Abstract
Maintenance of a reduced body weight is accompanied by a decrease in energy expenditure beyond that accounted for by reduced body mass and composition, as well as by an increased drive to eat. These effects appear to be due— in part—to reductions in circulating leptin concentrations due to loss of body fat. Gut microbiota have been implicated in the regulation of body weight. The effects of weight loss on qualitative aspects of gut microbiota have been studied in humans and mice, but these studies have been confounded by concurrent changes in diet composition, which influence microbial community composition. We studied the impact of 20% weight loss on the microbiota of diet-induced obese (DIO: 60% calories fat) mice on a high-fat diet (HFD). Weight-reduced DIO (DIO-WR) mice had the same body weight and composition as control (CON) adlibitum (AL) fed mice being fed a control diet (10% calories fat), allowing a direct comparison of diet and weight-perturbation effects. Microbial community composition was assessed by pyrosequencing 16S rRNA genes derived from the ceca of sacrificed animals. There was a strong effect of diet composition on the diversity and composition of the microbiota. The relative abundance of specific members of the microbiota was correlated with circulating leptin concentrations and gene expression levels of inflammation markers in subcutaneous white adipose tissue in all mice. Together, these results suggest that both host adiposity and diet composition impact microbiota composition, possibly through leptin-mediated regulation of mucus production and/or inflammatory processes that alter the gut habitat.
INTRODUCTION
Interactions between modern environments and strong biological mechanisms favoring energy storage have contributed to a dramatic increase in the prevalence of obesity over the past three decades (1). In humans and rodents, responses to weight reduction include reduced energy expenditure per unit of metabolic mass and increased hunger (2–6). These responses favor recidivism to obesity (7).
Recent studies in rodents and humans implicate gut microbiota in energy homeostasis (reviewed in ref. 8). Sequencebased studies have highlighted differences in gut microbial community composition between obese and lean humans (9) and mice (10,11). Altered gut microbial communities can impact host body weight in several ways. For example, compared to lean animals, mice rendered obese either by a highfat diet (HFD) or by leptin deficiency (ob/ob), harbor a gut microbiota enriched in the phylum Firmicutes, and depleted in Bacteroidetes (10,11). Metagenomic and biochemical analyses and microbiota transplantation experiments indicate that the obesity-associated microbiota has an enhanced ability to extract energy from a given diet (10,12). In this context, “extraction” means an increased amount of short chain fatty acids (a by-product of bacterial catabolism of dietary fiber; nonstarch polysaccharides, and other plant components) in the cecum and decreased fecal gross energy content (measured by bomb calorimetry) indicative of increased absorption of short chain fatty acids by the host (10). Finally, specific microbiota may trigger low grade inflammation that reduces insulin sensitivity and may affect body weight by reducing neuronal (e.g., hypothalamic) sensitivity to circulating hormones such as leptin and insulin (13,14).
Turnbaugh et al. reported strong effects of a HFD on the composition of the microbiota in mice (12). Since the switch to a HFD resulted in host weight gain, it is unclear if alterations in the gut microbiota were due to dietary changes, to host adiposity, or to interactions between diet and adiposity. To show that a HFD per se can cause an alteration in the microbiota, Hildebrandt and colleagues used RELMβ KO mice that become only slightly overweight when fed a HFD (15), yet still have significantly higher body weight and body fat content than low fat fed wild-type mice.
In the studies reported here we examined the effects of weight loss on the gut microbiota in the context of high and low fat diets (60% and 10% of calories derived from fat, respectively), while controlling for body weight. We compared the microbiotas of four groups of C57BL/6J mice: diet-induced obese mice (DIO-AL) and control (10% fat) diet-fed mice (CON-AL) given ad-libitum (AL) access to these diets, and mice weight-reduced to 20% below initial weight (weight-reduced DIO (DIO-WR) and weight-reduced control diet (CON-WR), respectively). The DIO-WR mice had body weights and body compositions similar to those of the CON-AL mice. This design allowed us to: (i) Compare diet effects on gut microbial community composition independent of body weight (DIO-WR vs. CON-AL); (ii) Compare the effects of weight loss in both lean and obese mice (DIO-WR vs. CON-WR); and (iii) Assess correlations between circulating leptin concentrations, inflammation marker expression levels in white adipose tissue, and the relative abundance of various gut bacteria.
METHODS AND PROCEDURES
Animals
The animals used in this study are described in detail in Ravussin et al. 2011 (6). Thirty-two 18-week old C57BL/6J-male mice were obtained from the Jackson Laboratory (Bar Harbor, ME); 16 (DIO) had been fed Research Diets,. D12492i (60% kcal fat, 20% kcal protein), and 16 (CON) had been fed Research Diets, D12450Bi (10% kcal fat, 20% kcal protein) from 6 weeks of age. Mice were individually housed upon arrival. Animals from both diet groups were randomized to remain on the AL diets or to be calorically restricted to decrease their body weight by 20% over a 1–2 week period by twice daily feeding of reduced (50%) quantities of their respective diets. The feeding regimen was then altered to keep each individual mouse weight stable 20% below their initial weight (WR). This reduced weight was maintained for 23 additional weeks to avoid “carryover” effects of the negative energy balance state required for weight loss, and to permit additional physiological analyses not reported here. All mice had AL access to water containing no bacterostatic agents throughout the entire experiment. Fat mass (FM) and fat free mass were assessed by time-domain-nuclear magnetic resonance (Minispec Analyst AD; Bruker Optics, Silberstreifen, Germany). Mice were killed after a 4 h fast during deep anesthesia. The cecum (among other organs) was removed from each mouse. Cecal content was aseptically removed, flash frozen, and stored at −80 °C until processing. The protocol was approved by the Columbia University Institutional Animal Care and Use Committee.
Inflammation markers
qRT-PCR in inguinal white adipose tissue
Inguinal fat pads were removed, flash frozen in liquid N2, and stored at −80 °C. RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA) (including the DNAse purification step) and reverse transcribed with the Transcriptor First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN) using random primers. To quantify transcript levels in the various organs, quantitative reverse transcriptase PCR (qRT-PCR) was performed on a Roche 480 LightCycler using Syber green (Roche) and normalized to cyclophilin b and presented as arbitrary units. Primers were as follows: Saa3 forward: AGCGATGCCAGAGAGGCTGTTC, reverse: AGCAGGTCGGAAGTGGTTGG; Pail forward: TCCTCATCCTGCCTAAGTTCTC, reverse: GTGCCGCDCTCGTTTACCTC; F4/80 forward: CTTTGGCTATGGGCTTCCAGTC, reverse: GCAAGGAGGACAGAGTTTATCGTG; Slc25α25 forward: GGGTGTCAAGATCTCGGAACA, reverse: GTAGTCCCTCCACTCGTTCCA; Angpl4 forward: TTCCAACGCCACCCACTTACA, reverse: ACCAAACCACCAGCCACCAGA; Tnfα, forward: CCAGACCCTCACTAGATCA, reverse: CACTTGGTGGTTTGCTACGAC; Il10, forward: GCTCTTACTGACTGGCATGAG, reverse: CGCAGCTCTAGGAGCATGTG; DioII, forward: GCTGCGCTGTGTCTGGAA, reverse: TGGAATTGGGAGCATCTTCAC; iNos, forward: AATCTTGGAGCGAGTTGTGG, reverse: CAGGAAGTAGGTGAGGGCTTG, Cdllc, forward: CCTACTTTGGGGCATCTCTTTG, reverse: GCACCTCTGTTCTCCTCCTCTC.
Leptin assay
Following a 4 h fast on the day of killing, mice were bled retro-orbit-ally. Blood for leptin assays was allowed to clot for lh at room temperature, centrifuged l0 min. at l,000g at 4°C, and serum was collected and frozen at −80 °C until time of assay. Leptin was assayed using the Quantikine ELISA kit (R&D Systems, Minneapolis, MN).
Cecal DNA extraction
Frozen cecal samples were ground under liquid N2; a subsample of ~100 mg was used for whole community DNA extraction (11): A 100 mg aliquot of each homogenized sample was suspended while frozen in a solution containing 500 µl of DNA extraction buffer ((200 mmol/l Tris (pH 8.0), 200 mmol/l NaCl, 20 mmol/l EDTA), 210 µl of 20% SDS, 500 µl of a mixture of phenol:chloroform:isoamyl alcohol (25:24:1)), and 500 µl of a slurry of 0.1-mm-diameter zirco-nia/silica beads (BioSpec Products, Bartlesville, OK). Microbial cells were then lysed by mechanical disruption with a bead beater (BioSpec Products) set on high for 2 min (22°C), followed by extraction with phenol:chloroform:isoamyl alcohol, and precipitation with isopropanol. The quantity and quality of purified DNA was assessed using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Eugene, OR) and a plate reader.
16S rRNA gene PCR amplification and sequencing
16S rRNA genes were amplified from each sample using a composite forward primer and a reverse primer containing a unique 12-base barcode (16) which was used to tag PCR products from respective samples (17). We used the forward primer 5′-GCCTTGCCAGCCCGCTCAGTCAGAGTTTGATCCTGGCTCAG-3′: the italicized sequence is 454 Life Sciences primer B, and the bold sequence is the broadly conserved bacterial primer 27E The reverse primer used was 5′-GCCTCCCTCGCGCCATCAGNNNNNNNNNNNNCATGCT GCCTCCCGTAGGAGT-3′: the italicized sequence is 454 Life Sciences primer A, and the bold sequence is the broad-range bacterial primer 338R. NNNNNNNNNNNN designates the unique 12-base barcode used to tag each PCR product, with “CA” inserted as a linker between the barcode and rRNA gene primer. PCR reactions consisted of HotMaster PCR mix (Eppendorf, Westbury, New York), 200 µmol/l of each primer, 10–100 ng template, and reaction conditions were 2 min at 95 °C, followed by 30 cycles of 20 s at 95 °C, 20 s at 52 °C, and 60 s at 65 °C on an Eppendorf thermocycler. Three independent PCRs were performed for each sample, combined and purified with Ampure magnetic purification beads (Agencourt Bioscience, Beverly, MA), and products visualized by gel electrophoresis. No-template extraction controls were analyzed for absence of visible PCR products. Products were quantified using Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen). A master DNA pool was generated from the purified products in equimolar ratios to a final concentration of 21.5 ng ml−1. The pooled products were sequenced using a Roche 454 FLX pyrosequencer at Cornell University’s Life Sciences Core Laboratories Center. Data have been deposited in GenBank under SRA022795.
statistical analysis of mouse phenotypes
Body weights, FM, leptin, and inflammation marker levels (Table 1) are expressed as arithmetic means ± s.e.m. Statistical analyses were performed using JMP (version 7; SAS, Cary, NC). Two-way ANOVAs were conducted using diet (DIO or CON) and treatment (WR or AL) as grouping variables with Tukey post-hoc ANOVA. T-tests were conducted when directly comparing phenotypes of DIO-WR and CON-AL mice using JMP (version 7; SAS, Cary, NC). All statistical tests were two-tailed.
Table 1.
Body weights, fat mass, leptin, and inflammation marker levels
| DIO diet |
CON diet |
Diet | Treatment | Diet × Treatment |
|||
|---|---|---|---|---|---|---|---|
| AL (n = 7) | WR (n = 8) | AL (n = 8) | WR (n = 8) | ||||
| Dataa | |||||||
| Body weight (g) | 52.0 ± 1.0c | 33.6 ± 1.0d | 32.2 ± 1.1d | 22.0 ± 0.6e | F = 292.5** | F = 243.9** | F = 20.2** |
| Fat-free mass (FFM, g) | 23.5 ± 0.5c | 20.0 ± 0.3d | 20.3 ± 0.3d | 15.1 ± 0.4e | F = 111.4** | F = 135.4** | F = 5.4* |
| Fat mass (FM, g) | 20.1 ± 1.0c | 7.0 ± 0.6d | 5.1 ± 0.7d | 1.8 ± 0.1e | F = 230.6** | F = 151.8** | F = 53.3** |
| Leptin (ng/ml) | 121.7 ± 14.9c | 25.1 ± 2.9d,* | 14.0 ± 3.6d | 9.0 ± 1.3d | F = 75.8** | F = 51.0** | F = 41.4** |
| mRNA levelsb | |||||||
| F4/80 | 1.06 ± 0.06c | 1.06 ± 0.09c | 1.0 ± 0.08c | 0.58 ± 0.05d | F = 14.4** | F = 8.9** | F = 9.2** |
| Slc25a25 | 1.08 ± 0.06c | 1.01 ± 0.06c | 0.63 ± 0.06d | 0.60 ± 0.04d | F = 40.9** | F = 0.6 | F = 0.1 |
| DioII | 1.34 ± 0.46c | 0.06 ± 0.03d | 0.87 ± 0.26c,d | 1.64 ± 0.19c | F = 4.1 | F = 0.9 | F = 14.3** |
| ll10 | 0.87 ± 0.14 | 1.63 ± 0.23 | 1.17 ± 0.24 | 0.99 ± 0.23 | F = 0.6 | F = 1.9 | F = 4.7* |
| Tnfα | 1.07 ± 0.23 | 1.64 ± 0.15 | 1.58 ± 0.14 | 1.36 ± 0.19 | F = 0.5 | F = 1.0 | F = 5.1* |
| iNos | 1.29 ± 0.10c | 0.79 ± 0.08d,e | 0.72 ± 0.03e | 1.06 ± 0.10c,d | F = 3.6 | F = 1.0 | F = 28.7** |
| Cd11c | 1.11 ± 0.16c | 0.86 ± 0.09d | 0.86 ± 0.08d | 0.71 ± 0.08d | F = 3.7 | F = 3.6 | F = 0.2 |
| Saa3 | 2.2 ± 0.55c | 0.89 ± 0.34d | 0.03 ± 0.02e | 0.02 ± 0.02e | F = 23.5** | F = 4.4* | F = 4.3* |
| Pai1 | 1.81 ± 0.29c | 1.25 ± 0.24c | 0.1 ± 0.05e | 0.18 ± 0.03e | F = 54.8** | F = 1.6 | F = 2.8 |
| Angptl4 | 1.80 ± 0.23c | 0.72 ± 0.08d,e | 1.07 ± 0.09d | 0.38 ± 0.12e | F = 15.6** | F = 42.6** | F = 2.1 |
Body weight, body composition, and circulating leptin concentrations (mean ± s.e.m.): previously published data (6).
mRNA levels (normalized to cyclophilin b and presented as arbitrary units) for inflammation markers measured by quantitative reverse transcriptase PCR in inguinal fat pads (mean ± s.e.m.). Data not marked by same letter are significantly different by two-way ANOVA with Tukey honestly significant difference post hoc analysis. Direct t-tests were conducted for body weight and body composition of weight-reduced diet-induced obese (DIO-WR) and ad-libitum control diet-fed mice (CON-AL) (*P < 0.05).
Significance of two-way ANOVA factors on the right hand side are denoted by *P < 0.05 and
P < 0.01.
16s rRNA gene sequence analysis
Sequences generated from pyrosequencing barcoded 16S rRNA gene PCR amplicons were quality filtered. Sequences were removed if they were shorter than 200 nucleotide, longer than 1,000 nucleotide, contained primer mismatches, ambiguous bases, uncorrectable barcodes, or homopolymer runs in excess of six bases. The remaining sequences were denoised (18) and analyzed using the open source software package Quantitative Insights Into Microbial Ecology (QIIME (19)). 16S rRNA gene sequences were assigned to operational taxonomic units (OTUs) using UCLUST with a threshold of 97% pair-wise identity, then classified taxonomically using the Ribosomal Database Project (RDP) classifier 2.0.1. Two highly abundant OTUs (716 and 303) were not classified beyond the phylum level (Firmicutes) with this method. For these two noteworthy OTUs, we used BLASTn against the NCBI nonredundant database, which yielded 98% and 97% ID matches to a 800 bp 16S rRNA gene sequence of a bacterium that has not been cultured (accession number FJ836349). This matched sequence was classified as belonging to the genus Allobaculum, family Erysipelotrichaceae (Firmicutes) with 100% confidence in RDP (the lack of a match with the shorter fragment is likely due to the many regions of low complexity in the short fragment which, when broken into random 7 base pairs words by the algorithm, leads to incongruent classifications).
For tree-based analyses, a single representative sequence for each OTU was aligned using PyNAST (20), then a phylogenetic tree was built using FastTree. The phylogenetic tree was used for measuring the α-diversity (phylogenetic diversity, PD) and β-diversity (using unweighted UniFrac (21)) of samples. Student’s t-tests were conducted and P values corrected for multiple comparisons. The “nearest shrunken centroid” method was used to identify OTUs that are specifically over (or under)-represented in a given category (diet, treatment, or diet-treatment combinations). The amount of shrinkage was chosen in order to minimize the overall misclassification error. The analysis was performed using the Predictive Analysis of Microarrays package under R software.
RESULTS
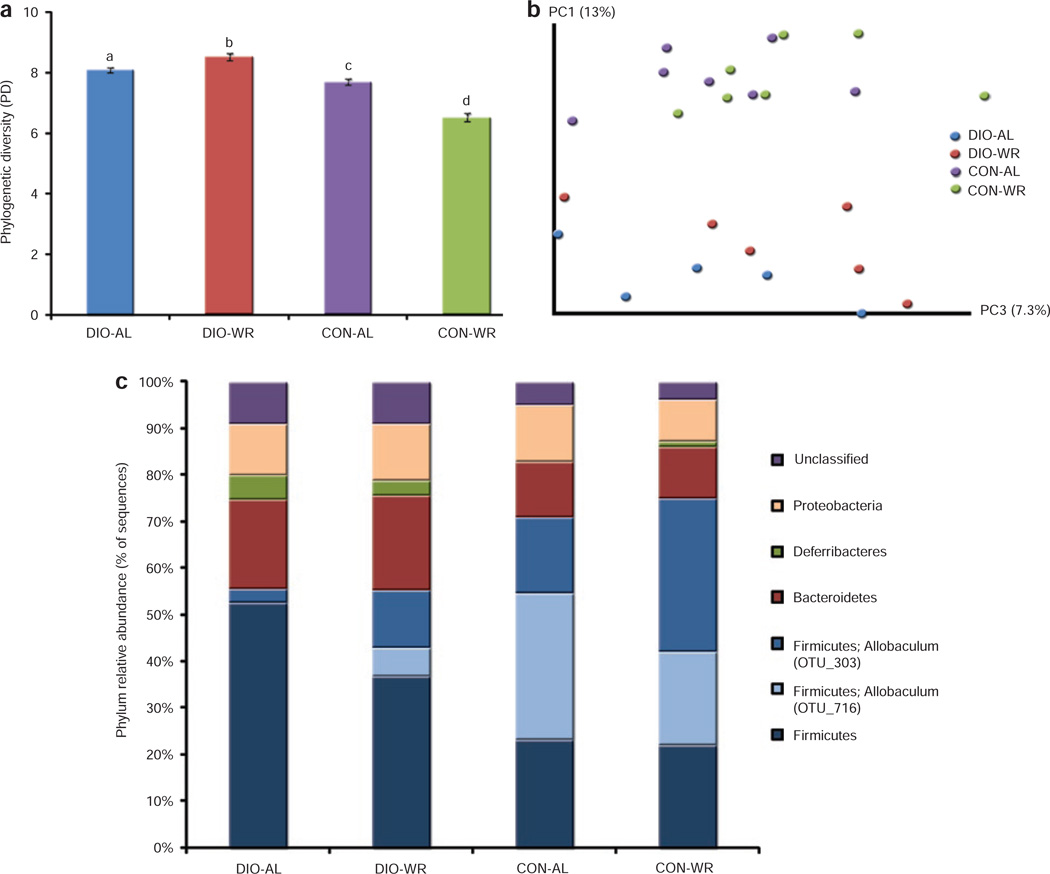
The denoised sequence library comprised 1,276 distinct OTUs (from >300,000 reads). Measures of α-diversity reflect phylogenetic richness in each sample; we measured phylogenetic diversity or PD, a phylogenetic-tree based measure of diversity calculated as the tree-branch length present in each sample (Figure 1a). The average PD of each treatment group was significantly different from the others, and DIO mice had higher PDs than the CON mice (P < 0.05; Student t-test, P values corrected for multiple comparisons). Bacterial communities of DIO-WR mice had the highest PD. Interestingly, the effect of weight reduction (i.e., WR vs. AL) had opposite effects on PD for the two diets: in the DIO mice the PD increased with weight reduction (t = 3, P value = 0.004), while the PD in CON mice declined with weight reduction (t = 6.7, P value = 7.31 × 10−10). Finally, DIO-WR mice microbiotas had a higher PD than CON-AL mice (t = 5.6, P value = 1.4 × 10−7) despite equivalent body weights and body composition.
Figure 1.
Effects of diet and weight reduction on the gut microbiota. (a) Phylogenetic Diversity (PD) of the cecal samples from the four groups of mice (mean ± s.e.m. compared by two-way ANOVA) and (b) Principal Coordinates Analysis (PCoA) plot of the unweighted UniFrac distances. PC1 and PC3 values for each mouse sample are plotted; percent variation explained by each PC is shown in parentheses: DIO-AL, diet-induced obese mice: blue; DIO-WR, weight-reduced DIO: red; CON-AL, control diet-fed mice: purple; CON-WR, weight-reduced CON: green. (c) Relative abundances of the different phyla in each of the groups. The phylum Firmicutes was broken down into OTU 303, OTU 716 (which are both classified as Allobaculum), and all other Firmicutes that did not fall into these two OTUs.
Overall effects of diet
We performed a Principal Coordinates Analysis (PCoA) on the unweighted UniFrac distances between samples to determine to what extent diet (i.e., DIO and CON) and treatment (i.e., WR or AL) affected gut microbial community diversity (21). Figure 1b shows a clear separation between the diets when principal coordinates 1 and 3 are plotted. In the DIO mice, the AL (blue dots) and WR (red dots) weight states can be distinguished, but such differences cannot be appreciated in the control mice between AL (purple dots) and WR (green dots) weight states. Globally, these results indicate that different diets promote different bacterial community diversity, and that weight reduction affects the gut community composition of DIO (60% fat) mice but not that of mice fed a 10% fat control diet (CON-WR).
Figure 1c summarizes the relative abundances of bacterial phyla in the different mouse groups. CON mice have greater abundance of Firmicutes than DIO mice: this difference reflects the dominance of two OTUs classified as the genus Allobaculum. Mice eating a HFD (DIO-AL and DIO-WR) have greater abundances of Firmicutes (excluding Allobaculum OTUs), and lower abundances of Allobaculum OTUs, when compared to animals fed the control diet (CON-AL and CON-WR). Bacteroidetes levels are elevated in all mice ingesting the high fat diet (DIO-AL and DIO-WR) when compared to CON-AL and CON-WR mice. DIO-AL and DIO-WR mice also have a higher abundance of Deferribacteres due to the presence of Mucispirillum.
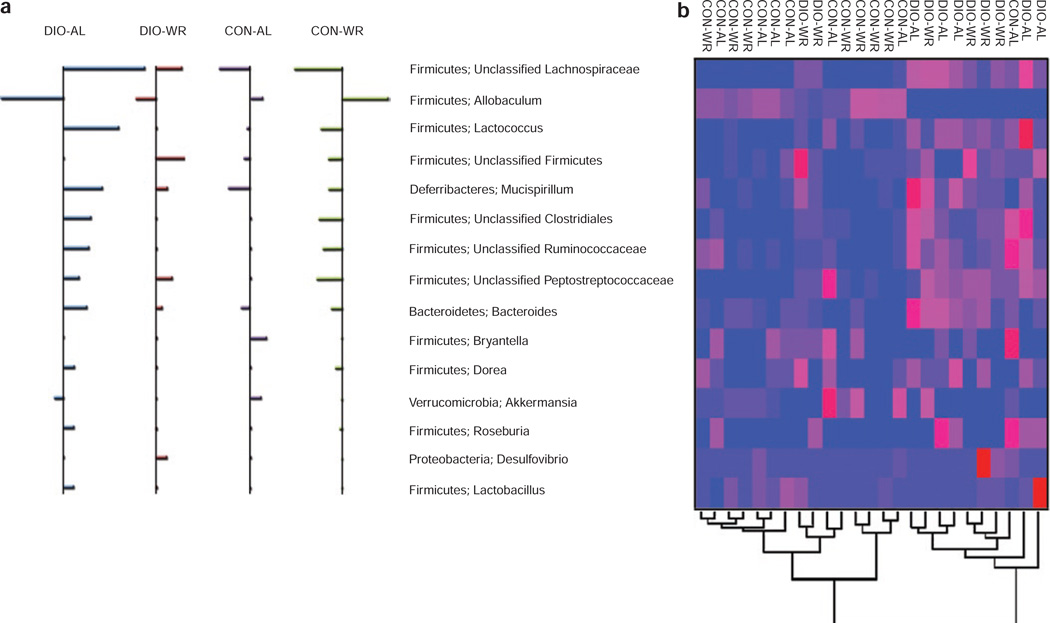
We performed a nearest shrunken centroid classification analysis to determine which OTUs account for differences in composition of the gut microbial community (22). In addition, this analysis assesses how well a mouse microbiota is assigned to its treatment group based on its composition. In this analysis as well, the two diets are very well separated: the class error rate between the two diets is very low (P = 0.08, two mice out of 25 are misclassified). However, when analyzing diet in the context of treatment, it is not possible to distinguish between CON-AL and CON-WR (misclassification error rate = 1) while DIO-AL and DIO-WR are readily distinguishable (only one out of five and one out of six respectively, were misclassified). The “classifying OTUs” (i.e., those driving the community differences) were retrieved from this analysis (Figure 2a) and an unsupervised hierarchical clustering was performed on their abundances. The resulting heat map (Figure 2b) shows an OTU (Firmicutes; Allobaculum) that is almost absent from all of the DIO samples and is present in all but one of the CON samples.
Figure 2.
Members of the microbiota that differ in abundance by diet composition and treatment (WR vs. AL). (a) Nearest shrunken centroid analysis of the 15 OTUs accounting for the differences among the four groups of mice. For each OTU listed in center, direction of the horizontal bars indicates relatively over-represented (right of vertical line) and under-represented (left of vertical line); the length of the bar indicates the strength of the effect. (b) Heat map of the “classifying” OTUs. Columns show, for each mouse, the abundance data of OTUs listed in center. The abundances of the OTUs were clustered using unsupervised hierarchical clustering (blue, low abundance; red, high abundance). The Phylum, Genus of each of the classifying OTUs is noted. AL, ad-libitum diets; OTU, relative operational taxonomic unit; WR, weight-reduced.
Nearest shrunken centroid classification revealed eight OTUs that discriminated between the two diets. Seven of the eight OTUs are under-represented in CON mice and over-represented in DIO. The OTU with the greatest contrast between CON and DIO was a member of the Lachnospiraceae family of the Firmicutes phylum: this OTU is under-represented in CON (score of −0.82) and over-represented in the DIO mice (score of +1.04). An OTU classified to the genus Allobaculum was over-represented in CON (score of +0.61) and underrepresented in DIO (score of −0.78). Members of an OTU classified as the genus Mucispirillum were also positively correlated with the DIO mice (score of +0.51) and negatively correlated with the CON mice (scores of −0.4).
Effect of weight reduction on composition of the microbial community
Te DIO-WR mice form a separate cluster from the DIO-AL group and are intermediate between the DIO-AL and the CON animals in the PCoA plot of unweighted UniFrac distances (Figure 1b). There is no significant difference in mean unweighted UniFrac distances within and between treatments (WR vs. AL, t = 0.82, P value = 0.41). The average unweighted UniFrac distances within and between diets is significantly different, indicating that diet type is a strong factor in bacterial diversity regardless of the abundances of specific types of bacteria (t = 9.47, P value = 9.13 × 10−19).
Nearest shrunken centroid analysis, which takes into account OTU abundances, indicated that four of the five DIO-AL mice, and five out of the six DIO-WR mice, could be correctly classified (overall error rate = 0.176). Five OTUs discriminated between the DIO-WR and DIO-AL. Allobaculum was enriched in the weight reduced mice and contributed most to the separation of these communities. Others, listed in order of effect size (Figure 2), are OTUs classified as members of the Ruminococcaceae and Lachnospiraceae families, and a member of the genus Lactococcus, all of which were enriched in AL; and an OTU classified as a Firmicute that was enriched in DIO-WR.
Unlike DIO mice, CON-AL and CON-WR microbiotas did not segregate in the unweighted UniFrac PCoA nor on the basis of shared OTU abundances. When comparing the relative OTU abundances and the effect of the weight reduction in DIO mice, there was an increase in the abundance of Allobaculum in leaner mice, but the overall abundance of Firmicutes was constant (Figure 1c). In the CON mice, the relative abundances of Firmicutes and Allobaculum stayed approximately the same between the AL and WR, but in the CON-WR mice there was an increase in the abundance of Allobaculum OTU_303, and a decrease in the abundance of Allobaculum OTU_716. We also noted a decrease in the relative abundance of members of the Proteobacteria phylum.
Mice of the same weight but ingesting different diets: comparison of DIO-WR and CON-AL
In the overall analysis using unweighted UniFrac, the DIO-WR formed an intermediate cluster between the DIO-AL and the CON. Predictive Analysis of Microarrays analysis comparing the microbiotas of mice of same body weights and body composition, but ingesting different diets (DIO-WR vs. CON-AL), identified five OTUs (Figure 2a) that accounted for differences between these two groups of mice: OTUs classified as members of the Lachnospiraceae family, the Firmicutes phylum, and the genera Bacteroides and Mucispirillum were found to be enriched in the DIO-WR mice, and Allobaculum was found to be enriched in the CON-AL mice. Abundances of OTUs belonging to the Lachnospiraceae and the Deferribacteraceae accounted for the majority of the differences between the DIO-WR and CON-AL mice. At the phylum level (Figure 1c), the DIO-WR mice harbored higher abundances of Bacteroidetes than the CON mice. Although the CON-AL mice had higher relative abundances of Firmicutes, this trend was driven exclusively by Allobaculum OTUs: when Allobaculum OTUs were excluded, the CON-AL mice showed lower Firmicutes abundance than the DIO-WR. The Deferribacteres (e.g., genus Mucispirillum), although present in low percentage in the DIO-WR, were absent from the CON-AL.
Circulating leptin, inflammation markers in inguinal fat, and bacterial community composition
Circulating leptin
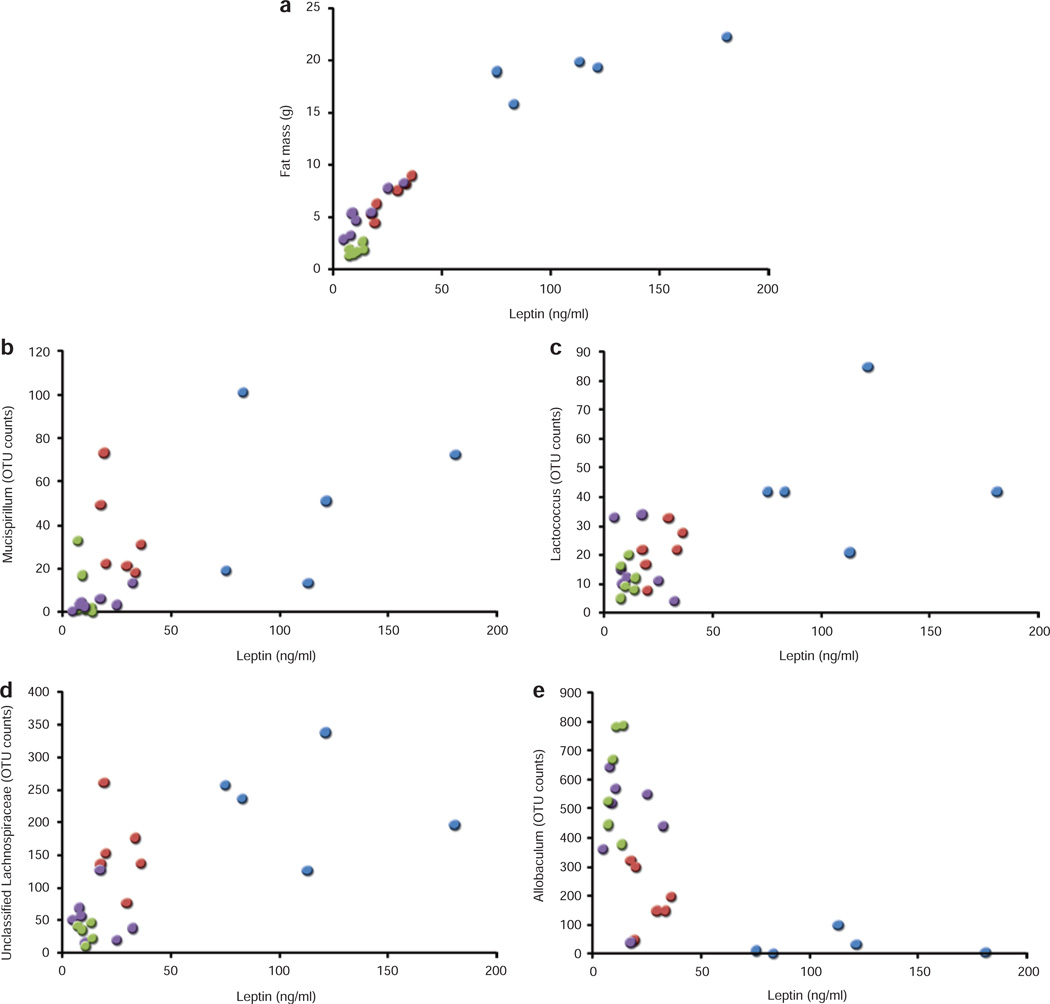
As expected, serum leptin concentrations were highly correlated with total FM (by nuclear magnetic resonance) (Figure 3a; r2 = 0.92, P < 0.0001), and there was no effect of weight loss per se on this relationship. DIO-WR mice lost significant amounts of FM and fat free mass (FM accounted for 65 ± 4% of weight loss), whereas CON-WR mice showed a significant decrease only in fat free mass that accounted for 87 ± 3% of lost weight. As a result, circulating leptin concentrations in DIO-WR mice were reduced about 80% compared to initial concentrations in DIO; whereas in CON animals, weight loss reduced leptin concentrations by only 12%. Consequently, DIO-WR mice had significantly higher circulating leptin concentrations, and slightly but not significantly higher FM, than CON-AL mice when these phenotypes are compared by direct t-test. These differences in absolute circulating concentrations of leptin reflected differences in FM only, i.e., were not due to differences in circulating leptin normalized to FM (Figure 3a). Figure 3 shows that circulating leptin concentration is positively correlated with OTU abundance of the genera of Mucispirillum, ρs = 0.61 P = 0.002, Lactococcus ρs = 0.52 P = 0.008, and an unclassified Lachnospiraceae, ρs = 0.63 P < 0.001, respectively (Figure 3b–d). Allobaculum abundance was negatively correlated (Figure 3e) with leptin concentration (ρs = −0.73 P = 0.001). No patterns were detected when comparing circulating concentrations of triiodothyronine, thyroxine, insulin, or adiponectin to relative abundances of the microbiota (data not shown).
Figure 3.
Associations between host serum leptin concentrations and gut microbiota. (a) Correlations of fat mass content (by nuclear magnetic resonance) with circulating leptin concentrations. (b, c, d, e) Correlations between leptin concentrations and the abundance of relative operational taxonomic units of interest. CON-WR, weight-reduced CON: green, CON-AL, control diet-fed mice: purple, DIO-AL, diet-induced obese mice: blue, DIO-WR, weight-reduced DIO: red.
Inflammation markers
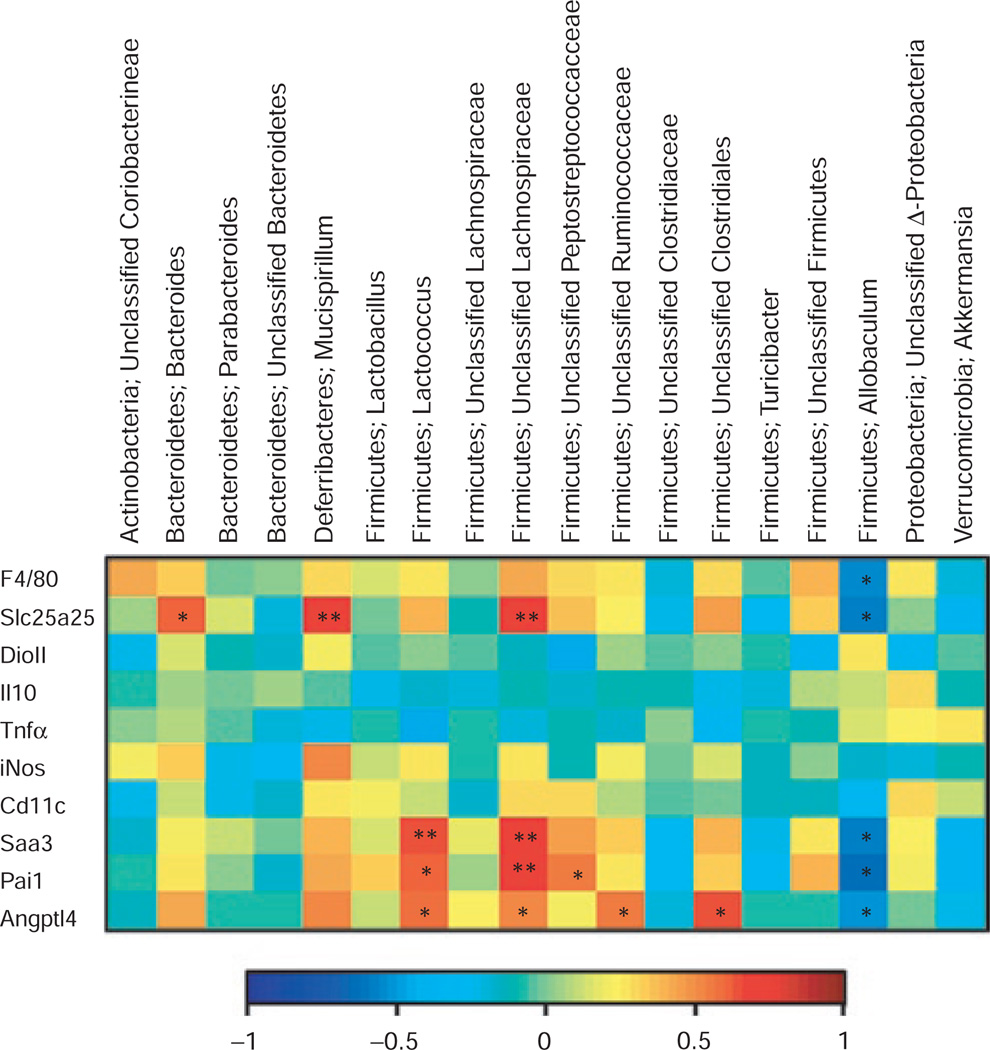
Expression levels of selected inflammatory markers and the solute carrier Slc25a25 were examined in inguinal fat pads (Table 1). Slc25a25 is a mitochondrial transporter that is believed to be involved in energy expenditure homeostasis; its gene expression in white adipose tissue correlates positively with diet composition and cold stress (L.P. Kozak, personal communication). Weight reduction was associated with significant decreases in Pai1 and Saa3 mRNA levels in DIO-WR compared to DIO-AL mice. Slc25a25 levels were higher in DIO mice than CON mice, regardless of weight reduction. F4/80 levels were significantly lower in CON-WR mice compared to all three other groups. Cd11c expression was significantly higher in DIO-AL than all other groups, although weight reduction per se, showed (by two-way ANOVA) near significance (P = 0.07). No significant differences in levels were seen for Il10 and Tnfα across all groups. Expression level of DioII, a gene that influences energy expenditure by peripheral tissue conversion of thyroxine to the more physiologically active triiodothyronine, was decreased in DIO-WR but increased in CON-WR animals. iNos expression was significantly decreased in DIO-WR animals but significantly increased in CON-WR animals. Figure 4 is a heatmap showing the correlations between the inflammation markers and the abundances of selected OTUs. Expression levels of Slc25a25 were strongly positively correlated with relative abundance of Bacteroides, Mucispirillum, and an unclassified Lachnospiraceae, and negatively correlated with Allobaculum. Saa3 and Pai1 were positively correlated with Lactococcus and a Lachnospiraceae. Allobaculum OTUs showed the opposite trend.
Figure 4.
Heat map describing the correlation of the abundances of different operational taxonomic units and transcription levels of inflammation-related genes in inguinal adipose tissue. The colors range from blue (negative correlation; −1) to red (positive correlation; 1). Significant correlations are noted by *P < 0.05 and **P < 0.01 (The computed false discovery rate is about 0.25 using the Benjamini Hochberg procedure (38)).
We also measured expression levels in adipose tissue of Angptl4 (also known as Fiaf) and found the WR mice to have lower levels of expression than the AL mice (Table 1). Figure 4 shows a very strong correlation between Angptl4 levels and an unclassified member of the Clostridiales. Angptl4 levels also correlated with relative abundance of Lactococcus and unclassified Ruminococcaceae and Lachnospiraceae OTUs.
DISCUSSION
In this study, diet composition per se had the biggest effect on the gut microbiota. Our DIO-WR and CON-AL groups had similar body composition and weights, allowing a comparison of their gut microbiotas without the confounding effects of weight/adiposity. The differences in relative bacterial abundances between DIO-WR and CON-AL corroborate those of Hildebrandt et al., who reported an effect of diet composition independent of host body weight, although the body weights in those experiments were not as closely matched as those in the present study and the mice in our study are not segregating for a monogenic mutation (RELMβ (15)). S ignificant differences have been demonstrated in the diversity of the microbiotas of rodents and humans ingesting high fat and low fat diets (13,15,23), but these studies do not adequately control for differences in body mass or body composition. In addition, these studies do not distinguish whether effects on gut microbiota are a result of increased caloric intake per se or the fact that the composition of the diet was higher in fat content. Our results confirm unambiguously that dietary fat content, and not increased caloric intake, affects gut microbiotas in animals of similar weights.
Weight reduction affects the composition of the gut microbial community in mice and humans (8,9,11,12,24). However, the weight loss in these studies resulted from changes in diet composition and/or changes in number of calories ingested, potentially confounding the respective contributions of diet composition, weight loss, and their interactions. We show here that, in mice fed a high fat (60%) diet, maintenance of a 20% reduced body weight affects the composition of the gut microbiota. This effect is not seen in weight-reduced mice fed a low fat (10%) diet. The different effects of weight reduction on the gut microbial community composition between these two groups of mice may reflect effects of diet, initial body weight/composition (and attendant biological consequences), and/or their interactions.
The changes in gut microbiota observed in weight-reduced mice on the DIO diet but not on the CON diet are intriguing. This difference may be attributable to the differential effects that weight loss has on absolute changes in leptin concentrations between the DIO and CON mice. Leptin concentrations are linearly correlated with FM (Figure 3a). DIO-WR mice lost significant amounts of FM (accounting for 65 ± 4% of weight loss), whereas CON-WR mice showed a significant decrease only in fat-free mass (accounting for 87 ± 3% of lost weight). As a result, leptin concentrations in DIO-WR mice were reduced about 80% compared to initial concentrations in DIO, whereas in CON animals, weight reduction lowered leptin levels by only 12%. Our results suggest that the effects of body weight change on the gut microbiota may be mediated, in part, by changes in circulating leptin concentrations.
A connection between circulating leptin concentrations and the composition of the microbiota is suggested by the following observations: (i) several OTUs have abundances that are correlated with circulating leptin concentrations, and (ii) some of these OTUs have been shown to interact with intestinal mucin, an important component of the intestinal milieu made up of heavily glycosylated proteins produced by endothelial cells. Mucin is important in creating micro-niches that are favored by some bacterial populations. For instance, Akkermansia and Allobaculum abundances and circulating leptin concentrations were negatively correlated, whereas Mucispirillum abundance was positively correlated with circulating leptin concentrations and showed highest relative abundance in the obese mice. These OTUs are also noteworthy because Akkermansia can subsist on mucin (25), and Mucispirillum is known to colonize the mucus layer (26). These relationships raise the question of whether leptin concentration affects mucin production and/or composition in the gut, which then in turn could influence the preponderance of specific populations of bacteria (27). Administration of leptin into the colon of rats strongly stimulates mucin production; and leptin stimulates mucous production in vitro in human intestinal mucin-producing cells (HT29-MTX (27,28)). Humans segregating for a single nucleotide polymorphism (Q223R) in the leptin receptor are more susceptible to infection by Entamoeba histolytica (29). Mice segregating for this same mutation, also showed increased susceptibility to Entamoeba histolytica infection, and increased apoptosis of cecal epithelium cells, suggesting that there is a direct link between leptin biology and mucosal immunity. Together, these results suggest that circulating leptin concentrations may affect the composition of the gut microbiota by affecting mucin production in the intestine. A decline in circulating leptin concentrations, as seen in the DIO-WR mice, could have a larger impact on the microbiota than a relatively small decrease in leptin concentrations observed in CON-WR mice.
The OTUs that account for the differences between DIO (WR & AL) and CON (WR & AL) mice, and are also negatively correlated to circulating leptin concentrations, belong to the genus Allobaculum, a member of the Firmicute family Erysipelotrichaceae. Interestingly, members of this family have been shown in several independent studies to change in abundance in response to changes in relative amounts of dietary fat intake (13,30). Furthermore, Allobaculum relative abundance has been reported to be positively correlated with plasma HDL concentrations in hamsters fed a diet supplemented with grain sorghum lipid extract (31). In our study, both diet composition (i.e., relative amount of dietary fat; DIO vs. CON) and body weight status (AL vs. WR) correlated with Allobaculum abundances, indicating that diet composition alone cannot account for changes in relative abundance, and that some metabolic or phenotypic change caused by maintenance of lower body weight must also be involved (see Figure 3e).
Does the composition of the microbiota itself contribute causally to host adiposity? Several studies suggest that the absence of micriobiota (gnotobiotic mice raised in a germ-free environment) is protective against diet-induced obesity (32–34), although perhaps not in all mouse strain/diet combinations (30). There are several ways in which the specific composition of microbiota might influence host adiposity (8,35). One is via the increased availability of short chain fatty acids produced by microbial breakdown of complex polysaccharides, giving the host access to more of the ingested calories (11). Another is by inducing inflammation, which can lead to insulin resistance and hyperphagia (15). A change in microbiota induced by a high fat diet can trigger metabolic inflammation when increased gut permeability allows lipopolysaccharides to enter the circulation (14,36). Specific changes in microbiota preponderance that are either increased (Allobaculum) or decreased (Lachnospiraceae) following maintenance of a WR state (irrelevant of diet composition), and correlated with hormones known to influence energy homeostasis (e.g., leptin), suggest that the specific composition of the microbiota may play a role in host energy balance in weight-perturbed individuals.
We observed a correlation between certain gut bacteria (e.g., Lactococcus and unclassified Lachnospiraceae) and gene expression levels in inguinal fat of inflammation markers (Saa3 and Pai1) and a mitochondrial transporter (Slc25a25). Contrary to what we anticipated, certain inflammation markers, such as Tnfα and F4/80, were not significantly elevated in DIO-AL mice when compared to CON-AL mice (Table 1). These discrepancies may be related to the fat pad (inguinal) in which gene expression was tested. Koren et al. (2011) have reported correlations between relative abundance of specific members of the gut microbiota (e.g., Lachnospiraceae) and circulating markers (e.g., low density lipoprotein concentrations) known to correlate with inflammation (37). Specific gut microbial communities induce low-grade inflammation in white adipose tissue: mice deficient in toll-like receptor 5 developed increased visceral fat, hyperlipidemia, hypertension, and decreased insulin sensitivity, an aggregate phenotype similar to that seen in humans with “metabolic syndrome;” this constellation of phenotypes can be transferred to germ-free wild-type recipients by microbial transplantation from affected animals (15). Thus, specific phylotypes observed in our study could be drivers of inflammation, although establishing a causative role will require further testing.
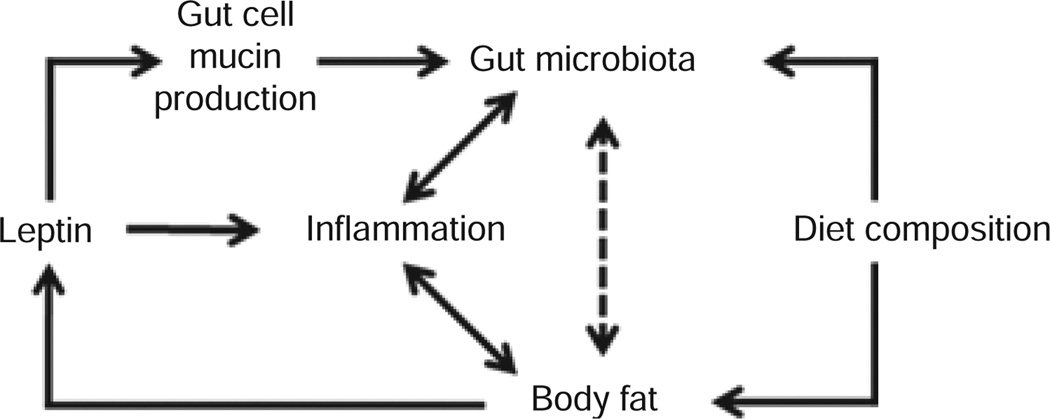
Host adiposity, diet composition, and gut microbiotas interact in complex, probably reciprocal ways. Figure 5 is a schematic of these possible interactions. Leptin concentration, both circulating and within the gut, and dietary fat, may interact to affect gut mucous production, the microbiota, and barrier integrity in ways that ultimately influence adiposity. The studies described here begin to disarticulate the effects of diet and weight perturbation, per se, on relative abundances of gut microbiota. The molecular mechanisms underlying these effects on gut microbiota, and the consequent roles of these bacteria in energy homeostasis and “metabolic inflammation” are clearly areas of clinical importance. Establishing the strength and direction of the relevant arrows of causality will require some relatively straightforward extensions of the studies and techniques reported here (Figure 5).
Figure 5.
Schematic depicting possible inter-relationships among diet composition, gut microbiota, circulating leptin, body fat, markers of inflammation, and gut mucin. Body fat directly determines leptin production and elevated body fat increases macrophage infiltration (with associated production/release of inflammatory molecules such as tumor necrosis factor α, serum amyloid A3, and chemokine (C-C motif) ligand 2 (MCP-1) in adipose tissue). The results presented here suggest that diet composition (fractional fat content) directly affects gut microbiota independent of effects mediated by body weight and body composition. Leptin promotes proliferation, differentiation, and survival of immune cells. Leptin also stimulates mucin production in mouse and human intestinal cells (27,28). Mucin affects local bacterial “micro-niches” in the gut by favoring the growth of some bacteria (25,26). Leptin can affect intestinal barrier function by inhibiting apoptosis and promoting regeneration of intestinal epithelium (39,40). These changes in epithelial composition may in turn affect microbiota populations in the gut. The dashed line between body fat and gut microbiota suggests biologically possible connection(s) that might be mediated by adipocytokines or other molecules secreted from adipose tissue.
ACKNOWLEDGMENTS
We thank Jeff Werner for bioinformatics support, Jeremy Koenig for initial discussions, and Nicholas Scalfone for Laboratory assistance. This work was supported by AstraZeneca, Howard Hughes Medical Institute, the Russell Berrie Foundation, NIH 5P30DK026687, 5R01DK052431, 5R01DK064773, DK78669, DK063608-08, HG4872, and HG4866 which have supported this work, as well as the NIH Human Microbiome Project Data Analysis and Coordination Center (O.K.), the Arnold and Mabel Beckman Foundation (R.L.), and The Hartwell Foundation (R.L.).
Footnotes
DISCLOSURE
The authors declared no conflict of interest.
REFERENCES
- 1.Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999–2008. JAMA. 2010;303:235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- 2.MacLean PS, Higgins JA, Johnson GC, et al. Enhanced metabolic efficiency contributes to weight regain after weight loss in obesityprone rats. Am J Physiol Regul Integr Comp Physiol. 2004;287:R1306–R1315. doi: 10.1152/ajpregu.00463.2004. [DOI] [PubMed] [Google Scholar]
- 3.Hambly C, Speakman JR. Contribution of different mechanisms to compensation for energy restriction in the mouse. Obes Res. 2005;13:1548–1557. doi: 10.1038/oby.2005.190. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbaum M, Sy M, Pavlovich K, Leibel RL, Hirsch J. Leptin reverses weight loss-induced changes in regional neural activity responses to visual food stimuli. J Clin Invest. 2008;118:2583–2591. doi: 10.1172/JCI35055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbaum M, Vandenborne K, Goldsmith R, et al. Effects of experimental weight perturbation on skeletal muscle work efficiency in human subjects. Am J Physiol Regul Integr Comp Physiol. 2003;285:R183–R192. doi: 10.1152/ajpregu.00474.2002. [DOI] [PubMed] [Google Scholar]
- 6.Ravussin Y, Gutman R, Diano S, et al. Effects of Chronic Weight Perturbation on Energy Homeostasis and Brain Structure in Mice. Am J Physiol Regul Integr Comp Physiol. 2011 doi: 10.1152/ajpregu.00429.2010. e-pub ahead of print 16 March 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leibel RL, Rosenbaum M, Hirsch J. Changes in energy expenditure resulting from altered body weight. N Engl J Med. 1995;332:621–628. doi: 10.1056/NEJM199503093321001. [DOI] [PubMed] [Google Scholar]
- 8.Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26:5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- 9.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 11.Ley RE, Bäckhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 14.Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724. doi: 10.1053/j.gastro.2009.08.042. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting abundance distributions. Nat Methods. 2010;7:688–699. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caporaso JG, Bittinger K, Bushman FD, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knights D, Kuczynski J, Koren O, et al. Supervised classification of microbiota mitigates mislabeling errors. ISME J. 2011;5:570–573. doi: 10.1038/ismej.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turnbaugh PJ, Ridaura VK, Faith JJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1 doi: 10.1126/scitranslmed.3000322. 6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 26.Robertson BR, O’Rourke JL, Neilan BA, et al. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol. 2005;55:1199–1204. doi: 10.1099/ijs.0.63472-0. [DOI] [PubMed] [Google Scholar]
- 27.El Homsi M, Ducroc R, Claustre J, et al. Leptin modulates the expression of secreted and membrane-associated mucins in colonic epithelial cells by targeting PKC, PI3K, and MAPK pathways. Am J Physiol Gastrointest Liver Physiol. 2007;293:G365–G373. doi: 10.1152/ajpgi.00091.2007. [DOI] [PubMed] [Google Scholar]
- 28.Plaisancie P, Ducroc R, El Homsi M, et al. Luminal leptin activates mucinsecreting goblet cells in the large bowel. Am J Physiol Gastrointest Liver Physiol. 2006;290:G805–G812. doi: 10.1152/ajpgi.00433.2005. [DOI] [PubMed] [Google Scholar]
- 29.Duggal P, Guo X, Haque R, et al. A mutation in the leptin receptor is associated with Entamoeba histolytica infection in children. J Clin Invest. 2011;121:1191–1198. doi: 10.1172/JCI45294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleissner CK, Huebel N, Abd El-Bary MM, et al. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br J Nutr. 2010:1–11. doi: 10.1017/S0007114510001303. [DOI] [PubMed] [Google Scholar]
- 31.Martínez I, Wallace G, Zhang C, et al. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl Environ Microbiol. 2009;75:4175–4184. doi: 10.1128/AEM.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding S, Chi MM, Scull BP, et al. High-fat diet: bacteria interactions promote intestinal inflammation which precedes and correlates with obesity and insulin resistance in mouse. PLoS ONE. 2010;5:e12191. doi: 10.1371/journal.pone.0012191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabot S, Membrez M, Bruneau A, et al. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010;24:4948–4959. doi: 10.1096/fj.10-164921. [DOI] [PubMed] [Google Scholar]
- 34.qaBäckhed F, Crawford PA, O’Donnell D, Gordon JI. Postnatal lymphatic partitioning from the blood vasculature in the small intestine requires fastinginduced adipose factor. Proc Natl Acad Sci USA. 2007;104:606–611. doi: 10.1073/pnas.0605957104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bäckhed F. Changes in intestinal microflora in obesity: cause or consequence. J Pediatr Gastroenterol Nutr. 2009;48(Suppl 2):S56–S57. doi: 10.1097/MPG.0b013e3181a11851. [DOI] [PubMed] [Google Scholar]
- 36.Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 37.Koren O, Spor A, Felin J, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discover rate: a practivcal and powerful approach to multiple testing. J R Statist Soc. 1994:289–300. [Google Scholar]
- 39.Ogunwobi OO, Beales IL. The anti-apoptotic and growth stimulatory actions of leptin in human colon cancer cells involves activation of JNK mitogen activated protein kinase, JAK2 and PI3 kinase/Akt. Int J Colorectal Dis. 2007;22:401–409. doi: 10.1007/s00384-006-0181-y. [DOI] [PubMed] [Google Scholar]
- 40.Sukhotnik I, Coran AG, Mogilner JG, et al. Leptin affects intestinal epithelial cell turnover in correlation with leptin receptor expression along the villus-crypt axis after massive small bowel resection in a rat. Pediatr Res. 2009;66:648–653. doi: 10.1203/PDR.0b013e3181be9f84. [DOI] [PubMed] [Google Scholar]