Abstract
While murine-based systems to identify cancer-promoting agents (carcinogens) are established, models to identify compounds that promote aging (gerontogens) have not been described. For this purpose, we exploited the transcription of p16INK4a, which rises dynamically with aging and correlates with age-associated disease. Activation of p16INK4a was visualized in vivo using a murine strain that harbors a knockin of the luciferase gene into the Cdkn2a locus (p16LUC mice). We exposed p16LUC mice to candidate gerontogens, including arsenic, high-fat diet, UV light, and cigarette smoke and serially imaged animals to monitor senescence induction. We show that exposure to a high-fat diet did not accelerate p16INK4a expression, whereas arsenic modestly augmented, and cigarette smoke and UV light potently augmented, activation of p16INK4a-mediated senescence. This work provides a toxicological platform to study mammalian aging and suggests agents that directly damage DNA promote molecular aging.
Introduction
Murine models have been critical to the identification of carcinogens. For example, commercially available models (i.e., BigBlue, MutaMouse, and the p53+/– strain; refs. 1–3) represent core tools in cancer toxicology. These reagents increase the efficiency and sensitivity of carcinogen identification, while reducing animal requirements and costs. In 1987, Martin proposed the concept of gerontogens (4): environmental agents that accelerate molecular aging, analogous to carcinogens promoting neoplasia. While murine models for carcinogen testing are advanced, in vivo tools for gerontogen testing do not exist.
Mammalian aging is complex, with distinct molecular processes contributing to age-related tissue dysfunction. While no single process underpins aging, several lines of evidence suggest senescence activation is an important contributor. Markers of cellular senescence dramatically increase with aging in humans and mice, including expression of the p16INK4a tumor suppressor gene (5–8). Moreover, activation of p16INK4a and senescence has been linked to replicative hypofunction in neural stem cells (9), hematopoietic progenitors (10), lymphocytes (11, 12), and pancreatic β cells (13). Altered regulation of the senescence-promoting CDKN2a/b locus, which encodes p16INK4a, p15INK4b, and ARF, has been linked to aging phenotypes, including atherosclerotic disease, type II diabetes, glaucoma, and several malignancies (14). Finally, pharmacologic and genetic approaches to manipulating the number of senescent cells in vivo can ameliorate age-associated phenotypes such as sarcopenia, kyphosis, and hyporeplication of T lymphocytes and pancreatic β cells (15–17). Here, we sought to determine whether in vivo activation of p16INK4a, measured using the recently described p16LUC reporter allele (18), could effectively assess the gerontogenic effects of environmental agents.
Results and Discussion
The p16LUC allele features firefly luciferase knocked in to exon 1α of the murine Cdkn2a locus (18), with expression under the control of the endogenous p16INK4a promoter and distant cis-regulatory elements (19, 20). Importantly, p16INK4a expression is negligible in healthy cells from young mice, but increases more than 300-fold on a per-cell basis with age-promoting stresses (7, 18). The large, dynamic range of p16INK4a expression makes p16LUC a valuable tool for in vivo imaging of senescence induction. To assess the effect of candidate gerontogens — high-fat diet (HFD), UV light, arsenic, and cigarette smoke (CS) — on senescence, we performed serial analysis of p16LUC cohorts.
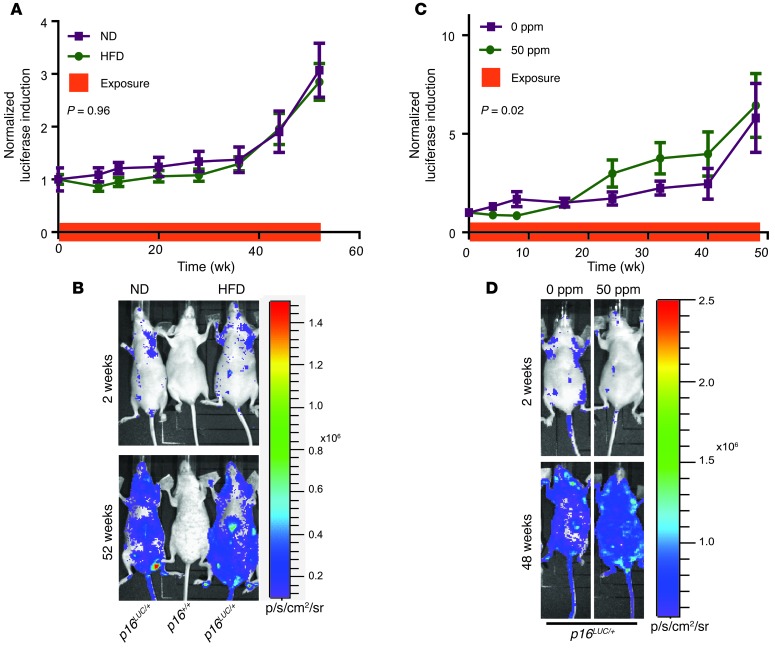
Prior studies on the effects of obesity and HFD on p16INK4a expression in vivo have yielded inconsistent results. HFD has been reported to increase senescence and/or p16INK4a expression in vascular cells (21, 22), whereas p16INK4a mRNA expression in peripheral blood T lymphocytes (PBTLs) does not correlate with body mass index, a marker of obesity, in humans (8). To examine the effects of HFD on whole-body senescence, littermate p16LUC mice were placed on a 42%-fat HFD or 4%-fat normal diet (ND) for 18 months starting at 8–10 weeks of age. HFD feeding produced a clear pharmacodynamic effect, causing significant weight gain and hepatic steatosis, although no differences in overall survival were noted (Supplemental Figure 1, A–C; supplemental material available online with this article; doi: 10.1172/JCI70960DS1). Consistent with prior reports (7, 18), we observed an exponential increase in p16INK4a expression with aging; however, the rate of increase, as assessed by total-body luciferase imaging (TBLI), was not different between cohorts (Figure 1, A and B). Due to the wide range of weight gain among HFD-fed mice, we analyzed the relationship between weight and luciferase intensity at 52 weeks and observed no correlation (Supplemental Figure 1D). Correspondingly, expression of p16INK4a mRNA was not altered in liver and spleen harvested from 18-month-old mice fed HFD versus ND (Supplemental Figure 1E). These data suggested that HFD does not accelerate p16INK4a expression in vivo, at least within the sensitivity of luciferase detection.
Figure 1. HFD does not increase p16INK4a expression, while arsenic moderately increases p16INK4a expression.
p16LUC mice were (A and B) fed ND or HFD or (C and D) exposed to 0 or 50 ppm arsenic. (A and C) Luciferase intensity, normalized to mean luciferase levels of the entire cohort at day 0 and graphed as a function of time. Exposure duration is shown by the orange bar on the x axis. Error bars indicate SEM. P values were determined by linear regression analysis. (B and D) Representative TBLI of experiments in A and C, respectively.
Arsenic is an environmental toxicant linked to human aging-associated phenotypes (i.e., several cancers, type II diabetes, and atherosclerosis; ref. 23). While not a direct clastogen, arsenic may indirectly affect DNA damage and repair (24–26). We examined the effect of chronic arsenic exposure on p16INK4a expression in vivo by administering 0 or 50 ppm arsenic to littermate p16LUC mice in the drinking water for 12 months starting at 8–10 weeks of age. After 48 weeks of exposure, arsenic-treated mice exhibited significant hepatic accumulation (Supplemental Figure 2A). Consistent with known effects of arsenic (27), treated mice showed small, structurally deranged pancreatic islets with an extreme decrease in total islet mass (Supplemental Figure 2, B and C). Overall survival rates were not different between cohorts (Supplemental Figure 2D). A modest but significant increase in p16INK4a expression was observed by 24 weeks of exposure using TBLI (P = 0.023; Figure 1, C and D). Additionally, expression of p16INK4a mRNA was higher in spleen, but not liver, of 1-year-old mice administered 50 versus 0 ppm (Supplemental Figure 2E). These data suggest that chronic arsenic exposure modestly accelerates the accumulation of p16INK4a-expressing cells.
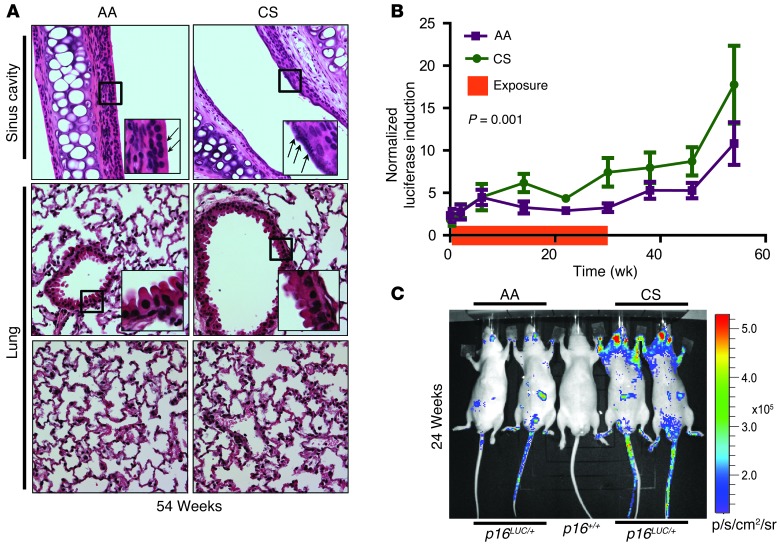
CS promotes DNA damage (28, 29) and age-related diseases such as emphysema, atherosclerosis, and several cancers (30, 31). Chronic CS exposure in humans is associated with increased p16INK4a expression in PBTLs (8, 32). To determine the effects of moderate CS exposure on cellular senescence in vivo, littermate p16LUC mice were exposed to CS or ambient air (AA) for 1 hour per day, 5 days per week for 6 months starting at 10–12 weeks of age. After 6 months of exposure, mice were further observed without CS up to 1 year of age. At the end of the study, histological analysis of the sinus cavity of CS-exposed mice showed particulate deposits along the epithelial layer of the nasal papilla, whereas no differences were noted in lung architecture between CS and AA cohorts at this tobacco dose (Figure 2A), which was also not associated with accelerated mortality (Supplemental Figure 3A). A significant increase in p16INK4a induction between the CS and AA cohorts, as measured by TBLI, was readily noted within weeks of CS exposure and persisted after CS was discontinued (Figure 2B). Consistent with histologic examinations of the nasal cavities and lungs (Figure 2A), luciferase induction was noted in the head and neck region of mice, but not the thoracic and abdominal regions (Figure 2C). Correspondingly, p16INK4a mRNA levels in the liver, lung, axillary lymph nodes, and spleen were similar in CS- and AA-exposed mice (Supplemental Figure 3B). These data suggest that low-dose CS exposure accelerates accumulation of senescent cells in tissues that receive the highest exposure (i.e., nasal epithelium).
Figure 2. CS increases p16INK4a expression. p16LUC mice were exposed to CS or AA.
(A) Upper sinus cavity (top), airway bronchioles (middle; airway epithelium peg cells highlighted), and alveolar sacs (bottom) after 54 weeks. Arrows indicate areas of particulate deposit along the epithelial layer. Original magnification, ×40, ×280 (insets). (B) Luciferase intensity, normalized to mean luciferase levels of the entire cohort at day 0 and graphed as a function of time. Exposure duration is shown by the orange bar on the x axis. Error bars indicate SEM. P value was determined by linear regression analysis. (C) Representative TBLI of experiments in B.
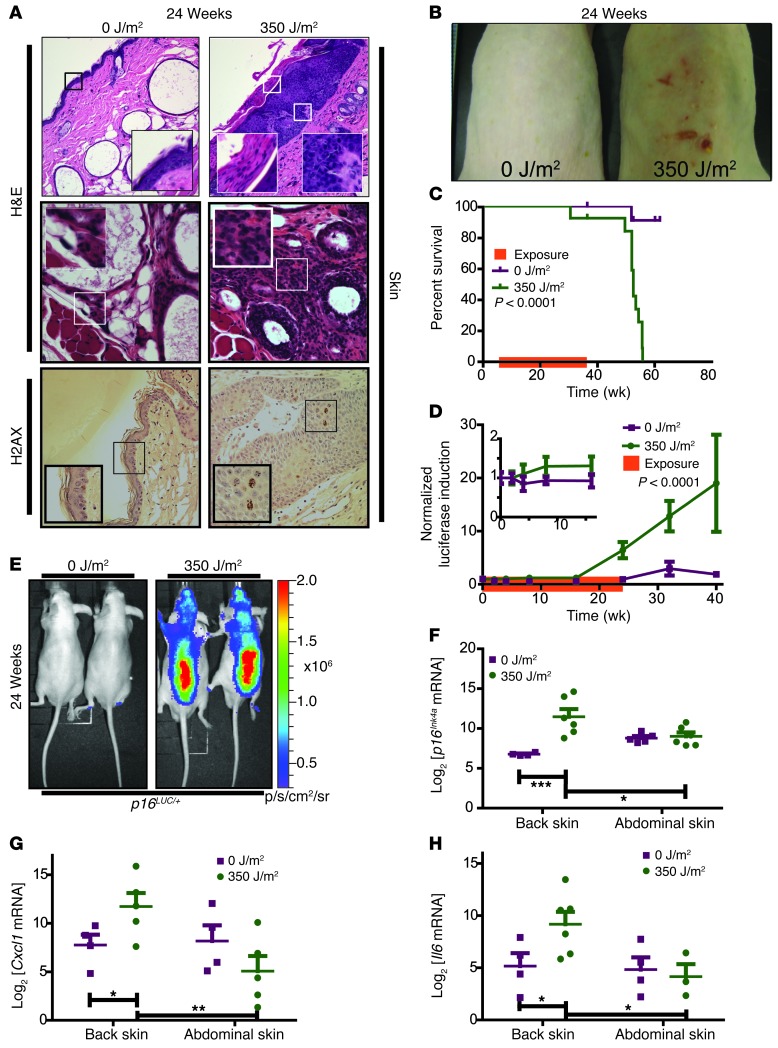
Chronic UVB exposure causes photoaging and skin cancer (33, 34). To directly assess whether chronic UVB induces senescence in vivo, the dorsal surface of p16LUC mice was exposed to 350 J/m2 UVB light 3 times a week for 6 months starting at 10–12 weeks of age. Littermate control mice were contemporaneously aged in the absence of UVB. After 6 months of exposure, mice were observed without further UVB exposure until 1 year of age. After 6 months of exposure, UV-exposed mice showed hyperkeratosis and metaplasia and inflammatory infiltration of the dermis and hypodermis compared with the nonexposed cohort (Figure 3A). The DNA damage marker phospho-H2AX was substantially increased in the epithelium of UV-exposed mice (Figure 3A and Supplemental Figure 4A), suggesting a persistent DNA damage response. Additionally, UV-exposed mice developed skin neoplasms within 5 months of exposure, which accelerated mortality (Figure 3, B and C). Luciferase expression was higher in p16LUC animals treated with UV than in unexposed controls prior to development of visible neoplasms, and TBLI was almost 8 times greater in UV-treated animals after 32 weeks of exposure (Figure 3D). In accord with the TBLI results (Figure 3E), p16INK4a mRNA levels from abdominal and dorsal skin from both cohorts confirmed the site-specific induction of p16INK4a expression (Figure 3F). Additionally, the UV-exposed dorsal skin exhibited higher expression than unexposed abdominal skin from the same mouse (Figure 3F).
Figure 3. UVB increases p16INK4a expression. p16LUC mice were exposed to 0 J/m2 (n = 13) or 350 J/m2 (n = 11) UVB light.
(A) Representative histopathological images of skin stained with H&E or phospho-H2AX. Original magnification, ×20 (top), ×40 (middle and bottom), ×200 (top, insets), ×120 (middle and bottom, insets). (B) Representative dorsal images. (C) Kaplan-Meier survival curves. Exposure duration is shown by the orange bar on the x axis. P value was determined by Gehan-Breslow-Wilcoxon test. (D) Luciferase intensity, normalized to mean levels of the entire cohort at day 0 and graphed as a function of time. Exposure duration is shown by the orange bar on the x axis. Inset highlights initial 20 weeks of exposure. Error bars denote SEM. P value was determined by linear regression analysis. (E) Representative TBLI of experiments in D. (F–H) Quantitative real-time PCR of p16INK4a (F), Cxcl1 (G), and Il6 (H) expression in back and abdominal skin at sacrifice. Error bars denote SEM. *P < 0.05, **P < 0.01, ***P < 0.005, Student’s t test.
To strengthen the association of p16INK4a induction with senescence, additional markers were analyzed in UV-exposed and unexposed skin, including mRNA of the senescence-associated (SA) cytokines Cxcl1 and Il6 and SA-β-galactosidase (SA-β-gal) staining. Cxcl1 and Il6 expression increased in UV-exposed versus nonexposed mice. Additionally, increased expression only occurred in skin directly exposed to UV light (back) and not in unexposed skin (abdomen) from the same animals (Figure 3, G and H). We observed a similar pattern of SA-β-gal staining (Supplemental Figure 4B). These data suggest that chronic DNA damage from UV light accelerates senescent cell accumulation.
Here, we demonstrated that p16LUC mice can detect the senescence-promoting effects of environmental exposures. Direct DNA-damaging agents (CS and UV light) appeared potently gerontogenic, and arsenic exposure produced a lesser, but significant, effect. We also identified anatomic gerontogenic effects of these agents, including a stronger effect of passively inhaled CS on nasal epithelia than lung parenchyma (Figure 2C) and effects of UVB on exposed rather than unexposed skin (Figure 3, E and F).
This system has some important limitations. First, although luciferase imaging is considerably more sensitive than optical methods, the p16LUC allele may be less useful for detecting senescence in deeper organs or in rare cellular subtypes. Therefore, while we observed no effect of HFD on TBLI, it is possible that HFD activates senescence in important cellular compartments (e.g., pancreatic β cells, ref. 35; and vascular cells, refs. 21, 22), but not to a level detectable using this system. Similarly, while we showed that gerontogen-induced p16INK4a expression occurred spatially near senescent cells, we have not explicitly demonstrated that p16INK4a-expressing cells are in all cases senescent. Finally, the p16LUC allele cannot be used to identify p16INK4a-independent gerontogens or aging processes.
In summary, we demonstrated using modest-sized cohorts of serially analyzed animals that the p16LUC allele reported in vivo activation of senescence markers and uncovered gerontogenic effects of environmental toxicants. This resource can supplement other rodent platforms used for toxicological assessments; specifically, it represents an in vivo system with which to assess a compound’s age-promoting activity. We believe this resource will provide an important tool for understanding the relationship between environmental exposures and molecular aging.
Methods
Further information can be found in Supplemental Methods.
Hairless SKH1-E p16LUC/+ mice were used for all experiments (18). Genotyping was performed as previously described (18). For histologic analysis, tissues were fixed with 10% formalin overnight, then transferred to 70% ethanol for paraffin blocking and staining. Quantitative TaqMan RT-PCR strategies for detection of p16INK4a were performed as previously described (7, 13).
Statistics.
Statistical significance was determined using 2-tailed Student’s t test or linear regression analysis for all comparisons, except survival analysis (Gehan-Breslow-Wilcoxon test). A P value less than 0.05 was considered statistically significant.
Study approval.
Experiments were performed under protocols approved by the University of North Carolina IACUC.
Supplementary Material
Acknowledgments
We thank H. Yuan, A. Deal, and S. Gaddameedhi for advice. This work relied on the UNC BRIC Small Animal Imaging Facility, Biostatistics Core, Microscopy Services Laboratory, Histopathology Core, Mouse Phase I Unit, and Animal Studies Facility. This work was supported by the NIA (AG024379 and AG036817), the NIEHS (T32 ES07126), and an HHMI training grant (Med into Grad Initiative).
Footnotes
Conflict of interest: Norman E. Sharpless has received financial support from MolecularMD and has an equity stake in G1 Therapeutics.
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Citation for this article: J Clin Invest. 2014;124(1):169–173. doi:10.1172/JCI70960.
References
- 1.Long GG, Morton D, Peters T, Short B, Skydsgaard M. Alternative mouse models for carcinogenicity assessment: industry use and issues with pathology interpretation. Toxicol Pathol. 2010;38(1):43–50. doi: 10.1177/0192623309354107. [DOI] [PubMed] [Google Scholar]
- 2.Goldsworthy TL, et al. Transgenic animals in toxicology. Fundam Appl Toxicol. 1994;22(1):8–19. doi: 10.1006/faat.1994.1002. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson-Kram D, Sistare FD, Jacobs AC. Use of transgenic mice in carcinogenicity hazard assessment. Toxicol Pathol. 2004;32(suppl 1):49–52. doi: 10.1080/01926230490424761. [DOI] [PubMed] [Google Scholar]
- 4.Martin GM. Interactions of aging and environmental agents: the gerontological perspective. Prog Clin Biol Res. 1987;228:25–80. [PubMed] [Google Scholar]
- 5.Nielsen GP, Stemmer-Rachamimov AO, Shaw J, Roy JE, Koh J, Louis DN. Immunohistochemical survey of p16INK4A expression in normal human adult and infant tissues. Lab Invest. 1999;79(9):1137–1143. [PubMed] [Google Scholar]
- 6.Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15(2):203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 7.Krishnamurthy J, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114(9):1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, et al. Expression of p16(INK4a) in peripheral blood T-cells is a biomarker of human aging. Aging Cell. 2009;8(4):439–448. doi: 10.1111/j.1474-9726.2009.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molofsky AV, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443(7110):448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janzen V, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443(7110):421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 11.Signer RA, Montecino-Rodriguez E, Witte ON, Dorshkind K. Aging and cancer resistance in lymphoid progenitors are linked processes conferred by p16Ink4a and Arf. Genes Dev. 2008;22(22):3115–3120. doi: 10.1101/gad.1715808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, et al. Expression of p16(INK4a) prevents cancer and promotes aging in lymphocytes. Blood. 2011;117(12):3257–3267. doi: 10.1182/blood-2010-09-304402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnamurthy J, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443(7110):453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 14.Jeck WR, Siebold AP, Sharpless NE. Review: a meta-analysis of GWAS and age-associated diseases. Aging Cell. 2012;11(5):727–731. doi: 10.1111/j.1474-9726.2012.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berent-Maoz B, Montecino-Rodriguez E, Signer RA, Dorshkind K. Fibroblast growth factor-7 partially reverses murine thymocyte progenitor aging by repression of Ink4a. Blood. 2012;119(24):5715–5721. doi: 10.1182/blood-2011-12-400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, et al. PDGF signalling controls age-dependent proliferation in pancreatic beta-cells. Nature. 2011;478(7369):349–355. doi: 10.1038/nature10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burd CE, et al. Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model. Cell. 2013;152(1–2):340–351. doi: 10.1016/j.cell.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visel A, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464(7287):409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang CY, et al. Obesity increases vascular senescence and susceptibility to ischemic injury through chronic activation of Akt and mTOR. Sci Signal. 2009;2(62):ra11. doi: 10.1126/scisignal.2000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Q, et al. Endothelial senescence after high-cholesterol, high-fat diet challenge in baboons. Am J Physiol Heart Circ Physiol. 2007;292(6):H2913–H2920. doi: 10.1152/ajpheart.01405.2006. [DOI] [PubMed] [Google Scholar]
- 23.Kapaj S, Peterson H, Liber K, Bhattacharya P. Human health effects from chronic arsenic poisoning--a review. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2006;41(10):2399–2428. doi: 10.1080/10934520600873571. [DOI] [PubMed] [Google Scholar]
- 24.Yamauchi H, Aminaka Y, Yoshida K, Sun G, Pi J, Waalkes MP. Evaluation of DNA damage in patients with arsenic poisoning: urinary 8-hydroxydeoxyguanine. Toxicol Appl Pharmacol. 2004;198(3):291–296. doi: 10.1016/j.taap.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 25.Hartwig A, Groblinghoff UD, Beyersmann D, Natarajan AT, Filon R, Mullenders LH. Interaction of arsenic(III) with nucleotide excision repair in UV-irradiated human fibroblasts. Carcinogenesis. 1997;18(2):399–405. doi: 10.1093/carcin/18.2.399. [DOI] [PubMed] [Google Scholar]
- 26.Andrew AS, Karagas MR, Hamilton JW. Decreased DNA repair gene expression among individuals exposed to arsenic in United States drinking water. Int J Cancer. 2003;104(3):263–268. doi: 10.1002/ijc.10968. [DOI] [PubMed] [Google Scholar]
- 27.Paul DS, Harmon AW, Devesa V, Thomas DJ, Styblo M. Molecular mechanisms of the diabetogenic effects of arsenic: inhibition of insulin signaling by arsenite and methylarsonous acid. Environ Health Perspect. 2007;115(5):734–742. doi: 10.1289/ehp.9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeMarini DM. Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat Res. 2004;567(2–3):447–474. doi: 10.1016/j.mrrev.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21(48):7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 30.Bernhard D, Moser C, Backovic A, Wick G. Cigarette smoke — an aging accelerator? Exp Gerontol. 2007;42(3):160–165. doi: 10.1016/j.exger.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 31.Ito K, Barnes PJ. COPD as a disease of accelerated lung aging. Chest. 2009;135(1):173–180. doi: 10.1378/chest.08-1419. [DOI] [PubMed] [Google Scholar]
- 32.Nelson JA, et al. Expression of p16(INK4a) as a biomarker of T-cell aging in HIV-infected patients prior to and during antiretroviral therapy. Aging Cell. 2012;11(5):916–918. doi: 10.1111/j.1474-9726.2012.00856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kligman LH. Photoaging. Manifestations, prevention, and treatment. Clin Geriatr Med. 1989;5(1):235–251. [PubMed] [Google Scholar]
- 34.Scharffetter-Kochanek K, et al. Photoaging of the skin from phenotype to mechanisms. Exp Gerontol. 2000;35(3):307–316. doi: 10.1016/s0531-5565(00)00098-x. [DOI] [PubMed] [Google Scholar]
- 35.Sone H, Kagawa Y. Pancreatic β cell senescence contributes to the pathogenesis of type 2 diabetes in high-fat diet-induced diabetic mice. Diabetologia. 2005;48(1):58–67. doi: 10.1007/s00125-004-1605-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.