Abstract
The Cancer Genome Atlas (TCGA) Research Network is an ambitious multi-institutional consortium effort aimed at characterizing sequence, copy number, gene (mRNA) expression, microRNA expression, and DNA methylation alterations in 30 cancer types. TCGA data have become an extraordinary resource for basic, translational, and clinical researchers and have the potential to shape cancer diagnostic and treatment strategies. DNA methylation changes are integral to all aspects of cancer genomics and have been shown to have important associations with gene expression, sequence, and copy number changes. This Review highlights the knowledge gained from DNA methylation alterations in human cancers from TCGA.
The Cancer Genome Atlas overview
The Cancer Genome Atlas (TCGA) Research Network is a multi-institutional consortium aimed at performing comprehensive molecular profiling of 10,000 primary tumors spanning 30 cancer types (Table 1). Molecular profiling includes whole-exome sequencing for mutation detection, RNA sequencing for gene expression profiling, microRNA sequencing for microRNA expression profiling, single nucleotide polymorphism arrays for determining somatic copy number alterations (SCNAs), and Illumina Infinium BeadArrays for DNA methylation profiling (1–3). The novelty of TCGA stems from large sample numbers, centralized pathological review, selection of samples with a high fraction of tumor nuclei, genomic characterizations using nucleic acids isolated from the same tissue samples, and data integration for high-level pathway interpretations. This Review summarizes the TCGA DNA methylation findings to date, focusing on the molecular features of CpG island methylator phenotypes (CIMPs) shared among cancer types, the relationship between DNA methylation profiles and chromatin modifiers, and epigenetic silencing of key driver genes of carcinogenesis.
Table 1.
Tumors selected by TCGA for genomic characterization

Overview of cancer epigenetics
The processes of DNA methylation, chromatin modifications, nucleosome positioning, and transcription factor binding shape the epigenome and are implicated in cellular differentiation, imprinting, and X chromosome inactivation (reviewed in refs. 4, 5). In mammalian cells, DNA methylation almost exclusively occurs at the C-5 position of cytosines in the sequence context of 5′-CpG-3′ (reviewed in ref. 6). Most CpGs are methylated but are located in low-density CpG regions. However, there are regions of the genome termed “CpG islands” that contain higher CpG and G:C content, are typically unmethylated in normal somatic tissues, and are frequently located in the promoter/5′ regions of genes (7).
Human cancers exhibit DNA hypomethylation in repetitive elements, low-density CpG regions (8–15), and lamin-associated domains (16–19). This reduction in DNA methylation occurs concomitant with locus-specific DNA hypermethylation in CpG islands (reviewed in ref. 20) and CpG island shores (21). DNA hypermethylation in gene promoter regions is a frequent event in every human cancer and can inversely correlate with gene expression (reviewed in refs. 4, 5, 22). Epigenetic silencing of tumor suppressor genes has been demonstrated in several cancer types, including the cyclin-dependent kinase inhibitor 2A (CDKN2A, also referred to as p16INK4A) and secreted frizzled-related protein genes in colorectal and lung cancers (23, 24), the breast cancer 1, early onset (BRCA1) gene in breast and ovarian cancers (25), and the von Hippel–Lindau tumor suppressor E3, ubiquitin protein ligase gene in kidney cancers (24).
There is also interest in understanding coordinated cancer-associated DNA methylation events across the genome. For example, polycomb repressive complex (PRC) gene targets in ES cells also exhibit cancer-specific DNA hypermethylation (26–28). PRCs repress expression of transcription factors and drivers of development and differentiation in ES cells (29). This enrichment of DNA hypermethylation at ES PRC targets suggests that crosstalk between PRC and DNA methylation machineries may occur early in tumorigenesis (28).
CIMPs in human cancers
In 1999, Issa and colleagues first described a distinct subset of human colorectal cancers with extensive DNA hypermethylation of a subset of CpG islands that remained unmethylated in the remaining colorectal tumors (30) and are therefore distinguished from general cancer-specific DNA methylation for a specific tumor type (Figure 1). These tumors were classified as positive for a CIMP. TCGA Research Network and others have identified CIMPs in breast, colorectal, and endometrial tumors as well as in glioblastomas and acute myeloid leukemias, but not in serous ovarian, lung squamous, or kidney renal cell cancers. TCGA has unveiled similar and unique characteristics between CIMPs of different tumor types that have potential implications for the development of novel cancer diagnostics and therapeutic agents.
Figure 1. CIMP in human cancer.
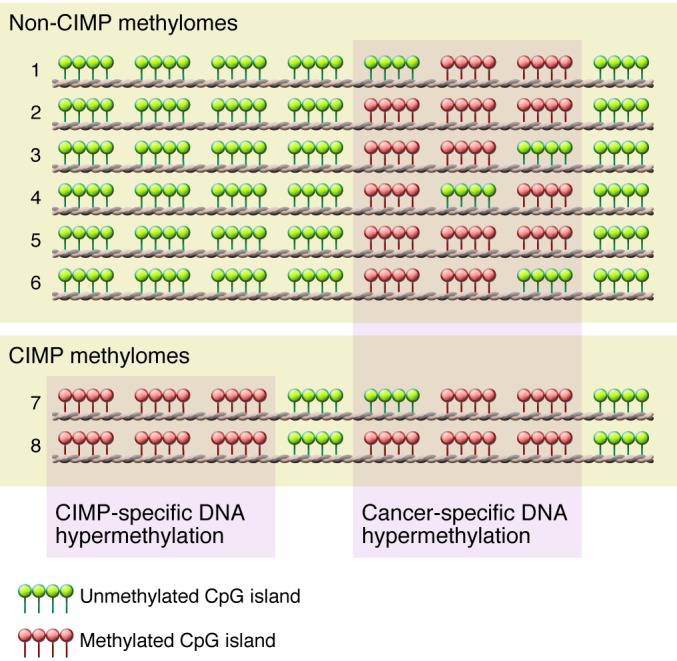
Eight individual methylomes are listed (numbered 1–8). Each row indicates an individual cancer methylome, in which clusters indicate individual CpG islands. CIMP-specific DNA hypermethylation is specific only for a proportion of tumors, while cancer-associated CpG islands are frequently methylated in both CIMP and non-CIMP tumors.
Colorectal CIMP is tightly associated with the BRAFV600E mutation.
Colorectal CIMP has been identified and characterized using candidate and genome-scale approaches in numerous reports after Issa first identified CIMP in colorectal cancer (30). Colorectal CIMP tightly associates with mutation of the v-raf murine sarcoma viral oncogene homolog B (BRAF) gene correlating with the V600 amino acid (BRAFV600E), DNA methylation of the mutL homolog 1 (MLH1) promoter region, microsatellite instability (MSI), location in the proximal colonic region, and female gender (31–35). Ogino et al. first described a CIMP-low (CIMP-L) subgroup as having an attenuated CIMP phenotype, and showed an association with mutations in the kirsten rat sarcoma viral oncogene homolog (KRAS) gene rather than BRAF (36). Similarly, Shen et al. identified the CIMP2 subgroup as displaying CIMP-associated DNA methylation, with enrichment in KRAS mutations (32). KRAS mutations are also enriched in non-CIMP tumors, but are strikingly absent in CIMP-high (CIMP-H) tumors (34–37). TCGA confirmed CIMP-H, CIMP-L, and two non-CIMP subgroups of colorectal cancer (34, 35). CIMP-H was present in approximately 15% of colorectal tumors, the majority of which also showed elevated mutation rates (hypermutated) and showed few SCNAs in contrast to the majority of colorectal tumors, which are non-CIMP, non-hypermutated, and microsatellite stable but which show substantial SCNAs (34).
The molecular mechanisms that explain the tight correlation between CIMP-H and the BRAFV600E mutation are not well understood. TCGA did not identify driver events, such as specific mutations or SCNAs in a trans-acting factor in CIMP-H and BRAFV600E colorectal tumors. While both the BRAFV600E mutation and DNA methylation changes are thought to occur early in colorectal tumorigenesis, it is unclear whether CIMP or BRAFV600E mutation is the initiating event. Hinoue and colleagues did not observe CIMP after introducing exogenous BRAFV600E into a wild-type BRAF, non-CIMP colorectal cancer cell line (38). However, Hinoue et al. did identify CIMP-specific epigenetic silencing of IGF-binding protein 7 (IGFBP7), which mediates BRAFV600E-induced apoptosis and cellular senescence. As BRAFV600E has been implicated in oncogene-induced senescence in melanomas and colorectal cancers (39), CIMP-specific silencing of IGFBP7 may create a favorable context for the generation of the BRAFV600E mutation in CIMP-positive tumors (38). However, additional experiments are needed to determine the molecular mechanism linking CIMP and BRAFV600E in colorectal cancer.
Isocitrate dehydrogenase mutations and DNA hypermethylation in glioma and AML.
Glioblastoma multiforme (GBM) was the first cancer selected by TCGA Research Network for molecular characterization (40). The majority of GBM tumors are considered primary or de novo, and approximately 5% are secondary GBMs which progress from lower-grade tumors. Parsons and colleagues used a comprehensive sequencing strategy and identified specific heterozygous somatic point mutations in the isocitrate dehydrogenase 1 (IDH1) gene (most often at amino acid residue R132) in 12% of GBM patients (41). Somatic mutations in IDH2 (amino acid R172) were also identified in gliomas. These mutations occur at a low frequency and are mutually exclusive with IDH1 mutations (42, 43).
TCGA Research Network sequenced 601 candidate genes for somatic mutations but did not sequence IDH1 or IDH2 in their initial report (40); however, in a recent follow-up report, TCGA identified IDH1 mutations through whole exome and whole genome sequencing approaches (44). In 2010, Noushmehr and TCGA network colleagues (45) convincingly showed an extremely tight correlation between GBM tumors with the IDH1R132H mutation and a glioma CIMP (G-CIMP). All primary GBM tumors with an IDH1 mutation were G-CIMP, but there were also a very small number of WT IDH1 (IDH1WT) G-CIMP tumors. G-CIMP tumors also showed attenuated SCNAs and TP53 alterations and correlated with younger patient age and improved survival. Moreover, G-CIMP is highly prevalent in recurrent and secondary GBM tumors and is inversely correlated with glioma stage (45).
Brennan and TCGA colleagues (44) confirmed the G-CIMP subgroup and its association with IDH1 and TP53 alterations. Including the G-CIMP subgroup, TCGA identified a total of six DNA methylation subgroups using unsupervised clustering analyses and showed enrichment of some individual DNA methylation groups with gene expression–based subgroups (neural, proneural, mesenchymal, and classical) identified previously (46), with G-CIMP tumors tightly associated with the proneural subgroup. Interestingly, one subgroup (cluster M6) was generally DNA hypomethylated and enriched for proneural non–G-CIMP tumors with IDH1WT. G-CIMP patients displayed longer survival times, whereas non–G-CIMP patients had shorter survival outcomes. Interestingly, even though the M6 tumors were proneural, patients with this tumor type did not show survival advantages, which suggests that the aberrant molecular features of G-CIMP may be important in conferring the survival advantage in G-CIMP patients.
Because of the expanded GBM tumor collection and exome sequencing depth, TCGA identified amplifications of MYC (v-myc avian myelocytomatosis viral oncogene homolog) in G-CIMP tumors. MYC is a transcription factor that is frequently altered in cancer and is involved in cell cycle progression, transformation, and apoptosis (reviewed in ref. 47). In addition, TCGA also identified ATRX (α-thalassemia/mental retardation syndrome X-linked) somatic mutations in G-CIMP tumors, which are highly correlated with IDH1 mutations. ATRX belongs to the SWI/SNF family of chromatin remodelers and functions as an ATPase and helicase that facilitates the substitution of variant histone H3.3 into chromatin at telomeres (48).
ATRX mutations are predominant in the alternative lengthening of telomeres (ALT), a process by which telomere length is maintained independent of telomerase in cancer cells (48, 49). Interestingly, TCGA identified telomerase (TERT) promoter mutations in 21 of 25 GBM tumors sequenced, and these mutations correlated with increased TERT gene expression. Notably, all four tumors without TERT mutations did not display elevated TERT gene expression, but instead contained ATRX alterations. Therefore, it is possible that GBM tumors maintain telomere length by either TERT mutations to reactivate TERT gene expression or via ATRX mutations in ALT.
The link between the IDH1R132H mutation and G-CIMP has generated tremendous attention from basic and translational scientists. Recent studies have shown that introducing exogenous IDH1R132H into immortalized cell lines with endogenous IDH1WT was sufficient to drive G-CIMP–based DNA methylation events and increased occupancy of histone H3 lysine 9 dimethylation (H3K9me2), histone H3 lysine 27 trimethylation (H3K27me3), and H3K36me3, which correlate with methylated DNA regions in the cancer genome (50, 51).
These epigenomic changes may occur as a result of the function of the mutant IDH proteins. IDH1WT functions as a dimer to catalyze the reduction of nicotinamide adenine dinucleotide phosphate (NADP+) to NADPH by converting of isocitrate to α-ketoglutarate (α-KG) (reviewed in ref. 52). However, the IDH mutant catalyzes the conversion of α-KG to D-2-hydroxyglutarate (2-HG) (53–55), resulting in elevated 2-HG levels (53, 56, 57). IDH1 mutant-mutant (IDH1MUT-MUT) homodimers or mutant-WT (IDH1MUT-WT) heterodimers have been identified in vitro (58). However, recent in vitro experiments have shown that the presence of the IDH1WT protein is associated with increased 2-HG levels (54), and that IDH1MUT-WT dimers show more enzymatic activity toward α-KG than IDH1MUT-MUT alone (59), suggesting that both forms may be required for 2-HG production.
2-HG inhibits the TET family of enzymes and Jumonji-C domain containing histone lysine demethylases, which normally utilize α-KG as a co-substrate (60, 61). Thus, the production of 2-HG by mutant IDH1-containing enzymes effectively inhibits TET activity (Figure 2A). TETs catalyze the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and ultimately to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (62, 63). 5fC and 5caC are substrates for the thymine DNA glycosylase-mediated base excision repair pathway that ultimately results in replacement with an unmethylated cytosine, effectively demethylating the locus (64–67) (Figure 2B).
Figure 2. DNA demethylation dynamics in human cancers.
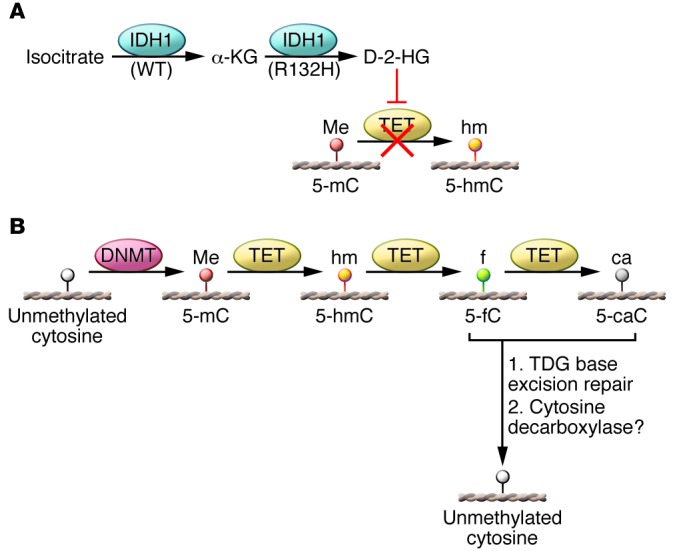
(A) Role of IDH1 in shaping the cancer methylome. WT IDH1 converts isocitrate to α-KG, but the mutant IDH1R132H enzyme catalyzes the conversion α-KG to 2-HG, which inhibits TET-mediated DNA demethylation. This mechanism is proposed to explain DNA hypermethylation in IDH1- and TET-mutated cancers. (B) Proposed mechanism of DNA demethylation by the TET family of DNA demethylases, followed by thymine-DNA glycosylase (TDG) base excision repair, resulting in unmethylated cytosines.
AML tumors also harbor IDH1 mutations (mostly at the R132 residue), IDH2 mutations (at residues R140 and R172) (68), and TET mutations. IDH1 and IDH2 mutations are mutually exclusive and occur in up to 30% of acute myeloid leukemias (69–74). Moreover, TET2 mutations are also mutually exclusive with IDH mutations. Figueroa and colleagues showed that AML tumors with TET2 mutations displayed a DNA hypermethylation signature that is similar to that of AML tumors with IDH mutations (69), suggesting that IDH and TET enzymes may have redundant roles in DNA demethylation. TCGA also demonstrated that AML tumors with IDH somatic mutations showed substantial gains in DNA methylation, and similarly, in TET2-mutated AML tumors (75).
The inhibition of DNA demethylation as a result of the IDH1 and TET mutations is consistent with the epigenomic landscapes identified in G-CIMP and AML tumors, but the basis for the target site specificity of the cancer-associated DNA methylation events observed in IDH1 mutant tumors is unclear. Two recent studies have identified IDAX (inhibition of the Dvl and Axin complex, also known as CXXC4) and early B cell factor 1 (EBF1) as potential TET2-interaction partners. IDAX can bind unmethylated promoter CpG islands as well as the TET2 catalytic domain, resulting in decreased 5hmC levels and TET2 degradation via caspase activation. EBF1, by binding to both DNA and TET2, may also regulate DNA demethylation in IDH1-mutant cancers in a tissue- and sequence-specific manner (76).
Clinical importance of MGMT DNA methylation in GBM.
The standard of care chemotherapeutic agent for treating GBM patients is temozolomide (TMZ), which acts as a methyl donor for alkylation of the N-7, O-3, and O-6 positions of nucleotide bases (reviewed in ref. 77). TMZ treatment initiates a DNA repair response, but it is believed that the mismatch repair machinery cannot effectively incorporate the correct base opposite to O-6-methylguanine lesions after the initial DNA strand-nicking step in the repair pathway. The nicks accumulate and are thought to promote an apoptotic response that results in cell death.
O-6-methylguanine methyltransferase (MGMT) removes methyl groups from the O-6 position of guanines, thereby rendering TMZ ineffective. Promoter DNA hypermethylation–based silencing of MGMT sensitizes GBM tumors to TMZ, and as a result has been used as a diagnostic barometer for selecting TMZ as a treatment option for GBM patients. TCGA recently identified MGMT DNA methylation in nearly 50% of GBM patients, and MGMT DNA methylation was more prevalent in G-CIMP tumors than in non–G-CIMP tumors (44). MGMT DNA methylation correlated with patient response in GBMs belonging to the classical gene expression subgroup only, and not in the proneural, neural, or mesenchymal groups.
Breast cancer CIMP.
Breast cancer is a complex and heterogeneous disease, and breast tumor subgroups have been proposed based on the expression status of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2; also known as ERBB2). TCGA identified five DNA methylation subgroups of breast tumors (78), with one subgroup exhibiting a CIMP-like DNA methylation signature (B-CIMP), as previously described by Fang and colleagues (79). B-CIMP tumors were positive for ER, PR, and HER2 expression and were enriched for the luminal B gene expression subgroup as well as epigenetic silencing of genes in the Wnt-signaling pathway (78), as has also been described for colorectal tumors (34).
Endometrial carcinoma CIMP.
Endometrial tumors are classified into two groups: the type I endometrioid tumors that are hormone receptor positive with good prognosis and the type II serous tumors that mostly occur in older women and correlate with poor outcome. TCGA identified four DNA methylation subgroups of endometrial tumors (80), with one subgroup displaying a CIMP-like (E-CIMP) DNA hypermethylation profile. E-CIMP was previously identified by Whitcomb and colleagues (81). Similar to colorectal CIMP tumors, the E-CIMP tumors were hypermutated, MSI positive due to MLH1 promoter DNA hypermethylation, and did not contain TP53 somatic mutations or extensive SCNAs (34, 80). However, E-CIMP tumors did not harbor BRAFV600E or IDH1 mutations, as described in colorectal and glioma CIMP tumors, respectively, pointing to an alternative mechanism of CIMP-specific DNA methylation in endometrial tumors.
Similarities and differences among CIMPs of individual human cancers
TCGA has confirmed the presence of CIMP in colorectal, breast, and endometrial cancers, while providing the first comprehensive view of G-CIMP. However, the molecular events that result in CIMP-specific DNA methylation events for each tumor type remain unclear and suggest that universal presentation of CIMP is not apparent across tumor types. For instance, colorectal CIMP is associated with BRAFV600E mutation, but G-CIMP is tightly linked to IDH1 mutations. However, B-CIMP and E-CIMP tumors are not associated with these gene mutations. Moreover, colorectal and endometrial CIMPs are associated with MSI via epigenetic silencing of MLH1, but this was not identified in CIMPs from other tumor types. Therefore, CIMP-specific DNA methylation targets may be largely non-overlapping with no consensus CIMP DNA methylation signature across tumor types, suggesting that CIMPs may be manifested by several molecular pathways and diverse sets of genomic alterations. However, formal analyses to determine the extent of common and unique CIMP-specific DNA hypermethylation signatures between tumor types are still needed and are currently in progress by TCGA researchers.
Normal-like DNA methylation subgroups of human cancers
Unsupervised analyses of TCGA breast and endometrial cancers each identified subgroups of tumors with normal-like DNA methylation profiles. In breast cancers, this subgroup showed enrichment with the basal-like (triple-negative) gene expression group, in which ER, PR, and HER2 were not expressed. In endometrial tumors, the normal-like DNA methylation subgroup was enriched in the serous-type tumors. Both subgroups displayed TP53 somatic mutations and extensive SCNAs and overall were similar to serous ovarian tumors (78, 80, 82). These findings indicate that these subtypes may share a common mechanism of epigenomic changes in tumorigenesis separate from their CIMP counterparts. CIMP-positive tumors generally present with genomic stability and the absence of TP53 mutations, with the exception of G-CIMP tumors, which show enrichment for TP53 mutations.
CDKN2A inactivation in lung squamous cell carcinoma
TCGA Research Network has recently reported the integrated genomic characterization findings of lung squamous cell carcinoma (LUSC) (83). Overall, LUSCs displayed increased rates of DNA sequence alterations compared with other tumor types. Among the genes that were significantly mutated, CDKN2A alterations are of interest. The CDKN2A locus encodes for two well-characterized cell cycle–regulating proteins, p16INK4A and p14ARF, that are encoded by alternate splicing of an overlapping set of exons in the locus. p16INK4A inactivates cyclin-dependent kinases that phosphorylate the retinoblastoma protein, resulting in a G1 phase arrest, while p14ARF stabilizes p53 by inducing MDM2 degradation and blocking its function (84). TCGA showed that CDKN2A inactivation occurs in over 70% of LUSCs by a mutually exclusive combination of epigenetic silencing, inactivating mutations and deletions, with each contributing to CDKN2A gene expression alterations, suggesting that multiple routes of CDKN2A inactivation are important to LUSC tumorigenesis.
BRCA1 epigenetic silencing in serous ovarian cancers
Integrative analyses of DNA methylation and gene expression data identified 168 epigenetically silenced genes in TCGA serous ovarian tumors (82). These genes display promoter DNA hypermethylation together with reduced gene expression. Among those is BRCA1, an important driver of carcinogenesis that encodes for a protein involved in repairing DNA double-strand breaks. Interestingly, TCGA demonstrated that BRCA1 was inactivated by mutation and epigenetic silencing in a mutually exclusive manner in ovarian tumors (82). Although neither BRCA1 mutation nor epigenetic silencing correlated with prognosis (85), BRCA1 epigenetic silencing was more abundant than somatic mutations in ovarian tumors, and patients with ovarian cancer with BRCA1 epigenetic silencing were younger than those with BRCA1 mutation inactivation, suggesting that epigenetic alterations of BRCA1 are important early events in ovarian tumorigenesis.
Chromatin modifier gene mutations affect the cancer methylome
Somatic mutations of chromatin-modifier genes have been reported in several human cancers (reviewed in ref. 5). While the effects of these mutations on the cancer methylome have not been completely explored, TCGA AML tumors displayed substantial levels of mutations in chromatin-modifier genes, with 30% of AML tumors harboring somatic mutations in chromatin modifier genes and 44% of tumors with mutations in DNA methylation–related genes. AML tumors with fusion events involving the myeloid/lymphoid or mixed-lineage leukemia (MLL) gene family or concordant mutations in nucleophosmin (NPM1), fms-related tyrosine kinase (FLT3), and DNA (cytosine-5-)-methyltransferase 3 alpha (DNMT3A) displayed DNA hypomethylation (75). The MLL gene family encodes histone lysine methyltransferases, and DNMT3A codes for a de novo DNA methyltransferase. These findings point to a strong link between cancer genetics and epigenetics that will be better understood by forthcoming mechanistic studies.
TCGA also identified somatic mutations of SET domain containing 2 (SETD2) in 11.5% of KIRC tumors (86). SETD2 is a H3K36 methyltransferase, and H3K36me3 marks are associated with DNA methylation at actively transcribed gene regions (87). Recent work from Chantalat and colleagues has also identified H3K36me3 marks in facultative and constitutive heterochromatin (88). TCGA demonstrated that KIRC tumors with SETD2 mutations displayed DNA hypomethylation at non-promoter regions that are marked by H3K36me3 and DNA hypermethylation at non-H3K36me3 occupied regions in normal kidney cells, suggesting a crosstalk between DNA methylation and H3K36 methylation in shaping the epigenome.
Future directions
Several interesting applications utilizing TCGA DNA methylation data are possible. For instance, TCGA data can be analyzed to delineate between driver and passenger DNA methylation events in carcinogenesis. Also of interest is determining an understanding of DNA methylation changes across several cancer types, not only in relation to CIMP, but also in identifying specific cancer-associated DNA methylation alterations for potential diagnostic purposes. Such DNA methylation–based biomarkers of disease can be used to identify primary tumors as belonging to specific subgroups and may have utility in personalized medicine. In addition, DNA methylation biomarkers can be used as tools for early detection of cancer in patient blood and a mechanism to track response to therapy.
TCGA has been and will continue to be a widely used discovery and validation resource for other genomics-based projects. TCGA will also continue to inspire the development of novel informatics approaches for integrative, high-level summaries of a wide range of molecular data types. With the tremendous volume of genomic data generated by TCGA efforts, functional experiments are needed to fully characterize cancer genomes. Currently, TCGA Research Network is completing genomic characterizations across several cancers to identify epigenomic signatures that will provide a powerful view into the molecular events that shape human carcinogenesis.
Acknowledgments
I thank Peter A. Jones and Peter W. Laird for helpful discussions and constructive comments.
Footnotes
Conflict of interest: Daniel J. Weisenberger is a consultant for Zymo Research Corporation, which distributes commercially available products for DNA methylation–based experiments. Zymo Research did not fund or in any way contribute to this Review.
Citation for this article: J Clin Invest. 2014;124(1):17–23. doi:10.1172/JCI69740.
References
- 1.Bibikova M, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Bibikova M, Fan JB. GoldenGate assay for DNA methylation profiling. Methods Mol Biol. 2009;507:149–163. doi: 10.1007/978-1-59745-522-0_12. [DOI] [PubMed] [Google Scholar]
- 3.Bibikova M, et al. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009;1(1):177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153(1):38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 7.Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40(1):91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 8.Ehrlich M, Wang RY. 5-Methylcytosine in eukaryotic DNA. Science. 1981;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- 9.Diala ES, Hoffman RM. Hypomethylation of HeLa cell DNA and the absence of 5-methylcytosine in SV40 and adenovirus (type 2) DNA: analysis by HPLC. Biochem Biophys Res Commun. 1982;107(1):19–26. doi: 10.1016/0006-291x(82)91663-1. [DOI] [PubMed] [Google Scholar]
- 10.Diala ES, Hoffman RM. DNA methylation levels in normal and chemically-transformed mouse 3T3 cells. Biochem Biophys Res Commun. 1982;104(4):1489–1494. doi: 10.1016/0006-291x(82)91419-x. [DOI] [PubMed] [Google Scholar]
- 11.Ehrlich M, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 13.Gama-Sosa MA, et al. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11(19):6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48(5):1159–1161. [PubMed] [Google Scholar]
- 15.Weisenberger DJ, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33(21):6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman BP, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44(1):40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43(8):768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hon GC, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22(2):246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 21.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki H, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36(4):417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 24.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hilton JL, Geisler JP, Rathe JA, Hattermann-Zogg MA, DeYoung B, Buller RE. Inactivation of BRCA1 and BRCA2 in ovarian cancer. J Natl Cancer Inst. 2002;94(18):1396–1406. doi: 10.1093/jnci/94.18.1396. [DOI] [PubMed] [Google Scholar]
- 26.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39(2):237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39(2):232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 28.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39(2):157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 29.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogino S, Kawasaki T, Ogawa A, Kirkner GJ, Loda M, Fuchs CS. Fatty acid synthase overexpression in colorectal cancer is associated with microsatellite instability, independent of CpG island methylator phenotype. Hum Pathol. 2007;38(6):842–849. doi: 10.1016/j.humpath.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 32.Shen L, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104(47):18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yagi K, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16(1):21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinoue T, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22(2):271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8(5):582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weisenberger DJ, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38(7):787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 38.Hinoue T, et al. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One. 2009;4(12):e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132(3):363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartmann C, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 43.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 48.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140(5):678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lovejoy CA, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8(7):e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duncan CG, et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res. 2012;22(12):2339–2355. doi: 10.1101/gr.132738.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 2013;13(5):345. doi: 10.1007/s11910-013-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin G, et al. Disruption of wild-type IDH1 suppresses D-2-hydroxyglutarate production in IDH1-mutated gliomas. Cancer Res. 2013;73(2):496–501. doi: 10.1158/0008-5472.CAN-12-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gross S, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207(2):339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sellner L, et al. Increased levels of 2-hydroxyglutarate in AML patients with IDH1-R132H and IDH2-R140Q mutations. Eur J Haematol. 2010;85(5):457–459. doi: 10.1111/j.1600-0609.2010.01505.x. [DOI] [PubMed] [Google Scholar]
- 58.Jin G, et al. 2-hydroxyglutarate production, but not dominant negative function, is conferred by glioma-derived NADP-dependent isocitrate dehydrogenase mutations. PLoS One. 2011;6(2):e16812. doi: 10.1371/journal.pone.0016812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bralten LB, et al. IDH1 R132H decreases proliferation of glioma cell lines in vitro and in vivo. Ann Neurol. 2011;69(3):455–463. doi: 10.1002/ana.22390. [DOI] [PubMed] [Google Scholar]
- 60.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336(6083):934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 64.Song CX, et al. Genome-wide Profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153(3):678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen L, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153(3):692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raiber EA, et al. Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase. Genome Biol. 2012;13(8):R69. doi: 10.1186/gb-2012-13-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hashimoto H, Hong S, Bhagwat AS, Zhang X, Cheng X. Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012;40(20):10203–10214. doi: 10.1093/nar/gks845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res. 2012;18(20):5562–5571. doi: 10.1158/1078-0432.CCR-12-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marcucci G, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28(14):2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paschka P, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28(22):3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 73.Tefferi A, et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia. 2010;24(7):1302–1309. doi: 10.1038/leu.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wagner K, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010;28(14):2356–2364. doi: 10.1200/JCO.2009.27.6899. [DOI] [PubMed] [Google Scholar]
- 75.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guilhamon P, et al. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat Commun. 2013;4:2166. doi: 10.1038/ncomms3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clin Cancer Res. 2000;6(7):2585–2597. [PubMed] [Google Scholar]
- 78.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fang F, et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med. 2011;3(75):75ra25. doi: 10.1126/scitranslmed.3001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whitcomb BP, Mutch DG, Herzog TJ, Rader JS, Gibb RK, Goodfellow PJ. Frequent HOXA11 and THBS2 promoter methylation, and a methylator phenotype in endometrial adenocarcinoma. Clin Cancer Res. 2003;9(6):2277–2287. [PubMed] [Google Scholar]
- 82.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haber DA. Splicing into senescence: the curious case of p16 and p19ARF. Cell. 1997;91(5):555–558. doi: 10.1016/S0092-8674(00)80441-9. [DOI] [PubMed] [Google Scholar]
- 85.Yang D, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA. 2011;306(14):1557–1565. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280(18):17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 88.Chantalat S, et al. Histone H3 trimethylation at lysine 36 is associated with constitutive and facultative heterochromatin. Genome Res. 2011;21(9):1426–1437. doi: 10.1101/gr.118091.110. [DOI] [PMC free article] [PubMed] [Google Scholar]


