstine et al., 2006; Zhu et al., 2012) in MEFs 2 days before Ascl1 infection. JmJD2d overexpression resulted in substantial demethylation and reduction of iN cells formed after 2 weeks (Figure 6H). The reduction of reprogramming may also be explained by other effects of JmjD2d. Finally H3K9me3 can undergo further dynamic changes during iN reprogramming (Figure S6F and S6G).
Ascl1 target genes are preferentially induced during reprogramming unlike Brn2 target genes
Next we wanted to explore the possible downstream mechanisms that mediate iN cell reprogramming initiated by expression of the BAM factors. To accomplish this, we first correlated Ascl1 binding with transcriptional changes. Compatible with the prevailing notion that Ascl1 acts primarily as transcriptional activator we found that most Ascl1 target genes are upregulated 48 hours after dox (Figure 7A and S7A). Of the 790 genes that significantly change expression by 48 hours, 143 genes are bound by Ascl1, and 133 of those are induced. In contrast, we did not find a similar correlation for genes associated with Brn2 target sites (Figure 7A). Only 42 genes of the 790 genes that change expression were bound by Brn2 in BAM-infected MEFs. Moreover, Ascl1 does not require Brn2 or Myt1l as a co-factor to induce its target genes, since its 1198 target genes are highly up-regulated on average with or without co-expression of Brn2 and Myt1l at 48h and, importantly, remain induced throughout the entire reprogramming process (Figure 7B). In contrast, Brn2 promotes fewer transcriptional changes with only subtle up-regulation of target genes (Figure 7A and 7C). In particular, we identified 454 MEF-specific Brn2 targets, which tend to be transiently induced but are eventually silenced in iN cells as well as in cortical neurons and NPCs (Figure 7C). From the small fraction of overlapping NPCs/MEFs Brn2 targets that actually contained a POU-motif we identified 87 genes that are upregulated faster, remain expressed during reprogramming and tend to be expressed in cortical neurons and NPCs (Figure 7C left panel). Thus, most Brn2 targets show only transient expression changes, but a small but distinct set of Brn2 target genes in MEFs does contribute to long term iN cell transcription.
iN cell reprogramming is mediated in part by the direct Ascl1 target gene Zfp238
Integrative analysis of our transcriptional data identified 25 transcription factors that were significantly upregulated by BAM factors and may mediate the iN cell reprogramming process. A literature survey revealed that as many as twelve of the 25 factors potentially play important roles in neural development. None of these twelve transcription factors could individually induce neuronal cells, nor improved iN efficiency when combined with Ascl1 (Figure 7D). However, when tested in combination with Myt1l one of the twelve factors, the Ascl1 target gene Zfp238, could induce neuron-like cells (Figures 2C, 7E–F). Although these cells expressed the neuronal markers Tuj1 and MAP2, their morphology appeared less mature, and the overall efficiency of their generation was highly reduced compared with Ascl1 and Myt1l (Figure 7E and 7F). Electrophysiological characterization showed that Zfp238/Myt1l-iN cells had less mature passive and active membrane properties than Ascl1/Myt1l-iN cells (Figure 7I and 7J). A smaller fraction of Zfp238/Myt1l-iN cells could generate single or repetitive action potentials (see pie charts in Figure 7I). While action potential thresholds and amplitudes were similar, the total numbers of action potentials in response to various amounts of current injections were significantly reduced in Zfp238/Myt1l-iN cells (Figure 7Ji-iii). Moreover, their resting membrane potential was depolarized, their capacitance decreased, and their input resistance increased compared to Ascl1/Myt1l-iN cells (Figure 7Jiv-vi). Na+ currents were not significantly different between the two conditions (Figure S7B and S7C). Thus, the single transcription factor Zfp238 mediates an important component of the transcriptional output of Ascl1 together with other critical downstream factors.
In order to identify these missing regulators, we measured the extent by which the remaining 11 candidates can improve the reprogramming of Zfp238/Myt1l-iN cells. We expressed Zfp238, Myt1l, and each of the 11 factors separately in MEFs and found that Lmo2, Rfx1, and Tcfl5 significantly improved the generation of iN cells (Figure 7D and 7G–H). These results suggest that these three transcription factors act in parallel to Zfp238 and together execute the BAM-induced program leading to iN cell formation (Figure S7D).
DISCUSSION
“On-target” pioneer factor activity of Ascl1
The dominant model of lineage reprogramming is transcription factor cooperativity based on factor co-occupancy and mutual positive feedback loops — a concept put forward based on iPS cell reprogramming (Jaenisch and Young, 2008). In fact a recent study confirmed this model by showing that Oct4, Sox2, Klf4, and c-Myc cooperatively bind to a vast number of the same enhancers in fibroblasts (Soufi et al., 2012). Three of the four factors were also shown to bind sites predominantly in a nucleosomal configuration, and hence were regarded pioneer factors. Importantly, though, the vast majority of binding sites were mislocalized as the proper targets appeared to be barred by large repressed chromatin domains enriched by H3K9me3 modification. The observed initial mistargeting of the reprogramming factors is a plausible explanation for the low efficiencies and slow kinetics of iPS cell reprogramming. By analogy it could be assumed that other types of lineage reprogramming also involve transcription factor cooperativity and positive feedback activation (Vierbuchen and Wernig, 2012).
In stark contrast, our investigation of transcription factor occupancy states in iN cell reprogramming has revealed a surprising asymmetry in the behavior of the three reprogramming factors. At the top of the hierarchy stands Ascl1, which we found could access nucleosomal DNA and – unlike Oct4 – also immediately bound its authentic neuronal target genes across the fibroblast genome. The term “pioneer factor” has been used to describe transcription factors that can bind nucleosomal DNA as single factors as opposed to regular transcription factors that require cooperation (Zaret and Carroll, 2011). Here we propose the term “on-target pioneer factor” to refer to pioneer factors like Ascl1, that bind their cognate lineage-specific targets whether genomic sites are freely available or nucleosome-bound in heterotypic cell types. Supported by structural models, classic pioneer factors have been proposed to access the DNA from one side leaving the opposite site available for histone interaction (Soufi et al., 2012; Zaret and Carroll, 2011). In contrast, Ascl1 – like any bHLH transcription factor – binds as heterodimer the two neighboring major groves of DNA in a fork-like pattern with its α-helical basic domain excluding the possibility of simultaneous nucleosome binding (Ma et al., 1994). Therefore the mechanism of nucleosomal DNA access must be different between pioneer factors reported to date. Assuming nucleosomes are not static and pioneer factors have a higher DNA affinity than histones, once target sites are bound, they might be barred from histone wrapping.
MyoD, one of the earliest reprogramming factors identified, is also a bHLH factor and was shown to create DNAse hypersensitive sites at myogenic promoters and eliminate nucleosome phasing in the Myogenin promoter in MEFs suggesting pioneer factor activity (Gerber et al., 1997). Whether MyoD can also act as “on target” pioneer factor requires more investigation. Its binding pattern in MEFs seems similar to muscle cells, but there is only a 30% overlap between binding in MEFs and the neurogenic P19 cell line after acute expression (Cao et al., 2010; Fong et al., 2012).
In contrast, Brn2 and Myt1l clearly do not have pioneer factor activity as they access mostly open chromatin and play a less dominant role during the early stages of iN cell reprogramming. Brn2 is predominantly localized to different sites than in NPCs. Surprisingly, this mislocalization could not simply be explained by differential chromatin accessibility as the “proper” neural Brn2 targets were overall in an accessible state in MEFs. At least a portion of the mislocalized targets could be explained since Ascl1 recruited Brn2 to many of its targets.
The “on-target” pioneer property of Ascl1 versus “off-target” character of the iPS cell factors may very well explain the dramatically higher iN cell reprogramming efficiency of ~20% within 2 weeks (Vierbuchen et al., 2010). “On-target” capability could become a valuable criterion to identify the most efficient reprogramming factors to induce other cell lineages especially if no candidate factor has reprogramming activity on its own. It would be interesting to explore whether the FoxA and GATA pioneer factors used for hepatocyte and cardiac reprogramming are also “on-target” factors (Ieda et al., 2010; Huang et al., 2011).
A chromatin signature predicts reprogramming capacity
While Ascl1 acted as on target pioneer factor in fibroblasts, it did not do so in keratinocytes. Therefore, chromatin features other than nucleosome occupancy must influence this factor’s access to chromatin. We discovered a unique “trivalent” chromatin signature - comprised of H3K4me1, H3K27ac, and H3K9me3 - that predicts permissiveness for Ascl1 occupancy and iN cell reprogramming among various non-neural cell types. This permissive chromatin signature should be highly useful in choosing recipient cells for neural reprogramming ex vivo or in vivo for therapeutic purposes.
The enrichment of a repressive mark was surprising, especially in light of the recent finding that H3K9 methylation sites are occluded for the iPS cell reprogramming factors (Soufi et al., 2012). However, the co-localization of H3K4me1 and H3K9me3 has been suggested a poised state for cell-type specific enhancers in ESCs that are later activated, similar to the H3K4me3 and H3K27me3 “bivalent” domains (Bernstein et al., 2006; Zhu et al., 2012). Hence, the trivalent signature described here may represent a remnant form of such early embryonic poised states.
While the discovery of a pre-existing defined chromatin configuration that is associated with Ascl1 binding is intriguing, it is also evident that this trivalent mark cannot explain all Ascl1 binding. Presumably non-chromatin factors, such as yet unknown co-factors may be expressed to different levels in different cell types impacting binding and transcriptional activity of the reprogramming factors. In addition, only a small fraction of the thousands of binding sites could be functionally relevant and the accessibility to those sites may explain the vast majority of the variation in reprogramming efficiencies between cell types despite similar global responses.
A distinct cascade of transcription factors mediating neuronal reprogramming
The interplay of Ascl1 and Zfp238 and other targets is yet another aspect the hierarchical organization of iN cell reprogramming. First, based on binding and transcriptional data Ascl1 is the direct upstream activator of Zfp238. Second, Zfp238 executes an important but only a part of the lineage-specification program of Ascl1 as we found three additional transcription factors that cooperate with Zfp238 to generate iN cells. Hence, out of the thousands of target genes, only a handful appear to be critical for neuronal reprogramming. This conclusion may be helpful to enable reprogramming of thus far resistant cell types such as keratinocytes. Collectively, our results suggest that iN cell reprogramming is a powerful tool to interrogate transcription factor function and to uncover transcriptional networks in neuronal lineage specification.
EXPERIMENTAL PROCEDURES
Cell derivation, culture and viral production
TauEGFP MEFs were isolated and lentivirus was produced as previously described (Vierbuchen et al., 2010). ES cell-derived NPCs (line NS5) were obtained from A. Smith, Cambridge, UK and cultured as reported (Conti et al., 2005). To establish Ascl1-inducible MEFs we first generated a Rosa26 CAGGS-M2rtTA-T2A-puro ES cell line by homologous recombination and infected this with a TetO-flagAscl1 lentivirus. A subclone showing robust induction was then injected into blastocysts to generate chimeric embryos for MEF isolation. More detailed information can be found in the supplemental information.
Expression analysis, Chromatin Immunoprecipitation and FAIRE-Seq
Total RNA was isolated and Poly-A-selected. Libraries were prepared and sequencing reads (100bp) were generated on Hi-Seq 2000 Illumina platforms. Paired-end reads were aligned to the mouse reference sequence NCBI Build 37/mm9. Expression levels of RefSeq annotated genes were calculated in unit of fragments per kilobase of exon model per million mapped fragments (FPKM). Differential expression analysis was performed using Students’ t-Test function “t.test” in R, and genes with p-value<0.05 and at least 2-fold expression change were defined as significant. Gene ontology analysis was performed using DAVID (david.abcc.ncifcrf.gov). ChIP-qPCR and ChIP-seq were carried out in Ascl1- and BAM-infected MEFs 48 hours after dox, and in uninfected or Ascl1-infected NPCs 18 hours after induction. Cells were infected with a pool of Ascl1, V5- or FLAG-tagged Brn2, and V5- or FLAG-tagged Myt1l, and the transgenes were induced with dox the day after. Antibodies used and detailed experimental procedures including ChIP co-IP and primary computational analysis can be found in the supplemental information. Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE-seq) was followed as previously described (Giresi et al., 2007).
Computational Analysis
To identify chromatin states we used the ChromHMM software (version 1.06) according to (Ernst and Kellis, 2012). A self-developed script was used to calculate the percentage of genomic bases within a +/−200bp window around the top 1000 transcription factor peak summits that were occupied by the given state. To obtain the normalized frequency of Ascl1 peaks in each state, the total number of Ascl1 peaks in each state was divided by the length of the corresponding state’s segmentation size, and multiplied by 1 Megabases (Mb). The Principal component analysis was performed using the PCA package in Microsoft Excel statistical add-in “XLSTAT”.
Electrophysiology
Whole-cell patches were established at room temperature using manipulators and amplifiers controlled by Clampex 10 Data Acquisition Software (Molecular Devices). Resting membrane potential, input resistance, and capacitance was recorded for every cell. The current clamp mode was used to record action potentials. We set the initial resting membrane potential (Vrest) to −60 mV using a small, constant holding current, and applied current-pulses of 10–50 pA with a step-size of 10 pA, to test the ability to generate action potentials. Statistical comparisons between cumulative plots were made using K-S (Kolmogorov-Smirnov) test.
Supplementary Material
HIGHLIGHTS.
Ascl1 has pioneer activity, accessing closed chromatin to allow other factors to bind
Unlike other pioneer factors Ascl1 binds its physiologic neural targets in fibroblast
A trivalent chromatin domain predicts iN reprogramming ability in other cell types
Zfp238 is a direct Ascl1 target and critical mediator of iN cell reprogramming
Acknowledgments
We would like to thank M.C. Tsai, M. Kareta, and P. Lovelace for invaluable initial help with sequencing protocols, cell culture, and cell sorting. We would also like to thank other members of the Chang and Wernig labs for discussions and support. This work was supported by NIH (RC4NS073015 to M.W., H.Y.C.), NSF (to O.L.W.), a predoctoral fellowship by CIRM (to T.V.). H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute. M.W. is a New York Stem Cell Foundation-Robertson Investigator.
Footnotes
ACCESSION NUMBERS
The GEO repository accession number for the ChIP- and RNA-sequenced tags reported in this paper is GSE43916. The accession numbers of publicly available datasets used in this study are reported in the supplemental information.
Supplemental information includes Extended Experimental Procedures and seven figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefroid EJ, Bourguignon C, Hollemann T, Ma Q, Anderson DJ, Kintner C, Pieler T. X-MyT1, a Xenopus C2HC-type zinc finger protein with a regulatory function in neuronal differentiation. Cell. 1996;87:1191–1202. doi: 10.1016/s0092-8674(00)81815-2. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DS, Guillemot F. Old and new functions of proneural factors revealed by the genome-wide characterization of their transcriptional targets. Cell Cycle. 2011;10:4026–4031. doi: 10.4161/cc.10.23.18578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, Gentleman RC, Tapscott SJ. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18:662–74. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, Sun Y, Sanzone S, Ying QL, Cattaneo E, Smith A. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez MH, Ayoub AE, Rakic P. POU-III Transcription Factors (Brn1, Brn2, and Oct6) Influence Neurogenesis, Molecular Identity, and Migratory Destination of Upper-Layer Cells of the Cerebral Cortex. Cereb Cortex. 2012 doi: 10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong AP, Yao Z, Zhong JW, Cao Y, Ruzzo WL, Gentleman RC, Tapscott SJ. Genetic and epigenetic determinants of neurogenesis and myogenesis. Dev Cell. 2012;22:721–735. doi: 10.1016/j.devcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11(4):436–50. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587– 594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475(7356):386–9. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Hudson LD. Novel member of the zinc finger superfamily: A C2-HC finger that recognizes a glia-specific gene. Mol Cell Biol. 1992;12:5632–5639. doi: 10.1128/mcb.12.12.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S, Trievel RC. Structural and functional analysis of JMJD2D reveals molecular basis for site-specific demethylation among JMJD2 demethylases. Structure. 2013;21:98–108. doi: 10.1016/j.str.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Lo LC, Johnson JE, Wuenschell CW, Saito T, Anderson DJ. Mammalian achaete-scute homolog 1 is transiently expressed by spatially restricted subsets of early neuroepithelial and neural crest cells. Genes Dev. 1991;5:1524–1537. doi: 10.1101/gad.5.9.1524. [DOI] [PubMed] [Google Scholar]
- Ma PC, Rould MA, Weintraub H, Pabo CO. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- Ma Q, Fode C, Guillemot F, Anderson DJ. Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marro S, Pang ZP, Yang N, Tsai MC, Qu K, Chang HY, Sudhof TC, Wernig M. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell. 2011;9:374–382. doi: 10.1016/j.stem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada Y, Hunsaker TL, Henke RM, Johnson JE. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus neuronal cell-type specification. Development. 2004;131:1319–1330. doi: 10.1242/dev.01008. [DOI] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, Fujita R, Yamashita T, Angulo S, Rhinn H, Rhee D, Doege C, Chau L, Aubry L, Vanti WB, et al. Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell. 2011;146:359–371. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and Impediments of the Pluripotency Reprogramming Factors’ Initial Engagement with the Genome. Cell. 2012 doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugitani Y, Nakai S, Minowa O, Nishi M, Jishage K, Kawano H, Mori K, Ogawa M, Noda T. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 2002;16:1760–1765. doi: 10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Wernig M. Molecular roadblocks for cellular reprogramming. Mol Cell. 2012;47:827–838. doi: 10.1016/j.molcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Wernig M. Direct lineage conversions: unnatural but useful? Nat. Biotechnol. 2011;29:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner JA, Chun J. Png-1, a nervous system-specific zinc finger gene, identifies regions containing postmitotic neurons during mammalian embryonic development. J Comp Neurol. 1997;381:130–142. doi: 10.1002/(sici)1096-9861(19970505)381:2<130::aid-cne2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.