Abstract
Alveolar rhabdomyosarcoma (aRMS) is an aggressive sarcoma of skeletal muscle characterized by expression of the paired box 3-forkhead box protein O1 (PAX3-FOXO1) fusion oncogene. Despite its discovery nearly two decades ago, the mechanisms by which PAX3-FOXO1 drives tumor development are not well characterized. Previously, we reported that PAX3-FOXO1 supports aRMS initiation by enabling bypass of cellular senescence checkpoints. We have now found that this bypass occurs in part through PAX3-FOXO1–mediated upregulation of RASSF4, a Ras-association domain family (RASSF) member. RASSF4 expression was upregulated in PAX3-FOXO1–positive aRMS cell lines and tumors. Enhanced RASSF4 expression promoted cell cycle progression, senescence evasion, and tumorigenesis through inhibition of the Hippo pathway tumor suppressor MST1. We also found that the downstream Hippo pathway target Yes-associated protein 1 (YAP), which is ordinarily restrained by Hippo signaling, was upregulated in RMS tumors. These data suggest that Hippo pathway dysfunction promotes RMS. This work provides evidence for Hippo pathway suppression in aRMS and demonstrates a progrowth role for RASSF4. Additionally, we identify a mechanism used by PAX3-FOXO1 to inhibit MST1 signaling and promote tumorigenesis in aRMS.
Introduction
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma of childhood and adolescence. Over the past 30 years, clinical trials in North America, Europe, and Australia have identified superior treatment strategies leading to the improved survival of discrete groups of RMS patients. A distinctly worse outcome is encountered for patients with the alveolar histologic variant of RMS (aRMS), who have a 5-year survival rate of less than 50% (1). Even more dismal is the survival for those whose tumors express the signature paired box 3-forkhead box protein O1 (PAX3-FOXO1) fusion gene; in the metastatic setting, their survival rate at 4 years is less than 10% (2). Although PAX3-FOXO1 (and the related fusion protein PAX7-FOXO1) was identified in the 1990s (3–5) and intensely studied in terms of its regulation, downstream targets, and cellular phenotypic effects, a unified understanding of how the fusion gene and its resulting oncoprotein contribute so profoundly to aRMS tumorigenesis remains obscure.
To understand the discrete molecular steps involved in aRMS tumorigenesis, we constructed a genetically defined model of aRMS based on the sequential introduction of a defined set of oncogenic cDNAs (PAX3-FOXO1, hTERT, MycN) into primary human skeletal muscle myoblasts (HSMMs). When implanted in immunodeficient mice, these transformed cells (abbreviated herein as HSMMPF+H+M) produce xenografts whose morphology and histologic markers mimic aRMS (6). The order of expression of the cDNAs is important for faithful generation of the model; most critically, PAX3-FOXO1 must be introduced first for the cells to transform in vivo. This suggests that PAX3-FOXO1 imparts critical cellular changes (genetic or epigenetic) that support subsequent tumorigenic steps and that investigation of these changes will provide needed insight into aRMS tumorigenesis. One such critical change required for tumorigenicity of primary HSMMs stably expressing PAX3-FOXO1 is proliferation past the senescence checkpoint (7). This observation suggests that PAX3-FOXO1 permits continued cell growth in an ordinarily prohibitive environment.
The Hippo signaling pathway is a highly conserved tumor suppressor cascade best known for its role in regulating organ size. The core components of the Hippo pathway are the MST1/2 and LATS1/2 tumor suppressor kinases, the RASSF, SAV1, and MOB1 adaptor proteins, the YAP transcriptional regulator, and the TEAD transcription factors. MST/RASSF, MST/SAV1, and LATS/MOB1 signaling complexes coordinate a tumor-suppressive signaling cascade that inhibits the nuclear localization and activation of YAP. YAP, when in the nucleus, regulates activation of the TEAD transcription factors, promoting the expression of pro-proliferative and survival genes (8). During normal growth and development, the Hippo pathway provides tumor suppressor functions at the intersection of cell proliferation, differentiation, and apoptosis. Not surprisingly, malignancies have evolved to corrupt this pathway. MST/LATS loss or YAP overexpression lead to tumorigenesis in mouse models (9–14), demonstrating that Hippo pathway inhibition is sufficient for tumorigenesis. Identifying the mechanisms of Hippo pathway inactivation in human cancer will be paramount in finding ways to exploit this pathway therapeutically.
In the current work, we have examined the transcriptional changes regulated by PAX3-FOXO1 in primary HSMMs and identified RASSF4 as a novel PAX3-FOXO1 transcriptional target. We found RASSF4 highly expressed in PAX3-FOXO1–positive aRMS and its expression necessary for aRMS cell proliferation, senescence evasion, and tumorigenesis. Mechanistically, we show that RASSF4 associates with MST1 kinase to inhibit downstream signaling in PAX3-FOXO1–positive aRMS. We also show that YAP is upregulated in both major RMS subtypes, suggesting that Hippo pathway dysregulation is an essential element of RMS tumorigenesis.
Results
PAX3-FOXO1 promotes transcriptional changes in primary human myoblasts.
We have shown previously that the expression of exogenous PAX3-FOXO1 in combination with p16INK4A loss in primary HSMMs is required for the generation of a genetic model of aRMS (6). PAX3-FOXO1 expression in these primary cells promotes senescence bypass, priming cells for transformation and tumor formation. Since the PAX3-FOXO1 protein is considered a hyperactive transcription factor (15), we reasoned that one important function of PAX3-FOXO1 was to regulate transcription of the genes necessary for senescence bypass. To this end, we compared global gene expression in presenescent vector–expressing control HSMMs, presenescent PAX3-FOXO1–expressing HSMMs, and postsenescent PAX3-FOXO1–expressing HSMMs using Affymetrix mRNA microarray analysis. Gene expression profiles of all cells were normalized by robust multiarray average (RMA) and zero transformed against the average expression levels of the same probe sets of the vector-expressing control HSMMs, as performed previously (16, 17).
When compared with vector-expressing control cells, 2,691 probe sets with at least two-fold changes impacted by PAX3-FOXO1 expression in at least two samples were selected and arranged by hierarchical clustering according to similarities in expression patterns (Figure 1A, left). Importantly, PAX3-FOXO1 expression affected several known PAX3-FOXO1 targets, including the upregulation of CXCR4 and FGFR4 (18, 19), and the downregulation of DUSP4 and SPRY4 (20). We also observed the upregulation of additional genes associated with aRMS including MYOD and MEF2A (21, 22). The altered expression of these genes was confirmed by semiquantitative RT-PCR (Figure 1A, right). We then examined this dataset for potential novel regulators of cell signaling that could be responsible for the senescence-suppressing effects of PAX3-FOXO1. One gene was of particular interest, namely Ras-association domain family member 4 (RASSF4), known as an effector of the small GTPase Ras and a regulator of the Hippo pathway (23–26). Because it is well established that Ras regulates senescence signaling in primary cells and that the Hippo pathway restrains cell proliferation and tumorigenesis (27–29), we hypothesized that the upregulation of RASSF4 inhibits senescence through suppression of Ras or Hippo signaling. Therefore, we initiated an investigation of RASSF4 regulation and function in aRMS.
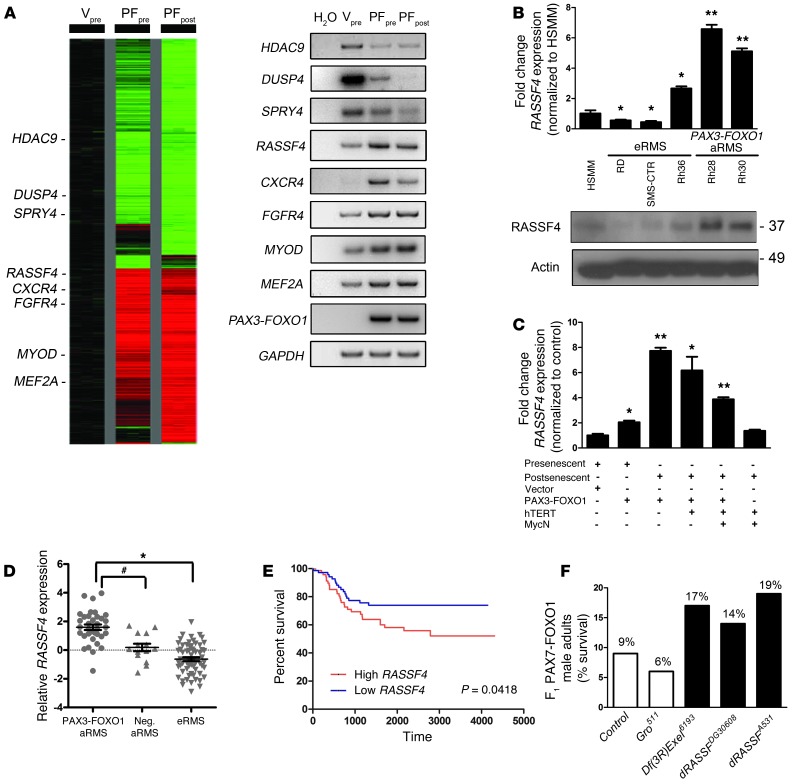
Figure 1. RASSF4 is upregulated in PAX3-FOXO1–positive aRMS cells and tumors.
(A) Left: Expression profile of HSMM control cells (Vpre) compared with PAX3-FOXO1–expressing HSMM presenescent (PFpre) or postsenescent (PFpost) cells. Right: Semiquantitative RT-PCR validation of select genes identified in the microarray. (B) PAX3-FOXO1–expressing aRMS cells expressed more RASSF4 than eRMS cells or HSMMs, as measured by qPCR and immunoblotting. *P < 0.05; **P < 0.005. Labels for cell lines correspond to qPCR and immunoblotting. Actin was used as a loading control. (C) HSMM-based model of aRMS displayed enhanced RASSF4 expression in a PAX3-FOXO1–dependent manner as measured by qPCR. *P < 0.05; **P < 0.005. (D) PAX3-FOXO1–positive primary human aRMS tumors expressed more RASSF4 than fusion-negative aRMS or eRMS. Error bars represent SEM. *P < 0.0001; #P = 0.0004; Mann-Whitney U test. Median-centered log2 values are shown, and microarray data were obtained from the Oncogenomics database. (E) RMS patient survival based on RASSF4 expression. The median RASSF4 expression value for RMS was the threshold for high versus low RASSF4 expression. High RASSF4, n = 73, Low RASSF4, n = 73. P value is based on log-rank test analysis. (F) dRASSF mutation genetically suppressed PAX7-FOXO1 pathogenicity in a Drosophila aRMS model. PAX7-FOXO1 expression in differentiating larval muscle causes semilethality, as PAX7-FOXO1 adults comprise only 9% of F1 adults (n = 170). (In Mendelian ratios, the F1 population should be composed of 50% wild-type and 50% PAX7-FOXO1 adults). The Df(3RExcel)6193 chromosomal deletion and dRASSFDG30608 and dRASSFA531 loss-of-function alleles suppressed PAX7-FOXO1–induced lethality. Groucho511 (Gro511) is an unrelated mutation included as a representative example of a nonsuppressor. Df(3R)Exel6193, n = 72; dRASSFA531, n = 130; dRASSFDG30608, n = 66; and Gro511, n = 116.
RASSF4 is upregulated in PAX3-FOXO1–positive aRMS cells and tumors.
Since PAX3-FOXO1–expressing HSMMs showed increased RASSF4 levels in microarray analysis, we next examined commonly used human RMS cell lines for RASSF4 expression. Compared with HSMMs, the PAX3-FOXO1–negative embryonal RMS (eRMS) cell lines RD and SMS-CTR displayed modest decreases in RASSF4, while Rh36 cells showed a small, but significant, increase in RASSF4. Strikingly, the PAX3-FOXO1–positive aRMS cell lines Rh28 and Rh30 displayed much higher levels of both RASSF4 mRNA and protein (Figure 1B). Cell lines used to generate our aRMS genetic model displayed a similar correlation between PAX3-FOXO1 status and RASSF4 expression (Figure 1C). To examine RASSF4 levels in a larger cohort of pediatric cancer cell lines and xenograft tumors, we queried the publicly available Oncogenomics database ( http://home.ccr.cancer.gov/oncology/oncogenomics/). Importantly, oncogenes associated with RMS, such as c-MET and FGFR4, displayed upregulation in RMS cell lines and xenograft tumors when compared with normal tissues. We observed similar upregulation of RASSF4 (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI67087DS1). However, when we examined the available data for other RASSF genes, we saw no such correlation with any other RASSF family members (RASSF1, 2, 5, 6, 7, or 8) (Supplemental Figure 1), suggesting that RASSF upregulation is specific to RASSF4 in aRMS.
Next, we used the same database to examine RASSF4 expression in primary human RMS tumors. When analyzed by subtype, PAX3-FOXO1–positive aRMS tumors exhibited significantly higher RASSF4 levels compared with fusion-negative aRMS or eRMS tumors (Figure 1D). Furthermore, when patient survival was examined based on RASSF4 expression, high RASSF4 expression was associated with decreased survival (Figure 1E). In summary, these data demonstrate a correlation between PAX3-FOXO1 fusion status and elevated RASSF4 expression in aRMS cell lines, xenografts, and primary human tumors.
dRASSF mutation dominantly suppresses PAX-FOXO1 pathogenicity in a Drosophila model of aRMS.
As an independent means of examining the relationship between PAX3-FOXO1 and RASSF4, we took advantage of a Drosophila model of aRMS pathogenicity. In this model, misexpression of PAX3/7-FOXO1 in differentiating muscle causes myoblast fusion defects that result in larval lethality, manifested as only about 10% of F1 adults being composed of PAX7-FOXO1 adults (30). Although tumorigenesis is not detectable due to lethality, the misfused PAX3/7-FOXO1 myogenic cells act aggressively to infiltrate nonmuscle tissue (30). Forward genetic screening for PAX-FOXO1 enhancers/suppressors can then be used to identify novel PAX3/7-FOXO1 gene targets and effectors (30, 31).
Using this Drosophila model, we first inspected the expression data for transcriptional changes promoted by PAX7-FOXO1. (Human PAX7 demonstrates a slightly higher sequence identity with Drosophila PAX3/7 than does human PAX3 and is used here.) We found dRASSF mRNA overexpressed 2.6-fold in PAX7-FOXO1 larval muscle (ArrayExpress database accession number E-MTAB-839), suggesting that similarly to the human myoblast model, this Drosophila model of aRMS shows upregulated RASSF.
We next examined whether dRASSF promotes PAX-FOXO1 phenotypes by testing the effect of Drosophila mutants that alter dRASSF levels or function. We initially focused on a Drosophila chromosomal deletion, Df(3R)Exel6193, that dominantly suppresses PAX7-FOXO1 semilethality (Figure 1F). Interestingly, Df(3R)Exel6193 removed segments 94D3-94E4 on chromosome 3, which includes dRASSF. We hypothesized that heterozygous deletion of dRASSF might account for Df(3R)Exel6193-mediated PAX7-FOXO1 suppression and that, consistent with PAX3-FOXO1 activation of RASSF4 in mammalian myoblasts, dRASSF might act as a PAX3/7-FOXO1 downstream target and effector. We next tested two publicly available transposable-element insertion loss-of-function dRASSF alleles, DG30608 and A531 (32), for suppression of PAX7-FOXO1. The DG30608 element inserts into the 5′UTR, which does not alter DRASSF protein structure but instead presumably alters dRASSF expression. The A531 insertion disrupts exon 5, resulting in a truncated dRASSF lacking the C-terminal SARAH domain, which mediates RASSF-MST1 physical association. Both dRASSF loss-of-function alleles ameliorated PAX-FOXO1 pathogenicity, with A531 acting as the stronger suppressor (Figure 1F). These data show that dRASSF and RASSF4 promote PAX-FOXO1 phenotypes in both Drosophila and mammalian models of aRMS.
PAX3-FOXO1 directly regulates a RASSF4 5′ enhancer.
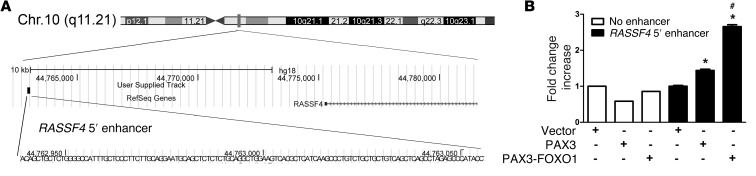
Recently, PAX3-FOXO1 binding sites were identified throughout the human genome and shown to act as distal enhancers (33). We queried the dataset from this published study and identified a PAX3-FOXO1 binding site 12 kb 5′ of the RASSF4 gene on chromosome 10 (Figure 2A). To determine whether this region is actively regulated by PAX3-FOXO1, we performed luciferase assays using this potential PAX3-FOXO1 enhancer region. RD cells (PAX3-FOXO1 negative) cotransfected with the putative RASSF4 5′ enhancer and PAX3-FOXO1 demonstrated elevated luciferase signal (Figure 2B), similar in amplitude to that of confirmed PAX3-FOXO1 enhancer regions (33). While considerably less than PAX3-FOXO1, we found that wild-type PAX3 had a small, but significant (P < 0.001), effect on enhancer activity, suggesting that PAX3 alone may play a role in regulating RASSF4 expression independently of the fusion protein. These data show that in aRMS, PAX3-FOXO1 can directly regulate RASSF4 expression through a 5′ genomic enhancer region.
Figure 2. PAX3-FOXO1 regulates a RASSF4 5′ enhancer.
(A) A putative PAX3-FOXO1 binding site 5′ to the RASSF4 gene on human chromosome 10. Figure derived from publically available data in ref. 33. (B) PAX3 and PAX3-FOXO1 directly regulated the RASSF4 5′ enhancer. RD cells were cotransfected with Renilla, a luciferase vector lacking an enhancer, or the RASSF4 5′ enhancer, along with vector, wild-type PAX3, or PAX3-FOXO1. *P < 0.001 compared with vector-expressing cells; #P < 0.005 compared with PAX3-expressing cells.
RASSF4 inhibits cellular senescence in PAX3-FOXO1–expressing HSMMs.
Since PAX-FOXO1 upregulated RASSF4 or dRASSF in two independent models of aRMS, we reasoned that RASSF4 influences human aRMS pathobiology. Therefore, we began to study the function of endogenous RASSF4 in HSMMs using RASSF4 RNAi loss of function. We generated five lentiviral shRNA plasmids independently targeting RASSF4. Three of these plasmids (RASSF4 sh2-4) were effective at suppressing RASSF4 protein expression in aRMS cells (Figure 3A). As additional validation, we used semiquantitative RT-PCR to measure endogenous RASSF4 mRNA, which demonstrated shRNA-mediated suppression of endogenous RASSF4 in primary HSMMs (Supplemental Figure 2A).
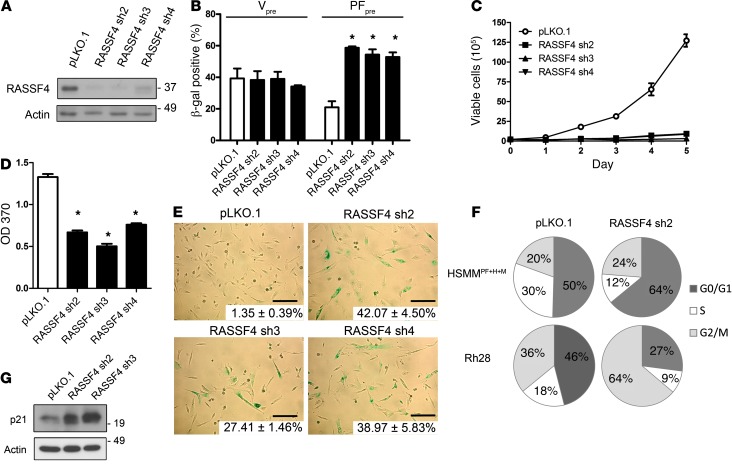
Figure 3. RASSF4 promotes cell proliferation and senescence inhibition in PAX3-FOXO1 aRMS cells.
(A) RASSF4 shRNA validation in HSMMPF+H+M cells. Knockdown was measured by immunoblot analysis for endogenous RASSF4 and actin, which was used as a loading control. pLKO.1 was used as a control vector. (B) Senescence induction in PAX3-FOXO1–expressing HSMM cells. Quantitation of β-gal staining of HSMMV or HSMMPF cells transduced with control vector or RASSF4 shRNAs. *P ≤ 0.01. (C) Loss of RASSF4 in HSMMPF+H+M cells caused deficient cell proliferation, as measured by hemocytometric counts over 5 days of growth in culture and (D) BrdU assay. *P < 0.0001. (E) RASSF4-deficient HSMMPF+H+M cells displayed cell shape change, elevated β-gal staining (insets, mean ± SD; scale bars: 125 μm), cell cycle arrest (F), and p21 upregulation (G). Error bars represent SD in B and D.
To address the role of RASSF4 in PAX3-FOXO1–mediated senescence bypass, we stably expressed RASSF4 shRNAs in vector- or PAX3-FOXO1–expressing HSMMs. To examine the effect of RASSF4 suppression on cellular senescence, we measured in situ β-gal activity, a biomarker of cellular senescence. In vector-expressing HSMMs, RASSF4 suppression did not change β-gal staining. In contrast, in PAX3-FOXO1–expressing cells, RASSF4 suppression induced a significant increase in the percentage of β-gal–positive cells, signifying that more cells were undergoing senescence (Figure 3B). These data demonstrate that in the context of PAX3-FOXO1 expression, RASSF4 prevents the onset of cellular senescence.
RASSF4 promotes cell proliferation and inhibits senescence in PAX3-FOXO1–positive aRMS cells.
Since RASSF4 suppression promotes senescence in PAX3-FOXO1–expressing HSMMs, we next examined the effect of reduced RASSF4 expression in our genetic model of aRMS, which is based on HSMMs expressing PAX3-FOXO1 (HSMMPF+H+M, Figure 1C). Stable expression of RASSF4 shRNAs in HSMMPF+H+M cells in culture caused a profound growth arrest (Figure 3C) and decreased BrdU incorporation (Figure 3D). Cells with reduced RASSF4 expression underwent substantial morphologic changes including enlargement, flattening, and increased granularity, reflecting senescence. In addition, β-gal staining was dramatically increased (Figure 3E). These results suggest that, similarly to HSMMs expressing PAX3-FOXO1, our genetic model of aRMS also relies on RASSF4 expression to promote cell proliferation and inhibit senescence.
We next examined RASSF4-deficient cells for changes in their cell cycle profile. Depending on the cell type, cells were arrested at either G0/G1 or G2/M checkpoints. This suggests that in aRMS, RASSF4 can promote cell cycle progression at both checkpoints or that loss of RASSF4 causes an overall slowing of the cell cycle (Figure 3F). Accordingly, we also observed elevation of p21 protein (Figure 3G), which participates in both the G0/G1 and G2/M checkpoints (34, 35). While we observed that RASSF4 knockdown cells were growth arrested for many days after transduction, they were ultimately not viable in culture after more than 1 week. However, the nonviable RASSF4 cells did not undergo apoptosis, as we observed no caspase 3 cleavage, annexin V positivity, or sub-G1 peak by flow cytometry (data not shown). These data suggest that RASSF4 promotes aRMS cell proliferation and viability by promoting cell cycle progression and evasion of cell senescence.
RASSF4 promotes aRMS tumorigenesis in vivo.
Together, in vitro studies point toward RASSF as a critical component of aRMS cell proliferation and viability. To investigate in vivo activity, we tested whether RASSF4 loss of function would alter tumorigenesis in an aRMS xenograft model. Since RASSF4 suppression caused growth arrest and loss of cell viability, we used a Tet-on lentiviral shRNA system (36) to induce RASSF4 knockdown upon doxycycline (Dox) exposure. We used the PAX3-FOXO1–positive Rh28 cell line to establish this dox-inducible system, first recapitulating prior RASSF4 knockdown data as demonstrated by decreased BrdU incorporation and increased p21 protein (Supplemental Figure 2, B–E).
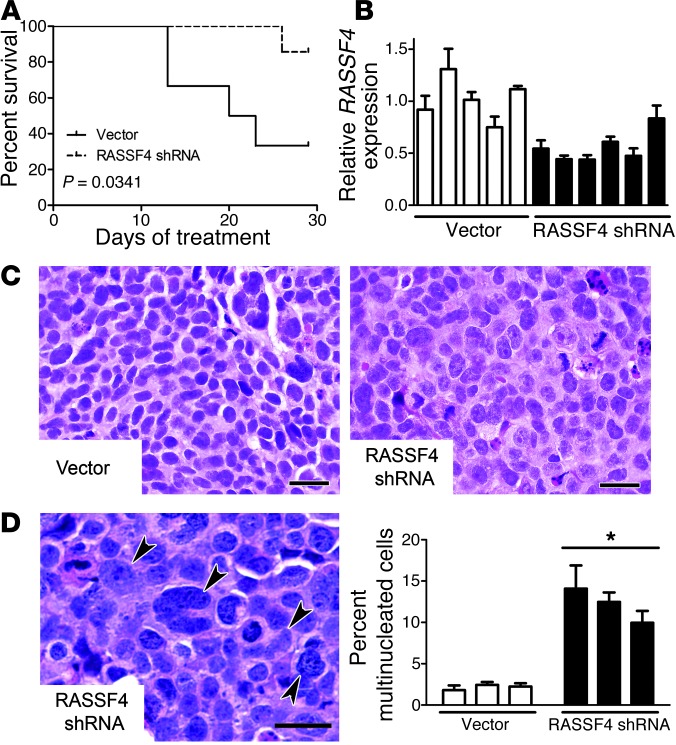
Next, we tested the effects of RASSF4 suppression in aRMS xenografts in vivo. With doxycycline treatment, Rh28 aRMS xenografts expressing RASSF4 shRNA exhibited a significant (P = 0.0341) delay in reaching maximum tumor burden when compared with control (Figure 4, A and B). Our examination of RASSF4-knockdown tumors for changes in the proliferative index using Ki67 immunostaining revealed no changes (data not shown). However, H&E-stained tumor sections revealed dramatic changes in cell morphology. We found that RASSF4-suppressed tumors contained large cells with increased cytoplasm and prominent nucleoli and an abundance of cells with large, irregularly shaped nuclei and multinucleated giant cells (Figure 4, C and D). These cellular morphologies suggest mitotic defects and growth arrest (37), which could explain the decreased tumor growth in RASSF4-knockdown tumors. Based on these data, we concluded that, similarly to in vitro studies, RASSF4 serves a pro-proliferative role in aRMS in vivo.
Figure 4. RASSF4 knockdown in aRMS xenografts inhibits tumor growth.
(A) RASSF4 knockdown tumors displayed a delay in tumor progression, as measured by their time to maximum tumor burden. (B) qPCR validation of RASSF4 knockdown in aRMS xenografts. Each bar represents an individual xenograft. RASSF4 knockdown in aRMS xenografts led to multinucleated cells and mitotic defects. Tumor cell morphologic changes were examined by H&E staining (C), and multinucleated cells (D, left, arrowheads) were quantified (D, right). Error bars represent SD. *P = 0.017. Scale bars: 25 μm.
RASSF4 associates with MST1 in aRMS cells.
Having demonstrated that RASSF4 is pro-proliferative in aRMS both in vitro and in vivo, we next examined the RASSF4 signaling mechanism in aRMS cells. RASSF proteins are typically described as scaffolding molecules, regulating protein complex formation to coordinate signaling cascades. Consistent with this notion, RASSF4, which possesses no identifiable catalytic domains, contains a Ras-association domain and a carboxy-terminal Salvador/RASSF/Hippo (SARAH) domain (Figure 5A). To identify proteins that associate with RASSF4, we immunopurified epitope-tagged RASSF4 protein complexes and probed for endogenous RASSF4-associating proteins. We were surprised to find no association of HA-RASSF4 with any of the canonical Ras isoforms (H-, K-, and N-Ras) in HSMMPF+H+M cells (Figure 5B, left). We observed identical results with the PAX3-FOXO1–positive Rh28 cell line (Figure 5B, right). Therefore, under these conditions, RASSF4 does not associate with Ras.
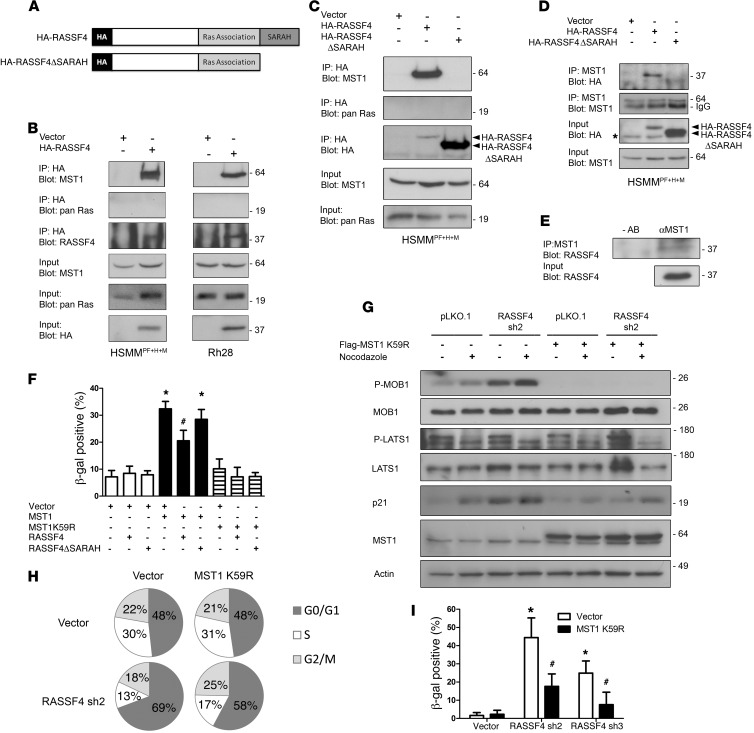
Figure 5. RASSF4 regulates MST1 to inhibit the Hippo pathway in aRMS.
(A) Domain architecture of HA-RASSF4 constructs. (B) RASSF4 associated with MST1 in aRMS cells. Anti-HA immunoprecipitates from aRMS cells expressing HA-RASSF4 or an empty vector were examined for coprecipitation of MST1 or pan H-, K-, and N-Ras by immunoblotting. (C) RASSF4-MST1 association was dependent on the RASSF4 SARAH domain. Anti-HA immunoprecipitates from HSMMPF+H+M cells expressing HA-RASSF4, HA-RASSF4ΔSARAH, or vector were used to examine the association with MST1 or pan Ras by immunoblotting. These results were confirmed by immunopurifying endogenous MST1 and by blotting for HA-RASSF4, HA-RASSF4ΔSARAH (D, top), or endogenous RASSF4 (E). (F) Exogenous MST1 expression induced aRMS cell senescence, which was partially inhibited by HA-RASSF4 expression. Error bars represent SD. *P < 0.0001 compared with vector expressed alone. #P = 0.0005 compared with MST1-expressing cells. (G) RASSF4 suppressed MST1 signaling to MOB1. HSMMPF+H+M cells expressing vector or MST1K59R, vector, or RASSF4 shRNA were cultured in the presence or absence of nocodazole. Protein lysates from these cells were analyzed by immunoblotting. (H) MST1 K59R partially blocked G0/G1 accumulation in RASSF4-deficient cells as measured by cell cycle analysis. (I) MST1K59R prevented senescence induction caused by RASSF4 loss as measured by β-gal assay. *P < 0.00005 compared with vector control cells; #P < 0.005 compared with cells with RASSF4 knockdown alone.
RASSF4 also contains a carboxy-terminal SARAH domain (Figure 5A), which was predicted to associate with SARAH domain–containing proteins of the Hippo pathway. SARAH domain hetero- and homodimerizations play critical roles in the regulation of the Hippo pathway at the level of MST kinases, which also contain SARAH domains. As other members of the RASSF family have been shown to interact with MST1, we tested whether RASSF4 does as well. In both HSMMPF+H+M and Rh28 lysates, we found coprecipitated, endogenous MST1 with HA-RASSF4 (Figure 5B). To determine whether the SARAH domain of RASSF4 regulated the association with MST1, we generated a RASSF4 construct lacking the SARAH domain (HA-RASSF4ΔSARAH) (Figure 5A) and stably expressed this plasmid in HSMMPF+H+M cells. Deletion of the SARAH domain from RASSF4 was sufficient for a loss of association with MST1 (Figure 5C). Similarly to previous experiments, Ras was not found to co-precipitate with HA-RASSF4 or HA-RASSF4ΔSARAH (Figure 5C). We then performed the reverse experiment by immunopurifying endogenous MST1 from aRMS cells and immunoblotting for copurified HA-RASSF4 or HA-RASSF4ΔSARAH. We observed a similar association between MST1 and HA-RASSF4, but not HA-RASSF4ΔSARAH (Figure 5D). Additional studies were performed using an epitope-tagged RASSF4 SARAH domain to examine the sufficiency of RASSF4 interaction with MST1, but these experiments were inconclusive, as the SARAH domain was not expressed at detectable levels. This may be due to protein insolubility, which has been observed with the RASSF1 SARAH domain (38). Last, we validated the endogenous MST1-RASSF4 protein complex using MST1 immunoprecipitation (Figure 5E). Altogether, we found that in aRMS cells, RASSF4 associates with MST1, and this association is dependent on the RASSF4 SARAH domain.
RASSF4 inhibits MST1 signaling.
As a core component of the Hippo signaling pathway, MST1 plays a critical role in promoting the phospho-relay cascade that results in growth arrest, differentiation, or apoptosis (12, 13, 39). Therefore, we hypothesized that RASSF4 associates with MST1 as an inhibitory complex to block activation of the Hippo pathway. If this hypothesis were true, then artificial activation of MST1 in aRMS cells should phenocopy RASSF4 loss. Therefore, we generated stable aRMS cell lines expressing wild-type MST1, kinase-dead MST1 (MST1K59R), or vector control. We found that MST1 expression in aRMS cells was not tolerated, as cells were not viable in culture beyond two to three passages. This effect was dependent on the kinase activity of MST1, since expression of MST1K9R had no effect on aRMS cell growth and proliferation in culture (data not shown). To test whether activation of the Hippo pathway through overexpression of MST1 resulted in senescence induction, as seen with RASSF4 knockdown, we performed a senescence assay on HSMMPF+H+M cells expressing MST1, MST1K59R, or vector alone. Cells expressing MST1, but not MST1K59R, had significantly higher β-gal staining than did vector-expressing control cells (Figure 5F), suggesting that MST1 signaling in aRMS cells is sufficient to cause growth arrest and senescence.
To determine the effect of RASSF4 on the senescence-promoting activity of MST1, we generated HSMMPF+H+M cell lines expressing either MST1 or MST1K59R in combination with HA-RASSF4 or HA-RASSF4ΔSARAH. Coexpression of MST1 with HA-RASSF4 blunted the senescence-inducing effects of MST1, but expression of MST1 with HA-RASSF4ΔSARAH, which cannot bind MST1, induced senescence similarly to cells expressing MST1 alone (Figure 5F). Based on these data, we conclude that Hippo pathway signaling by MST1 overexpression induces growth arrest and senescence in aRMS cells. RASSF4 expression blocks this function of MST1, suggesting that RASSF4 may act as a Hippo pathway inhibitor.
We next tested the hypothesis that RASSF4 inhibits MST1-mediated signaling in aRMS. As a readout for MST1 activation, we examined the phosphorylation status of LATS1 and MOB1. We performed these experiments in the context of nocodazole treatment, as MST1 has been shown to be activated under these conditions and to promote phosphorylation of LATS1 and MOB1 (40). Surprisingly, we observed a reduction in phosphorylated LATS1 in nocodazole-treated cells, which was similar between control and RASSF4-knockdown cells (Figure 5G). Although we did not see changes in signaling to LATS1 in aRMS cells with RASSF4 loss, we did observe higher basal and nocodazole-induced phosphorylated MOB1. To determine whether these effects were due to MST1 signaling, we performed RASSF4 knockdown in cells expressing kinase-dead MST1 K59R. Again, we did not observe a significant change in the phosphorylation of LATS1 in cells expressing MST1 K59R (Figure 5G). However, the expression of MST1 K59R completely blocked the induction of phosphorylated MOB1 in RASSF4-knockdown cells. Further, MST1 K59R prevented the induction of p21 in RASSF4-deficient cells. To determine whether MST1 K59R expression reversed the RASSF4-knockdown phenotype, we measured senescence induction and cell cycle progression. MST1 K59R expression ameliorated the senescence induction and cell cycle arrest caused by RASSF4 loss (Figure 5, H and I). Based on these data, we conclude that aRMS cells promote senescence evasion through RASSF4-mediated suppression of MST1 signaling.
As described above, the expression of kinase-dead MST1 K59R did not impact aRMS cell growth or viability. However, even without the ability to activate downstream signaling, MST1 K59R can still be regulated like wild-type MST1 through phosphorylation by upstream kinases and caspase cleavage. Since endogenous phospho-MST1 levels and MST1 cleavage products were in low abundance in aRMS cells and were difficult to detect, we used cells expressing MST1K59R to boost MST1 expression, while maintaining cell viability. We stimulated cells with staurosporine (STS), a well-established activator of MST1, and examined the role of RASSF4 in MST1 phosphorylation and cleavage (41). We used immunoblotting to measure STS treatment in MST1 K59R–expressing HSMMPF+H+M cells and found that it led to increased phosphorylation of full-length MST1. In RASSF4-deficient cells, phospho-MST1 levels were higher in response to STS and also exhibited an approximately 35-kDa cleavage fragment (Supplemental Figure 3A). This is the reported size of the MST1 cleavage fragment believed to be regulated by caspases (41–43). We found that RASSF4-deficient, STSA-treated aRMS cells displayed increased cleaved caspase 3 species in cells expressing MST1 K59R (data not shown) or in cells with RASSF4 knockdown alone (Supplemental Figure 3B). In summary, RASSF4 may act as an endogenous inhibitor of the Hippo pathway in aRMS cells by preventing apoptotic signaling to caspase 3, blocking MST1 phosphorylation, and/or restraining MST1-mediated signaling pathways.
Hippo signaling is suppressed in aRMS.
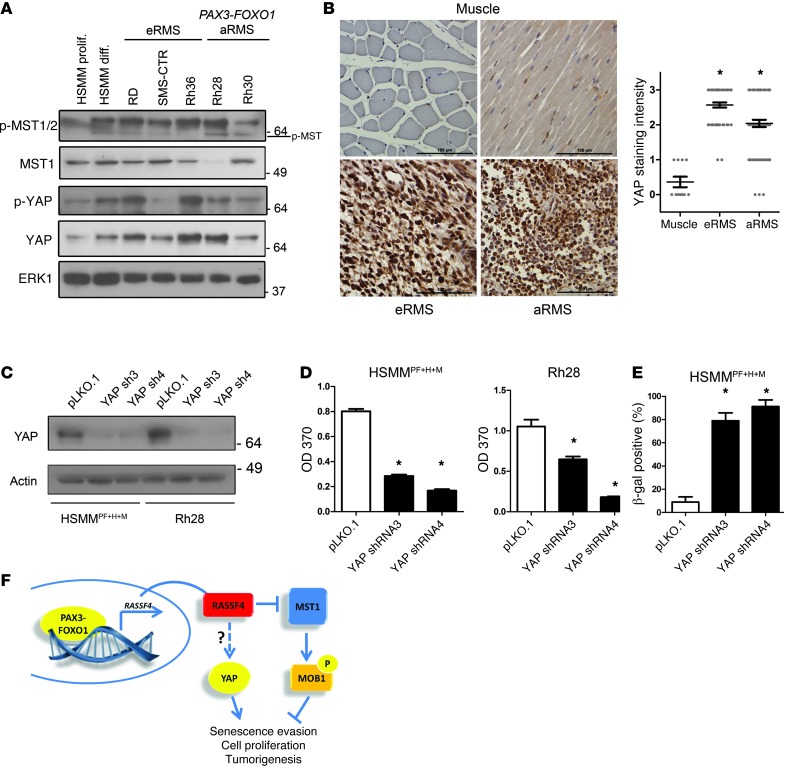
The PAX3-FOXO1/RASSF4/MST1 signaling axis identified in aRMS cells suggests that Hippo pathway suppression may play a critical role in aRMS biology. Since the Hippo pathway has not been examined in the context of RMS, we next examined the status of core Hippo signaling components in RMS cell lines and tumors. First, we examined the levels of phospho-YAP, YAP, phospho-MST1/2, and total MST1 in RMS cell lines by immunoblot analysis. We compared these levels with primary, proliferating HSMMs and in vitro differentiated HSMMs. To examine phosphorylated MST1, we used commercially available antibodies against phospho-MST1/2 (MST1 T183, MST2 T180). The resulting immunoblot had a predominant immunoreactive band that was much larger than the predicted size of phosphorylated MST1/2 (Figure 6A). A less dominant band was evident at the correct size (~60 kDa) and showed variable abundance in the HSMM and RMS cell lines. The identity of the larger molecular weight band is not known, however, phosphorylated PAK1 (~70 kDa) and phosphorylated MST1/2 are known to cross-react on immunoblots (44, 45). Phospho-YAP (S127) was induced with in vitro differentiation of myoblasts, as has been observed previously (46). We observed that phospho-YAP levels were highly variable in RMS cell lines and did not correlate with histological subtype. However, when we examined total YAP, there appeared to be upregulation of YAP protein in many of RMS cell lines (Figure 6A). Therefore, while phosphorylation of MST1 and YAP in RMS cell lines was variable, YAP protein was upregulated, suggestive of a role for YAP in RMS.
Figure 6. YAP is upregulated and pro-proliferative in aRMS.
(A) Phosphorylated MST1/2 (MST1 T183, MST2 T180), MST1, phosphorylated (S127) YAP, and YAP levels in HSMMs (proliferating and differentiated) and RMS cell lines as measured by immunoblot analysis. ERK1 was used as a loading control. (B) Left: Representative images from RMS TMAs immunostained for YAP protein. Scale bars: 100 μm. Right: Quantitation of YAP-immunostained RMS TMAs. Muscle, n = 11; eRMS, n = 58; aRMS, n = 72. Error bars represent SEM. *P < 0.0001, Mann-Whitney test. (C) YAP1 shRNA knockdown validation in HSMMPF+H+M and Rh28 cell lines as measured by immunoblot analysis. YAP1 knockdown aRMS cells were defective in proliferation, as measured by BrdU incorporation (D). YAP1 loss induced senescence in aRMS cells as measured by β-gal staining (E). (F) Model of RASSF4-mediated suppression of the Hippo pathway in aRMS. PAX3-FOXO1 transcriptionally upregulates RASSF4 through a 5′ enhancer region. Subsequent RASSF4 protein associates with MST1 and inhibits signaling to MOB1. While YAP expression is elevated in aRMS cells and tumors, a connection between RASSF4 and YAP remains to be elucidated. Both MST1 inhibition and YAP expression support cell proliferation and senescence evasion to promote tumorigenesis in aRMS cells.
We then examined the status of YAP in human RMS tumors. Using an RMS tissue microarray (TMA), we performed immunohistochemistry on 356 tissue samples representing 130 individual RMS tumors (or normal muscle) for YAP protein expression. In cross sections of normal muscle, YAP was rarely detectable, and in longitudinal sections, YAP was only weakly stained in the cytoplasm (Figure 6B). In both cases, there was no significant nuclear YAP staining, suggesting that in normal muscle, YAP was not playing an active, transcriptional role. Conversely, we found that both eRMS and aRMS samples showed high expression of cytoplasmic and nuclear YAP protein (representative samples, Figure 6B). Therefore, similarly to other cancers, RMS malignancies may upregulate the YAP oncoprotein for tumor growth and survival.
We next examined the role of YAP itself in aRMS cells. YAP-deficient aRMS cells were significantly less proliferative than control cells (Figure 6, C and D) and displayed a dramatic increase in senescence-associated β-gal staining (Figure 6E). Since YAP has been shown to regulate transcriptional complexes, we examined the effect of YAP loss on the YAP target gene CTGF. Surprisingly, CTGF levels in aRMS cells were much lower than in nontransformed HSMMs (Supplemental Figure 4A). Additionally, YAP knockdown, while causing growth arrest and senescence in aRMS cells, had no effect on CTGF expression (Supplemental Figure 4B). Together, these data suggest that either CTGF is not a YAP target gene in aRMS cells, or that YAP functions in aRMS in a manner distinct from that observed in other human cancers.
Finally, we examined whether there was a connection between RASSF4 and YAP. Compared with control aRMS cells, phospho-YAP and total YAP protein levels in RASSF4-knockdown aRMS cells were significantly reduced (Supplemental Figure 5A). Since YAP protein is turned over by sequential phosphorylation and ubiquitin-mediated degradation (47), we attempted to pharmacologically block YAP protein turnover in RASSF4-deficient cells using MG132 or IC 261. Despite these treatments, phospho-YAP and total YAP levels were still lower in RASSF4-deficient cells (data not shown), suggesting alternative mechanisms of YAP regulation. When examined at the transcriptional level, we observed a decreasing trend in YAP mRNA levels that only reached significance with one RASSF4 shRNA (Supplemental Figure 5B). Since RASSF4 loss of function was causing growth arrest and senescence, we reasoned that these effects on YAP could be caused by the growth arrest itself and RASSF4 loss. Similarly, YAP S127A add-back experiments in RASSF4-knockdown cells did not fully reverse cell viability in RASSF4-deficient cells. Instead, cells expressing YAP S127A and RASSF4 shRNA had more pronounced mitotic defects (Supplemental Figure 6, A–C). Therefore, while we cannot identify a clear connection between RASSF4 and YAP in aRMS at this time, Hippo signaling is clearly dysregulated at multiple levels in RMS through RASSF4-MST1– and YAP-mediated mechanisms (Figure 6F).
Discussion
Although the presence of the PAX3-FOXO1 fusion gene in aRMS has been known for almost 20 years, how its expression promotes poor outcome in aRMS patients remains unclear. Our previous studies in primary HSMMs showed that PAX3-FOXO1 plays an important role in senescence bypass, which is a key event in the stepwise progression toward malignant transformation, and that PAX3-FOXO1–expressing HSMMs can be precursor cells for modeling aRMS. In the current work, we present a gene expression profile of these “aRMS precursor cells,” which identifies PAX3-FOXO1–regulated genes that are involved in priming cells for aRMS tumorigenesis. An important member of this expression profile is RASSF4. Since RASSF4 has appeared in several PAX3-FOXO1 gene signatures (48–50), we predicted that it was a direct PAX3-FOXO1 target gene. Through a 5′ RASSF4 gene enhancer region, we show that this is indeed the case and that PAX3-FOXO1 can transcriptionally activate this locus. Therefore, we show that PAX3-FOXO1 itself can control an important senescence-regulating gene, RASSF4.
To the best of our knowledge, the current work is the first to study the function of endogenous RASSF4 expression. In aRMS, this role is clearly pro-proliferative. To date, upregulation of RASSFs in cancer has only been described for RASSF7, which is in a different subclass of RASSFs (N-terminal RASSFs) (51). While RASSF7 has been shown to associate with MST proteins, its role in MST signaling is not known (52). RASSF4 is most similar to RASSFs 2 and 6 (26, 53), which have also been proven to inhibit Hippo signaling (54). However, this function is divergent from many of the other RASSF family members. Indeed, in vitro and in vivo evidence has firmly established most RASSFs as tumor suppressors through their effects on many different signaling pathways including Ras and Hippo (26). As tumor suppressors, many RASSFs have been shown to be downregulated in cancer (26, 55, 56); even RASSF4 itself has been found to be methylated and downregulated in nasopharyngeal and squamous cell carcinomas, suggesting a tumor-suppressive role (57, 58). Similarly, RASSF4 expression has been shown to induce apoptosis upon expression in MCF-7 breast cancer cell lines (59). More studies are needed to understand the role of RASSFs in normal and pathologic settings.
Although this progrowth function of RASSF4 in aRMS may be distinct from the presumed canonical role of RASSFs in cancer, it does resolve a conflict between RASSF function in mammals versus Drosophila. While there are 10 RASSF genes in human cells, the Drosophila genome contains only two orthologs, known as dRASSF and dRASSF8 (53, 60). Of these genes, dRASSF is most similar to the C-terminal RASSF1-6, and most similar to RASSF4 within the Ras-association and SARAH domains. In contrast to the majority of mammalian RASSFs, dRASSF promotes cell proliferation and inhibits Hippo pathway activation through recruitment of phosphatases or competition with the SAV1 ortholog Salvador (44, 53). While RASSF4 and DRASSF can inhibit Hippo signaling, our experiments to rescue the RASSF4-knockdown phenotype with dRASSF expression were not successful (data not shown). Therefore, RASSF4 is likely not a true ortholog of dRASSF. However, our observations of the effects of RASSF4 on aRMS may reflect one evolutionarily conserved role of RASSFs in cell proliferation.
More importantly, we have revealed the importance of Hippo pathway suppression in aRMS. This work adds to the increasing body of data implicating dysregulated Hippo signaling in human malignancy (61). Indeed, inhibition of the Hippo pathway has been noted in adult cancers occurring through downregulation of Hippo pathway tumor suppressors or upregulation of the YAP oncogene (62). In murine models, homozygous loss of Lats1 leads to spontaneous soft tissue sarcomas in 14% of female mice, and 83% of Lats1–/– mice develop soft tissue sarcomas in response to carcinogenic treatments (11). In human primary soft tissue sarcomas, epigenetic downregulation has been observed in STK3/4 and LATS1 (63, 64). Although we have not observed RASSF4 regulation of MST1 signaling to LATS1, our data suggest that RASSF4 suppresses MST1 signaling to MOB1. A predominant substrate for MST1, MOB1 has been shown to be a critical mediator of cell cycle progression (40). Importantly, recent studies show that MOB1 deficiency in mice leads to spontaneous tumor susceptibility in many tissue types, including the induction of osteosarcomas and fibrosarcomas (65). While it is increasingly clear that the regulation of Hippo signaling is critical for tumor initiation and progression, surprisingly, there have been few examples of mutation of the core components of the Hippo pathway in human tumors (29). Therefore, the identification and characterization of Hippo pathway regulators have become important aspects in understanding the Hippo pathway in human disease. In the case of aRMS, a unique mechanism of Hippo pathway regulation has evolved through increased expression of the endogenous MST1 inhibitor RASSF4.
While this work identifies what we believe to be a novel mechanism of Hippo suppression in aRMS by RASSF4, it also shows that YAP is upregulated in RMS tumors. The mechanism remains to be determined, as YAP was not transcriptionally upregulated in the expression profile of PAX3-FOXO1–expressing HSMMs (data not shown). YAP promotes cell proliferation and senescence evasion, but does not promote the expression of CTGF in aRMS cells. While YAP expression has been shown to be sufficient for CTGF transcription in many models (66–68), we found that CTGF levels were very low in RMS cell lines, and loss of YAP had no effect on CTGF levels in aRMS cells. Interestingly, we observed that CTGF was a downregulated gene in PAX3-FOXO1–expressing HSMMs, suggesting that it may have a prosenescence role (data not shown). Indeed, some models of senescence show CTGF expression to be increased (69–71), suggesting that CTGF expression can be modulated by YAP-independent mechanisms. Nevertheless, YAP is clearly involved in senescence regulation. We found that YAP protein was downregulated in senescent, RASSF4-deficient cells and that YAP loss was sufficient for senescence induction in aRMS cells. As similar functions for YAP have been observed in other cell types (72), this may reflect a conserved role for YAP. Further studies are needed to define the mechanism of YAP regulation in cell proliferation and senescence in RMS.
In conclusion, we have identified a pro-proliferative signaling axis that is regulated by PAX3-FOXO1 in aRMS. RASSF4 is a PAX3-FOXO1 target gene that promotes aRMS cell growth, proliferation, and evasion of senescence. This is due in part to the inhibition of MST1 signaling by association with the MST1 protein complex. Further, the YAP oncoprotein is upregulated in the two major subtypes of RMS, implying that RMS relies on dysregulated Hippo signaling at both RASSF4-MST1 and YAP levels. To the best of our knowledge, this work is the first to establish a role for Hippo pathway suppression in RMS, provides insight into one of the many mechanisms that PAX3-FOXO1 uses to promote tumorigenic properties in aRMS, and suggests that Hippo pathway–directed therapeutics may be beneficial in this aggressive pediatric cancer.
Methods
Microarray analysis.
RNA was isolated using an RNeasy kit (QIAGEN) according to the manufacturer’s protocol. Affymetrix U133A arrays were hybridized by the Duke University Microarray Facility according to the manufacturer’s instructions. CEL files of all samples were normalized by RMA using Expression Console software (Affymetrix), zero transformed against the average expression levels of the same probe sets of the vector-expressing control HSMMs, filtered by indicated criteria, clustered using Cluster 3.0 software ( http://bonsai.hgc.jp/~mdehoon/software/cluster/), and displayed with TreeView as previously described (16, 17). All the microarray data are MIAME-compliant and have been submitted to the Gene Expression Omnibus database (GEO accession number GSE40543).
BrdU incorporation.
Cells were plated at equal density in 96-well plates. After 48 hours of growth, cells were labeled with BrdU for 3 hours, and levels of BrdU incorporation were measured using the BrdU Colorimetric kit (Roche) according to the manufacturer’s protocol.
Senescence assays.
Cells were plated at equal density in 6-well plates. The following day, cells were assayed for β-gal staining using an X-Gal–based senescence detection kit (Calbiochem) according to the manufacturer’s protocol.
Quantitative PCR and semiquantitative PCR.
RNA isolation and conversion to cDNA were performed as previously described (6). The iQ SYBER Green system (Bio-Rad) was used for quantitative PCR (qPCR) according to the manufacturer’s specifications. Semiquantitative PCR was performed as previously described (6). The primers used for semiquantitative and qPCR are listed in Supplemental Table 1. qPCR data were quantified using the equation: R = (Etarget)(ΔCt target(control – sample) / (EGAPDH)ΔCt GAPDH(control – sample), where R is the fold expression of the target gene in the sample compared with control, and Ct is the threshold cycle. Assay efficiency (E) was calculated for each primer set from the standard curve of the linear regression of a 5-point ×10 dilution series. The samples were run in triplicate.
Flow cytometry.
Cells were prepared as previously described (73) and analyzed at the Duke University Microscopy Facility.
Mouse xenograft studies.
Rh28 cells stably expressing Tet-pLKO-puro RASSF4 shRNAs were implanted s.c. into SCID/beige mice. When tumors were palpable, water was supplemented with 5% sucrose and 1 mg/ml doxycycline (Sigma-Aldrich) or 5% sucrose alone as a control. Mice were monitored twice weekly. Tumor volume was estimated by external caliper measurements and calculated as [(width)2 × length] / 2. Mice were sacrificed when tumors met IACUC-defined tumor burden or ill-health thresholds. Portions of tumors were either placed in RNAlater (QIAGEN) for RNA isolation or formalin fixed for immunohistochemical analysis.
Luciferase reporter assay.
The PAX3-FOXO1 binding site was PCR amplified from human genomic DNA using the following primers (capitalized text designates the primer region for binding genomic DNA): forward, 5′-taatcccccgggTGTAACAAACCTGCACGTTGTG-3′; reverse, 5′-cgcggatccGAAAGCTTGAAATCTTGGGCA-3′. The resulting PCR product was digested with SmaI and BamHI and cloned into a modified pGL3 control vector (lacking the SV40 enhancer) at the HpaI/BamHI sites. Plasmids were verified by DNA sequencing. Enhancer assays were performed using the Dual Luciferase Assay System (Promega) to allow normalization for transfection efficiency (Renilla). Experiments were performed similarly to those described previously (33) using a reporter-to-activator plasmid ratio of 1:1. Samples were run in triplicate, and experiments were performed at least three times.
YAP immunohistochemistry.
YAP immunohistochemistry was performed on RMS TMA slides (see Study Approval section below). Endogenous peroxidase activity was blocked by incubating deparaffinized tissue sections with 3% hydrogen peroxide (Fisher Scientific) for 20 minutes, and antigen retrieval was performed by boiling in 10 mM sodium citrate buffer for 20 minutes (pH 6). Sections were blocked for 1 hour in TBS containing 0.05% Tween 20 (Fisher Scientific), 10% normal goat serum (5425; Cell Signaling Technology), and incubated overnight at 4°C with anti-YAP primary antibody (4912, 1:40 dilution; Cell Signaling). Samples were incubated for 45 minutes with biotinylated anti-rabbit secondary antibody (1:200 dilution; Vector Laboratories), followed by catalyzed signal amplification for 30 minutes using the Vectastain Elite ABC kit (Vector Laboratories). Signal was visualized using diaminobenzidine as a substrate (4 minutes; Vector Laboratories) before sections were counterstained with hematoxylin.
The TMA was scored by two independent observers who were blinded to the annotated histologic type. For each core, YAP staining was given an integer score of 0 (no staining) to 3 (strong staining), based on staining intensity within the nucleus and cytoplasm and the percentage of YAP-positive tumor cells. In the case in which there were replicate cores from a single tumor, the scores were averaged and rounded to the nearest integer.
Statistics.
All statistical analysis (with the exception of microarray analysis) was performed using GraphPad Prism software (GraphPad Software). A P value of 0.05 was considered significant. Data represent the mean ± SD unless otherwise noted.
Study approval.
All animal studies were performed under Duke University IACUC-approved protocols. RMS TMAs generated from tumor tissue collected with informed consent through the Children’s Oncology Group (COG) were obtained from the Biopathology Center at Nationwide Children’s Hospital (Columbus, Ohio, USA). The samples were deidentified and approved for use by the IRB of Duke University.
Information regarding cell lines, plasmids, immunoprecipitation, immunoblotting, immunohistochemistry, immunofluorescence, and Drosophila genetics and expression profiling can be found in the Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Gerard Blobe, Bernard Mathey-Prevot, and Dan Wechsler for helpful discussions; Karthikeyan Mythreye for microscopy support; Katie Horowitz and Elaine Justice for technical support; and the staff of the Duke University Microarray, Flow Cytometry, and PhotoPath facilities. This research was supported by NIH grants R01 CA122706 (to C.M. Linardic); T32 CA059365 (to L.E.S. Crose); R01 CA125618 (to J.T. Chi); an Alex’s Lemonade Stand Foundation Young Investigator Award (to K.A. Galindo) and an “A” Award (to R.L. Galindo); a Burroughs Wellcome Fund Career Award for Medical Scientists (to R.L. Galindo); a grant from the Liddy Shriver Sarcoma Initiative (to C.M. Linardic); and a grant from The Hartwell Foundation (to L.E.S. Crose). COG research was supported by the Chair’s Grant CA 98543-08 and Statistics and Data Center Grant CA98413-08 from the National Cancer Institute (NCI), NIH (Bethesda, Maryland, USA). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI or the NIH. A complete listing of grant support for research conducted by the Children’s Cancer Group (CCG) and the Pediatric Oncology Group (POG) before initiation of the COG grant in 2003 is available online at: http://applications.childrensoncologygroup.org/admin/grantinfo.htm.
Footnotes
Conflict of interest: Lisa E.S. Crose, Jen-Tsan Ashley Chi, and Corinne M. Linardic disclose that they have submitted a US provisional patent application describing the development of therapeutic agents that target RASSF4.
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Citation for this article: J Clin Invest. 2014;124(1):285–296. doi:10.1172/JCI67087.
References
- 1.Ognjanovic S, Linabery AM, Charbonneau B, Ross JA. Trends in childhood rhabdomyosarcoma incidence and survival in the United States, 1975–2005. Cancer. 2009;115(18):4218–4226. doi: 10.1002/cncr.24465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorensen PH, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children’s oncology group. J Clin Oncol. 2002;20(11):2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro DN, Sublett JE, Li B, Downing JR, Naeve CW. Fusion of PAX3 to a member of the forkhead family of transcription factors in human alveolar rhabdomyosarcoma. Cancer Res. 1993;53(21):5108–5112. [PubMed] [Google Scholar]
- 4.Galili N, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5(3):230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 5.Davis RJ, D’Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54(11):2869–2872. [PubMed] [Google Scholar]
- 6.Naini S, et al. Defining the cooperative genetic changes that temporally drive alveolar rhabdomyosarcoma. Cancer Res. 2008;68(23):9583–9588. doi: 10.1158/0008-5472.CAN-07-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linardic CM, et al. The PAX3-FKHR fusion gene of rhabdomyosarcoma cooperates with loss of p16INK4A to promote bypass of cellular senescence. Cancer Res. 2007;67(14):6691–6699. doi: 10.1158/0008-5472.CAN-06-3210. [DOI] [PubMed] [Google Scholar]
- 8.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27(4):355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130(6):1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu L, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A. 2010;107(4):1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.St John MA, et al. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet. 1999;21(2):182–186. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- 12.Zhou D, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16(5):425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou D, et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci U S A. 2011;108(49):E1312–E1320. doi: 10.1073/pnas.1110428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song H, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U S A. 2010;107(4):1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredericks WJ, et al. The PAX3-FKHR fusion protein created by the t(2;13) translocation in alveolar rhabdomyosarcomas is a more potent transcriptional activator than PAX3. Mol Cell Biol. 1995;15(3):1522–1535. doi: 10.1128/mcb.15.3.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang X, et al. Functional interaction between responses to lactic acidosis and hypoxia regulates genomic transcriptional outputs. Cancer Res. 2012;72(2):491–502. doi: 10.1158/0008-5472.CAN-11-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JL, et al. Lactic acidosis triggers starvation response with paradoxical induction of TXNIP through MondoA. PLoS Genet. 2010;6(9):e1001093. doi: 10.1371/journal.pgen.1001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomescu O, et al. Inducible short-term and stable long-term cell culture systems reveal that the PAX3-FKHR fusion oncoprotein regulates CXCR4, PAX3, and PAX7 expression. Lab Invest. 2004;84(8):1060–1070. doi: 10.1038/labinvest.3700125. [DOI] [PubMed] [Google Scholar]
- 19.Crose LE, et al. FGFR4 blockade exerts distinct anti-tumorigenic effects in embryonal versus alveolar rhabdomyosarcoma. Clin Cancer Res. 2012;18(14):3780–3790. doi: 10.1158/1078-0432.CCR-10-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66(14):6936–6946. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 21.Weintraub M, Kalebic T, Helman LJ, Bhatia KG. Disruption of the MyoD/p21 pathway in rhabdomyosarcoma. Sarcoma. 1997;1(3–4):135–141. doi: 10.1080/13577149778218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen SL, Wang SC, Hosking B, Muscat GE. Subcellular localization of the steroid receptor coactivators (SRCs) and MEF2 in muscle and rhabdomyosarcoma cells. Mol Endocrinol. 2001;15(5):783–796. doi: 10.1210/me.15.5.783. [DOI] [PubMed] [Google Scholar]
- 23.Stieglitz B, et al. Novel type of Ras effector interaction established between tumour suppressor NORE1A and Ras switch II. EMBO J. 2008;27(14):1995–2005. doi: 10.1038/emboj.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khokhlatchev A, et al. Identification of a novel Ras-regulated proapoptotic pathway. Curr Biol. 2002;12(4):253–265. doi: 10.1016/S0960-9822(02)00683-8. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol Cell Biol. 2004;24(11):4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avruch J, et al. Rassf family of tumor suppressor polypeptides. J Biol Chem. 2009;284(17):11001–11005. doi: 10.1074/jbc.R800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferbeyre G, et al. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol Cell Biol. 2002;22(10):3497–3508. doi: 10.1128/MCB.22.10.3497-3508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/S0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 29.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13(4):246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 30.Galindo RL, Allport JA, Olson EN. A Drosophila model of the rhabdomyosarcoma initiator PAX7-FKHR. . Proc Natl Acad Sci U S A. 2006;103(36):13439–13444. doi: 10.1073/pnas.0605926103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avirneni-Vadlamudi U, Galindo KA, Endicott TR, Paulson V, Cameron S, Galindo RL. Drosophila and mammalian models uncover a role for the myoblast fusion gene TANC1 in rhabdomyosarcoma. . J Clin Invest. 2012;122(1):403–407. doi: 10.1172/JCI59877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bellen HJ, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167(2):761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao L, et al. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010;70(16):6497–6508. doi: 10.1158/0008-5472.CAN-10-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20(15):1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 35.Fang L, Igarashi M, Leung J, Sugrue MM, Lee SW, Aaronson SA. p21Waf1/Cip1/Sdi1 induces permanent growth arrest with markers of replicative senescence in human tumor cells lacking functional p53. Oncogene. 1999;18(18):2789–2797. doi: 10.1038/sj.onc.1202615. [DOI] [PubMed] [Google Scholar]
- 36.Wiederschain D, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8(3):498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 37.Vakifahmetoglu H, Olsson M, Zhivotovsky B. Death through a tragedy: mitotic catastrophe. Cell Death Differ. 2008;15(7):1153–1162. doi: 10.1038/cdd.2008.47. [DOI] [PubMed] [Google Scholar]
- 38.Hwang E, et al. Structural insight into dimeric interaction of the SARAH domains from Mst1 and RASSF family proteins in the apoptosis pathway. Proc Natl Acad Sci U S A. 2007;104(22):9236–9241. doi: 10.1073/pnas.0610716104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avruch J, Zhou D, Fitamant J, Bardeesy N. Mst1/2 signalling to Yap: gatekeeper for liver size and tumour development. Br J Cancer. 2011;104(1):24–32. doi: 10.1038/sj.bjc.6606011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol. 2008;18(5):311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graves JD, Draves KE, Gotoh Y, Krebs EG, Clark EA. Both phosphorylation and caspase-mediated cleavage contribute to regulation of the Ste20-like protein kinase Mst1 during CD95/Fas-induced apoptosis. J Biol Chem. 2001;276(18):14909–14915. doi: 10.1074/jbc.M010905200. [DOI] [PubMed] [Google Scholar]
- 42.Cheung WL, et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113(4):507–517. doi: 10.1016/S0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 43.Praskova M, Khoklatchev A, Ortiz-Vega S, Avruch J. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem J. 2004;381(pt 2):453–462. doi: 10.1042/BJ20040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ribeiro PS, et al. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol Cell. 2010;39(4):521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 45.Lian JP, Toker A, Badwey JA. Phosphorylation of the activation loop of gamma p21-activated kinase (gamma-Pak) and related kinases (MSTs) in normal and stressed neutrophils. J Immunol. 2001;166(10):6349–6357. doi: 10.4049/jimmunol.166.10.6349. [DOI] [PubMed] [Google Scholar]
- 46.Watt KI, et al. Yap is a novel regulator of C2C12 myogenesis. Biochem Biophys Res Commun. 2010;393(4):619–624. doi: 10.1016/j.bbrc.2010.02.034. [DOI] [PubMed] [Google Scholar]
- 47.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 2010;24(1):72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ebauer M, Wachtel M, Niggli FK, Schafer BW. Comparative expression profiling identifies an in vivo target gene signature with TFAP2B as a mediator of the survival function of PAX3/FKHR. Oncogene. 2007;26(51):7267–7281. doi: 10.1038/sj.onc.1210525. [DOI] [PubMed] [Google Scholar]
- 49.Wachtel M, et al. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64(16):5539–5545. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- 50.Ren YX, et al. Mouse mesenchymal stem cells expressing PAX-FKHR form alveolar rhabdomyosarcomas by cooperating with secondary mutations. Cancer Res. 2008;68(16):6587–6597. doi: 10.1158/0008-5472.CAN-08-0859. [DOI] [PubMed] [Google Scholar]
- 51.Underhill-Day N, Hill V, Latif F. N-terminal RASSF family: RASSF7-RASSF10. Epigenetics. 2011;6(3):284–292. doi: 10.4161/epi.6.3.14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan JJ, Flatters D, Rodrigues-Lima F, Yan J, Thalassinos K, Katan M. Comparative analysis of interactions of RASSF1-10. Adv Biol Regul. 2013;53(2):190–201. doi: 10.1016/j.jbior.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Polesello C, Huelsmann S, Brown NH, Tapon N. The Drosophila RASSF homolog antagonizes the hippo pathway. Curr Biol. 2006;16(24):2459–2465. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikeda M, et al. Hippo pathway-dependent and -independent roles of RASSF6. Sci Signal. 2009;2(90):ra59. doi: 10.1126/scisignal.2000300. [DOI] [PubMed] [Google Scholar]
- 55.Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796(2):114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 56.van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta. 2007;1776(1):58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chow LS, Lo KW, Kwong J, Wong AY, Huang DP. Aberrant methylation of RASSF4/AD037 in nasopharyngeal carcinoma. Oncol Rep. 2004;12(4):781–787. [PubMed] [Google Scholar]
- 58.Steinmann K, Sandner A, Schagdarsurengin U, Dammann RH. Frequent promoter hypermethylation of tumor-related genes in head and neck squamous cell carcinoma. Oncol Rep. 2009;22(6):1519–1526. doi: 10.3892/or_00000596. [DOI] [PubMed] [Google Scholar]
- 59.Eckfeld K, Hesson L, Vos MD, Bieche I, Latif F, Clark GJ. RASSF4/AD037 is a potential ras effector/tumor suppressor of the RASSF family. Cancer Res. 2004;64(23):8688–8693. doi: 10.1158/0008-5472.CAN-04-2065. [DOI] [PubMed] [Google Scholar]
- 60.Langton PF, Colombani J, Chan EH, Wepf A, Gstaiger M, Tapon N. The dASPP-dRASSF8 complex regulates cell-cell adhesion during Drosophila retinal morphogenesis. Curr Biol. 2009;19(23):1969–1978. doi: 10.1016/j.cub.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 61.Bao Y, Hata Y, Ikeda M, Withanage K. Mammalian Hippo pathway: from development to cancer and beyond. J Biochem. 2011;149(4):361–379. doi: 10.1093/jb/mvr021. [DOI] [PubMed] [Google Scholar]
- 62.Chan SW, et al. The Hippo pathway in biological control and cancer development. J Cell Physiol. 2011;226(4):928–939. doi: 10.1002/jcp.22435. [DOI] [PubMed] [Google Scholar]
- 63.Hisaoka M, Tanaka A, Hashimoto H. Molecular alterations of h-warts/LATS1 tumor suppressor in human soft tissue sarcoma. Lab Invest. 2002;82(10):1427–1435. doi: 10.1097/01.LAB.0000032381.68634.CA. [DOI] [PubMed] [Google Scholar]
- 64.Seidel C, et al. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Mol Carcinog. 2007;46(10):865–871. doi: 10.1002/mc.20317. [DOI] [PubMed] [Google Scholar]
- 65.Nishio M, et al. Cancer susceptibility and embryonic lethality in Mob1a/1b double-mutant mice. J Clin Invest. 2012;122(12):4505–4518. doi: 10.1172/JCI63735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22(14):1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao B, Kim J, Ye X, Lai Z-C, Guan K-L. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 2009;69(3):1089–1098. doi: 10.1158/0008-5472.CAN-08-2997. [DOI] [PubMed] [Google Scholar]
- 68.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474(7350):179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 69.Kim KH, et al. Expression of connective tissue growth factor, a biomarker in senescence of human diploid fibroblasts, is up-regulated by a transforming growth factor-beta-mediated signaling pathway. Biochem Biophys Res Commun. 2004;318(4):819–825. doi: 10.1016/j.bbrc.2004.04.108. [DOI] [PubMed] [Google Scholar]
- 70.Capparelli C, et al. CTGF drives autophagy, glycolysis and senescence in cancer-associated fibroblasts via HIF1 activation, metabolically promoting tumor growth. Cell Cycle. 2012;11(12):2272–2284. doi: 10.4161/cc.20717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lopez-Bermejo A, et al. Characterization of insulin-like growth factor-binding protein-related proteins (IGFBP-rPs) 1, 2, and 3 in human prostate epithelial cells: potential roles for IGFBP-rP1 and 2 in senescence of the prostatic epithelium. Endocrinology. 2000;141(11):4072–4080. doi: 10.1210/en.141.11.4072. [DOI] [PubMed] [Google Scholar]
- 72.Xie Q, et al. YAP/TEAD-mediated transcription controls cellular senescence. Cancer Res. 2013;73(12):3615–3624. doi: 10.1158/0008-5472.CAN-12-3793. [DOI] [PubMed] [Google Scholar]
- 73.Fang Y, Linardic CM, Richardson DA, Cai W, Behforouz M, Abraham RT. Characterization of the cytotoxic activities of novel analogues of the antitumor agent, lavendamycin. Mol Cancer Ther. 2003;2(6):517–526. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.