Abstract
Centrins are members of the EF-hand family of calcium-binding proteins, which are highly conserved among eukaryotes. Centrins bind to several cellular targets, through a hydrophobic triad. However, the W1xxL4xxxL8 triad in XPC (Xeroderma Pigmentosum Group C protein) is found in the reverse orientation, as in the L8xxxL4xxW1 triad in Sfi1 (Suppressor of Fermentation-Induced loss of stress resistance protein 1). As shown by previous NMR studies of human centrin 2 in complex with XPC or Sfi1, the E148 residue of human centrin 2 is in contact with XPC but is pushed away from the triad of Sfi1. We corroborated these findings using site-directed mutagenesis to generate mutations in Scherffelia dubia centrin (SdCen) and by using isothermal titration calorimetry to analyze the binding affinity of these mutants to XPC and Sfi1. We mutated the F109 residue, which is the main residue involved in target binding regardless of triad orientation, and the E144 residue, which was thought to be involved only in XPC binding. The F109L mutation reduced the binding of SdCen to XPC and Sfi1 and the negative effect was greater upon temperature increase. By contrast, the E144A mutation reduced the binding to XPC but had no effect on Sfi1 binding. The F109L-E144A mutation enhanced the negative effect of the two single mutations on XPC binding. Sfi1 proteins from Ostreococcus lucimarinus and Ostreococcus tauri, which belong to the same clade as S. dubia, were also investigated. A comparative analysis shows that the triad residues are more conserved than those in human Sfi1.
Keywords: Centrins, Scherffelia dubia, XPC, Sfi1, Binding, ITC
Abbreviations: SdCen, Scherffelia dubia centrin; C-SdCen, C-terminal domain of Scherffelia dubia centrin; XPC, Xeroderma Pigmentosum group C protein; Sfi1, Suppressor of Fermentation-Induced loss of stress resistance protein 1; ITC, isothermal titration calorimetry; OstLu, Ostreococcus lucimarinus; OstTa, Ostreococcus tauri; CID, proximal CTD-Interacting Domain; HCA, hydrophobic amino acids clusters analysis
Highlights
-
•
We used isothermal titration calorimetry to study how algal centrin binds to its targets.
-
•
Centrin with the mutation F109L reduced binding to both XPC and Sfi1.
-
•
Centrin with the mutation E114A reduced binding to XPC but not to Sfi1.
-
•
The double mutation F109L–E144A further reduced XPC binding compared to the single mutations.
1. Introduction
Centrins are small (∼20 kDa), acidic, Ca2+-binding proteins that belong to the EF-hand superfamily. Centrins were first identified in the unicellular green algae Tetraselmis striata [1] and Chlamydomonas reinhardtii as components of the contracting fibers associated with the basal bodies [2,3]. Centrins have also been found in many eukaryotes, including higher plants [4], yeast [5], invertebrates [6] and humans [7,8]. Only one isoform has been found in lower eukaryotes such as unicellular algae and yeast, whereas four isoforms have been found in higher eukaryotes. Even more isoforms have been identified in the unicellular ciliated protozoan Paramecium tetraurelia [9]. Purified centrins have been characterized, and algal centrin exhibits calcium-dependent filaments formation [10].
Centrins have been reported to interact with several cellular targets, such as centrosomal proteins, XPC, Sfi1, Sac3 and transducin. Involvement of centrin in cell division has been demonstrated in yeast. Three temperature-sensitive alleles of yeast Cdc31 that contain single point mutations (i.e., E97G, A101T and F105L) induce arrest in the G2/M transition of the cell cycle [11]. Centrins are concentrated in the microtubule organizer centers and are required for the cell cycle. When centrin is absent or non-functional, microtubule organizer center duplication is blocked, resulting in perturbed mitosis [12,13].
Centrins are also involved in several other cellular processes, for example, modulation of homologous recombination [14]. Centrin participates in the first step of nucleotide excision repair, in particular the global genome repair pathway, which involves centrin binding to XPC in concert with Rad23B. This heterodimer plays a key role in the initial phase of DNA-damage recognition by recruiting other factors, including TFIIH [15]. Because of the coupling of transcription, pre-mRNA processing, and the transport of mature RNA from the nucleus to the cytoplasm, interactions between the transcription machinery and the nuclear pore complex are necessary. Thus, proteins involved in this interaction play an important role in the process. In yeast, Sac3 acts in concert with Sus1 and Cdc31 to mediate interactions between the whole transcription machinery and the nuclear pore complex [16]. Experiments using immunoelectron microscopy have shown that centrin and transducin α co-localize in the connecting cilium of mouse rod photocells. In addition, co-immunoprecipitation, centrifugation, and size-exclusion chromatography experiments have demonstrated that transducin β as well as the heterotrimer transducin αβγ binds centrin 1 [17,18]. Sfi1, (suppressor of fermentation-induced loss of stress resistance 1), has been identified as an interacting partner for Cdc31 [19]. Furthermore, immunoelectron microscopy experiments revealed that Cdc31 and Sfi1 are localized to the spindle pole body. Deletion of Sfi1 causes G2 arrest in Saccharomyces cerevisiae [20]. In all these processes, centrin binds to a hydrophobic triad W1xxL4xxxL8 found in its cellular targets. Although this triad corresponds to the centrin-binding motif found in XPC, the centrin-binding motif of other targets (Sfi1, Sac3, Transducin β) is in the opposite orientation L8xxxL4xxW1.
Several structures of centrin alone or in complex with cellular targets have been solved. Centrins are composed of two relatively independent domains, each one possessing two Ca2+ binding sites. Experiments using microcalorimetry have demonstrated that the C-terminal domain of centrin is sufficient for target binding. However, for algal centrins, the N-terminal domain also binds targets, but only in the presence of calcium [21,22]. The structures of the N-terminal domain of C. reinhardtii [21] and human centrins have been solved by NMR [23].
Two structures of full-length and truncated HsCen2 in complex with an XPC peptide have been solved by X-ray crystallography [24,25], and one structure of the C-terminal domain of HsCen2 in complex with XPC peptide in solution has been solved by NMR [26]. The structure from [24] shows that only residues in the C-terminal domain of HsCen2 are involved in the interaction with the XPC peptide. Nine non-polar residues of HsCen2 (L133, L112, M145, F113, M166, L126, A109, E105 and V129) form contacts with the residues of the XPC peptide. The F113 and M145 residues form the pocket where the W residue of XPC binds. The Y172 residue of HsCen2, which interacts with L9 of the XPC peptide, is located near the S170, which can be phosphorylated and is reported to have a functional role [27].
The structure of CID region (proximal CTD-Interacting Domain) of Sac3 in complex with Sus1 and Cdc31 has been solved by X-ray crystallography [28]. In the complex, the W802 residue of Sac3 appears to play a central role in the interaction with the hydrophobic cavity of the centrin Cdc31. The F105 residue of Cdc31 in turn plays an important role in the association with Sac3.
A crystal structure of centrin Cdc31 in complex with truncated Sfi1 has been solved by X-ray crystallography [29]. Both the C-terminal and the N-terminal domains of Cdc31 make contact with Sfi1. The structure also reveals several centrin–centrin interactions. These interactions suggest that a filament of several centrin molecules is formed by centrin–centrin interactions and that this filament is stabilized by Sfi1 through Sfi1 repeat–centrin interactions.
An NMR structure of human centrin 2 in complex with the R17-Sfi1 peptide has been solved [30]. Comparison of this structure with superimposed with the NMR structure of the C-terminal domain of human centrin 2 in complex with either peptide-P17-XPC peptide [26] led to the proposal that the centrin 2 E148 residue discriminates between XPC and Sfi1 [30]. Although the helix dipoles of the XPC and Sfi1 peptides are reversed, their W residues are embedded in the hydrophobic cavity containing the centrin F113 residue. For Sfi1, this requires a rotation of the W residue and a slight translation of the peptide within the cavity. The carboxyl group of E148, which faces the positive pole of the XPC helix, establishes a hydrogen bond with W residue of XPC. By contrast, if E148 faces the negative pole of the Sfi1 helix, it is pushed away from the tryptophan leading to the disruption of hydrogen bonds.
We used site-directed mutagenesis to mutate the phenylalanine residue that is important for target binding as well as the glutamate residue that discriminates between XPC and Sfi1 targets. We also used isothermal titration calorimetry (ITC) to assess the thermodynamic parameters of the binding reaction. The role of the phenylalanine residue in binding was first established using temperature-sensitive mutants in S. cerevisiae, thus we examined the effects of temperature on the association between centrin and two targets, XPC and Sfi1. The E148 residue is conserved among centrins. Because there is only one centrin in unicellular green algae, the same centrin isoform interacts with all cellular targets regardless of the centrin-binding motif orientation. We used the Scherffelia dubia centrin to investigate the role of the E144 residue (which corresponds to E148 residue in human centrin) in the interaction with XPC and Sfi1. We also retrieved sequences for Sfi1 and XPC homologues protein from databases and analyzed their centrin-binding motifs.
2. Results
2.1. The centrin targets Sfi1 and XPC are found in green algae
A single centrin is found in S. dubia that must interact with all cellular targets expressed in this unicellular green alga. Sequences of centrins have been found in several other green algae genomes using human centrins as query sequence. Several of the green algae genomes that were investigated contain only one centrin gene (data not shown), and these algal centrins are conserved (Fig. S1). The unique centrin sequences from S. dubia (SdCen) and Ostreococcus lucimarinus (OlCen) were aligned with each other and with human centrins using the ClustalW program (Fig. 1). The C-terminal domain of SdCen (M93-F168) has 100% identity with OlCen. Residues involved in the interactions with XPC, Sfi1, and Sac3 (E105, A109, and F113 in human centrin) are conserved among the centrins from green algae as is the E148 residue specific for XPC binding (Fig. S1).
Fig. 1.

Sequence alignment of centrins. (A) Scherffelia dubia with Ostreococcus lucimarinus (full-length); (B) C-terminal domain of S. dubia with human centrin1; and (C) C-terminal domain of S. dubia with human centrin 2. Sequences were aligned using ClustalW ((*) identity, (:) strongly similar, (.) weakly similar). Amino acids are colored based on their implication in binding to centrin targets. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
S. dubia is a unicellular green alga of the order Chlorodendrales that belongs to the clade of Prasinophyceae. Unfortunately, the genome of S. dubia has not been sequenced yet. However, the genomes of two species of Mamiellales, O. lucimarinus (OstLu) and Ostreococcus tauri (OstTa), which belong to the same clade as S. dubia, have been sequenced and analyzed.
A BLAST search using Sfi1 from both human and S. cerevisiae as query sequences identified Sfi1 homologues in the genomes of two other green algae, namely O. lucimarinus (Sfi1 containing 1315 residues) and O. tauri (Sfi1 containing 1370 residues). We identified centrin-binding motifs using a sequence alignment of human and algae Sfi1 (Table 1) as well as the hydrophobic cluster analysis program HCA. Fig. S2 shows that the hydrophobic clusters identified by HCA are composed of the same residues identified by the sequence alignment (Table 1). Similar to human and yeast Sfi1, Sfi1 homologues from green algae are long molecules composed of 28 and 22 centrin-binding motifs for OstLu and OstTa, respectively. Using the protein secondary prediction programs SOPMA and GOR4, we found that Sfi1 proteins from both green algae are composed of 70 to 90–93% of α-helices similar to Sfi1 found in other organisms. We compared the hydrophobic triad L8xxxL4xxW1 that forms the centrin-binding motif of Sfi1 proteins from OstLu, OstTa, and human (Table 1). We found that the W residue in position 1 is absolutely conserved except for an L residue found in one repeat in human Sfi1. By contrast, several other residues replace W in S. cerevisiae Sfi1 (data not shown). In algal Sfi1, an F residue is absolutely conserved in position 4 with the exception for a Val residue in one repeat of OstTa Sfi1; however, in other organisms, L, W or F residues are found in position 4. Similar to Sfi1 homologues from other organisms, an L residue is mainly found at position 8, with a basic residue at position 7. Thus, the hydrophobic triad found predominantly in algal Sfi1 is L8xxxF4xxW1.
Table 1.
Sfi1 and Rad4 centrin-binding motifs.
 |
Using a BLAST search with human XPC sequence as the query sequence, we retrieved homologous sequences for both green algae, namely a Rad4 protein containing 637 residues in OstLu and a Rad4 protein containing 1122 residues in OstTa. The centrin-binding motifs are located at the C-terminus of the Rad4 protein as in other organisms. They are identical in both green algae with an I residue found at position 8 in place of an L residue (W1RL4LLGAI8) and a basic residue near the W residue. Thus, the motif in Rad4 has a strong similarity to human XPC (Table 1).
2.2. C-SdCen has more affinity for P17-XPC than for R18-Sfi1
Because the S. dubia genome has not been sequenced yet, we did not have access to Sfi1 and XPC sequences. We used an R18-Sfi1 and a P17-XPC peptide derived from human targets (see Material and Methods) that have been used with human centrin 2 and SdCen in previous work [26,30] (Fig. 2). Algal Sfi1 and XPC (Rad4) share a conserved hydrophobic triad with their human homologues. The ITC experiments reported here were performed with the C-terminal domain (M93-F168) of SdCen (C-SdCen) (Tables 2 and 3).
Fig. 2.
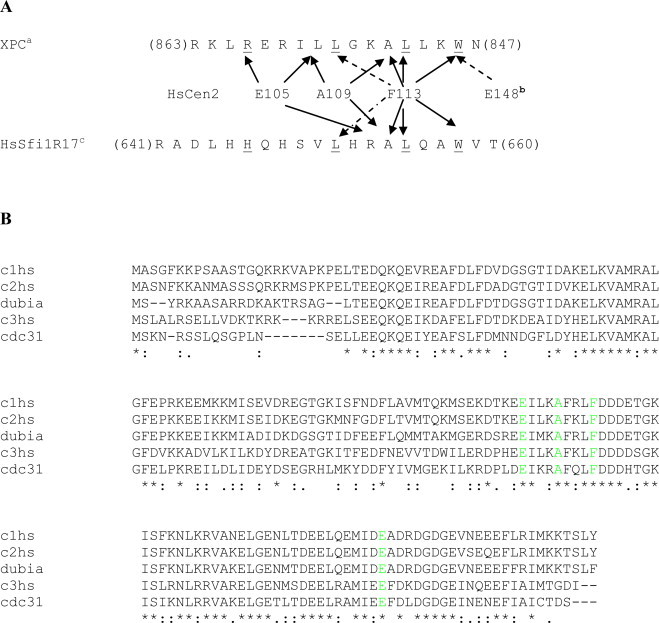
The centrin residues involved in target binding. (A) Graph showing the main human centrin 2 residues interacting with targets XPC and Sfi1. The F113 residue is central to the interaction and makes contacts with the three conserved residues (underlined) that form the hydrophobic triad of the centrin-targets. (B) Sequence alignment of centrins. The three human centrins (HsCen1, HsCen2, HsCen3) are aligned with yeast centrin (Cdc31) and centrin from Scherffelia dubia (SdCen). Amino acids are colored based on their implication in binding to centrin targets. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Thermodynamic parameters of wild-type and mutant SdCen variants binding to XPC and Sfi1.
| Protein C-SdCen | Ligand | Ca2+ | N | Ka (error) (106 M−1) | ΔH (error) (kcal/mol) | TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|---|---|
| WT | P17-XPC 30 °C | + | 0.95 | 59.4(9.10) | −23.4(0.16) | −12.63 | −10.77 |
| 30 °C | − | 1.02 | 30.2 (1.2) | −33.8 (0.07) | −23.39 | −10.41 | |
| 37 °C | + | 1.02 | 73.2(7.0) | −23.36(0.08) | −12.18 | −11.18 | |
| 37 °C | − | 1.04 | 10.6 (0.4) | −40.6 (0.12) | −29.94 | −10.66 | |
| F109L | P17-XPC 30 °C | + | 1.02 | 67.1(5.0) | −15.63(0.09) | −4.78 | −10.85 |
| 30 °C | − | 1.33 | 2.02 (0.07) | −20.6 (0.09) | −11.88 | −8.72 | |
| 37 °C | + | 1.23 | 37.8(5.02) | −16.97(0.10) | −6.20 | −10.77 | |
| 37 °C | − | 1.05 | 0.62 (0.01) | −28.96 (0.2) | −20.27 | −8.63 | |
| E144A | P17-XPC 30 °C | + | 1.02 | 95.6(15.6) | −20.44(0.12) | −9.36 | −11.08 |
| 30 °C | − | 1.15 | 10.1 (0.5) | −29.5 (0.13) | −19.78 | −9.72 | |
| 37 °C | + | 1.01 | 107(11.5) | −25.17(0.62) | −13.76 | −11.41 | |
| 37 °C | − | 1.03 | 2.49 (0.08) | −38.4 (0.17) | −28.66 | −9.74 | |
| F109L-E144A | P17-XPC 30 °C | + | 1.07 | 32.7(2.65) | −14.11(0.06) | −3.69 | −10.42 |
| 30 °C | − | 1.0 | 0.43 (0.01) | −22.2 (0.2) | −14.39 | −7.81 | |
| 37 °C | + | 1.01 | 29(1.92) | −17.19(0.06) | −6.60 | −10.59 | |
| 37 °C | − | 1.06 | 0.24 (1.24) | −21.2 (0.5) | −13.21 | −7.99 | |
| WT | R18-Sfi1 30 °C | − | 1.12 | 4.5 (0.1) | −31.5 (0.08) | −22.24 | −9.26 |
| 37 °C | − | 1.15 | 1.23 (0.02) | −37.09 (0.1) | −27.78 | −9.31 | |
| F109L | R18-Sfi1 30 °C | − | 1.17 | 0.77 (0.01) | −23.2 (0.1) | −14.96 | −8.24 |
| 37 °C | − | 1.05 | 0.23 (0.01) | −30.4 (1.0) | −22.3 | −8.10 | |
| E144A | R18-Sfi1 30 °C | − | 1.19 | 4.64 (0.08) | −30.3 (0.06) | −21.06 | −9.25 |
| 37 °C | − | 1.12 | 1.38 (0.04) | −36.2 (0.17) | −26.88 | −9.34 | |
| F109L-E144A | R18-Sfi1 30 °C | − | 1.17 | 0.66 (0.02) | −24.4 (0.16) | −16.33 | −8.07 |
| 37 °C | − | 1.04 | 0.29 (0.01) | −26.4 (0.3) | −18.24 | −8.19 |
The ITC experiments were conducted at 30 °C or 37 °C in buffer (MOPS 50 mM, NaCl 100 mM, EDTA 2 mM or Ca2+ 2 mM pH 7.5). N was the stoichiometry of binding. NB means no binding. The standard deviations of the fitted parameters are given in parenthesis.
Table 3.
Thermodynamic parameters of wild-type and mutant SdCen variants binding to XPC and Sfi1.
| ProteinC-SdCen | Ligand | Ca2+ | ΔH (error) (kcal/mol) | ΔΔHa (kcal/mol) | ΔG (kcal/mol) | ΔΔGb (kcal/mol) |
|---|---|---|---|---|---|---|
| WT | P17-XPC 30 °C | – | −33.8 (0.07) | – | −10.41 | – |
| 37 °C | – | −40.6 (0.12) | – | −10.66 | – | |
| F109L | P17-XPC 30 °C | – | −20.6 (0.09) | −13.2 | −8.72 | −1.69 |
| 37 °C | – | −23.16 (0.1) | −17.44 | −8.63 | −2.03 | |
| E144A | P17-XPC 30 °C | – | −29.5 (0.13) | −4.3 | −9.72 | −0.69 |
| 37 °C | – | −38.4 (0.17) | −2.2 | −9.74 | −0.67 | |
| F109L–E144A | P17-XPC 30 °C | – | −22.2 (0.2) | −11.6 | −7.81 | −2.6 |
| 37 °C | – | −21.2 (0.5) | −19.4 | −7.99 | −2.67 | |
| WT | R18-Sfi1 30 °C | – | −31.5 (0.08) | – | −9.26 | – |
| 37 °C | – | −37.09 (0.1) | – | −9.31 | – | |
| F109L | R18-Sfi1 30 °C | – | −23.2 (0.1) | −8.3 | −8.24 | −1.02 |
| 37 °C | – | −30.4 (1.0) | −6.69 | −8.1 | −1.21 | |
| E144A | R18-Sfi1 30 °C | – | −30.31 (0.06) | −1.19 | −9.25 | −0.01 |
| 37 °C | – | −36.22 (0.17) | −0.87 | −9.34 | +0.02 | |
| F109L–E144A | R18-Sfi1 30 °C | – | −24.4 (0.16) | −7.1 | −8.07 | −1.19 |
| 37 °C | – | −26.43 (0.3) | −10.66 | −8.19 | −1.12 |
The ITC experiments were conducted at 30 °C or 37 °C in buffer (MOPS 50 mM, NaCl 100 mM, EDTA 2 mM pH 7.5). N was the stoichiometry of binding. NB means no binding. The standard deviations of the fitted parameters are given in parenthesis.
The difference in binding enthalpy between the interaction of SdCen wt and P17-XPC and the interaction of SdCen mutants and P17-XPC, as well as for the SdC-R18-Sfi1 complexes.
The difference in free energy between the interaction of SdCen wt and P17-XPC and the interaction of SdCen mutants and P17-XPC, as well as for the SdC-R18-Sfi1 complexes.
C-SdCen binds P17-XPC and R18-Sfi1 in the absence of calcium; however, the presence of calcium enhanced the binding to both targets (Table 2). Because the effect of mutations in C-SdCen on target binding was more pronounced in the absence of calcium, the figures reported here show only the binding experiments performed in the absence of calcium (i.e., in the presence of EDTA).
The affinity of C-SdCen for P17-XPC and R18-Sfi1 was measured by ITC in the presence of EDTA at two temperatures, 30 °C and 37 °C. For both targets, the change in enthalpy (ΔH) was negative indicating that the binding reaction was exothermic. The stoichiometry of binding was one, meaning that the C-terminal domain of SdCen had only one site for P17-XPC as well as for R18-Sfi1 (Figs. 3 and 4). The two targets also exhibited unique behaviors. C-SdCen exhibited a higher affinity for P17-XPC than for R18-Sfi1 by a factor of 7 at 30 °C and by a factor of 10 at 37 °C (Table 2). However, the change in enthalpy (ΔH) differed only by 3 kcal/mol, and the change in entropy (TΔS) differed by 1 kcal/mol. Thus, for both targets the interaction is enthalpically driven.
Fig. 3.
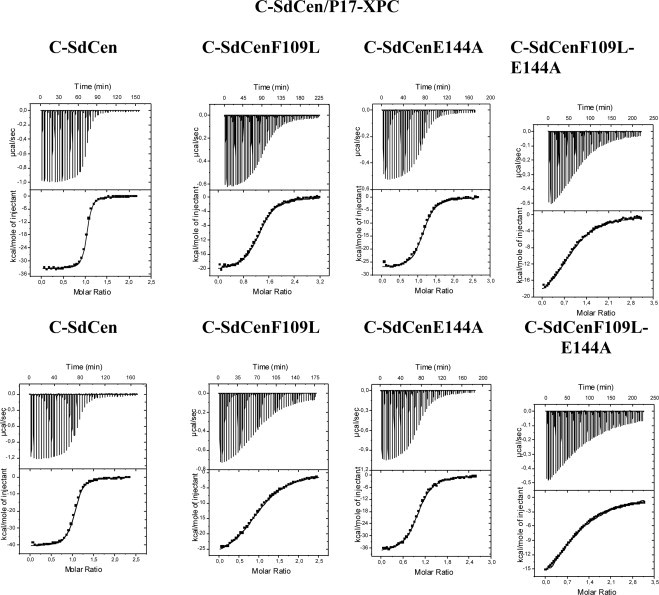
Temperature effect of wild-type C-SdCen and mutant C-SdCen variants binding to P17-XPC in the absence of calcium as measured by ITC. The ITC measurements show the C-SdCen variants binding to XPC at 30 °C (upper graph) and at 37 °C (lower graph). For each graph, the upper panels show the raw data, and the lower panels show the binding isotherms fit to a one-site model. Peptide solutions (100 μM) were injected into the protein (10 μM) equilibrated in buffer (50 mM MOPS, 100 mM NaCl, 2 mM EDTA, pH 7.5) at 30 °C or 37 °C in the calorimeter cell.
Fig. 4.
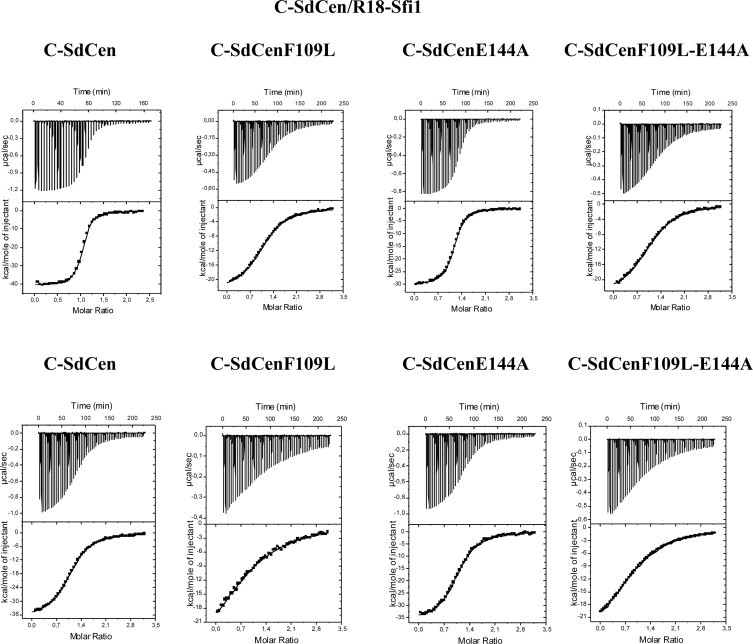
Temperature effect of wild-type and mutant C-SdCen variants binding to R18-Sfi1 in the absence of calcium as measured by ITC. The ITC measurements show the C-SdCen variants binding to XPC at 30 °C (upper graph) and at 37 °C (lower graph). For each graph, the upper panels show the raw data, and the lower panels show the binding isotherms fitted to a one-site model. Peptide solutions (100 μM) were injected into the protein (10 μM) equilibrated in buffer (50 mM MOPS, 100 mM NaCl, 2 mM EDTA, pH 7.5) at 30 °C or 37 °C in the calorimeter cell.
The effect of temperature increase on the binding of C-SdCen to P17-XPC and R18-Sfi1 was measured by ITC in the absence of calcium (with EDTA) at two temperatures, 30 °C and 37 °C. Increasing the temperature from 30 °C to 37 °C decreased the binding of C-SdCen to P17-XPC by a factor of 3 and decreased the binding to R18-Sfi1 by a factor of 4 (Figs. 3 and 4). The change in enthalpy (ΔH) decreased by a factor of 1.2, becoming more negative for both targets and indicating that the binding reaction was more exothermic (Table 2). The change in entropy (TΔS) also decreased with the temperature by a factor of 1.3, becoming more negative. However, increasing temperature did not change the binding reaction mode, which is still enthalpically driven.
2.3. Mutating phenylalanine 109 to leucine decreases the C-SdCen affinity for P17-XPC as well as for R18-Sfi1
The F109 residue of SdCen corresponds to the F113 residue of human centrin 2, which has been shown to interact with several residues of XPC as well as Sfi1, specifically the W1, L4 and L8 residues of the conserved hydrophobic triad. In the absence of calcium, the binding reaction of the C-SdCenF109L variant to P17-XPC as well as to R18-Sfi1 was exothermic (ΔH negative) with a binding stoichiometry of one (Figs. 3 and 4). Mutating F109 to leucine decreased the binding of C-SdCen to P17-XPC as well as to R18-Sfi1 but to different extents. The binding constant for the SdCenF109L-P17-XPC interaction was reduced by a factor of 15 at 30 °C, and the same ratio was observed at 37 °C (Table 2). However, the binding constant for the SdCenF109L-R18-Sfi1 interaction was reduced by a factor of 6 at 30 °C, and the same ratio was observed at 37 °C. The decrease in binding was accompanied by an increase in change in enthalpy (ΔH) for P17-XPC by a factor of 1.7 at 30 °C and by a factor of 1.4 at 37 °C. Similarly, the change in enthalpy for R18-Sfi1 increased by a factor of 1.4 at 30 °C and 37 °C. Thus, mutating F109 to leucine resulted in a change in enthalpy (ΔH) that was less negative than that observed for the wild-type C-SdCen for both targets. Furthermore, the binding reaction of C-SdCenF109L to both targets was still enthalpically driven. The increase in the change in enthalpy (ΔΔH) was greater (10 kcal/mol) than the corresponding increase in binding free energy (ΔΔG) (less than 2 kcal/mol) (Table 3). Consequently there is a compensating gain in entropy of binding (10 kcal/mol) (Table 2).
2.4. Mutating glutamate 144 to alanine decreases the affinity of C-SdCen for P17-XPC but has no effect on R18-Sfi1 binding
The E144 residue corresponds to the E148 residue of HsCen2, which has been shown to interact with XPC, in particular with the W1 residue of the triad (Fig. 1). In the absence of calcium the binding of the C-SdCenE144A variant to P17-XPC was exothermic (ΔH negative) with a binding stoichiometry of one (Fig. 4). Mutating E144 to alanine decreased the affinity of C-SdCen for XPC by a factor of 3 at 30 °C and by a factor of 4 at 37 °C. However, the change in enthalpy (ΔH) was the same as for the wild type (ΔH −30 kcal/mol at 30 °C and −38 kcal/mol at 37 °C) but the change in entropy increased (ΔG of −9.72 kcal/mol at 30 °C as well as at 37 °C). The change in free energy (ΔG) for C-SdCenE144A was intermediate between that of wild-type C-SdCen and C-SdCenF109L (Table 2).
In the absence of calcium, the binding reaction of C-SdCenE144A to R18-Sfi1 was exothermic (ΔH negative) and enthalpically driven with a binding stoichiometry of one (Fig. 4). In contrast to what we observed with P17-XPC, the E144A mutation had no effect on the binding of C-SdCen to R18-Sfi1 and the SdCenF109L-E144A variant exhibited the same behavior as wild-type SdCen at both temperatures. The affinity of C-SdCenE144A for R18-Sfi1 was almost identical to the value found for wild-type C-SdCen, as were the changes in enthalpy and entropy values (Table 2).
2.5. Both F109 and E144 act in concert to stabilize the binding of C-SdCen to P17-XPC, but the effect of the double F109L-E144A mutation on R18-Sfi1 binding is similar to that of the single F109L mutation
Because both single mutations had a negative effect on the binding of C-SdCen to P17-XPC, a variant with the F109L-E144A double mutation was constructed and analyzed. In the absence of calcium, the binding reaction of SdCenF109L-E144A to P17-XPC was exothermic (ΔH negative) with a binding stoichiometry of one (Fig. 3). The F109L-E144A double mutation decreased the binding of C-SdCen to P17-XPC more than the single F109L and E144A mutations did. The Ka values were 30 × 106 M−1, 10 × 106 M−1, 2 × 106 M−1, 0.4 × 106 M−1 for wild-type C-SdCen, SdCenE144A, SdCenF109L, and SdCenF109-E144A, respectively, at 30 °C. The Ka values at 37 °C were lower (10 × 106 M−1, 2.5 × 106 M−1, 0.6 × 106 M−1, 0.25 × 106 M−1) (Table 2). The double mutation led to an increase in the change in enthalpy (ΔH) (ΔH of −22.2 kcal/mol at 30 °C) and an increase in the change of free energy (ΔG) (ΔG of −7.81 kcal/mol at 30 °C) with a compensatory effect in the change in entropy (TΔS) (TΔS of −14.4 kcal/mol), which increased twofold compare to wild-type SdCen.
By contrast, the effect of the double F109L-E144A mutation R18-Sfi1 binding was similar to the single F109L mutation. In the absence of calcium, the binding reaction was exothermic (ΔH negative) with a binding stoichiometry of one. The binding reaction of C-SdCenF109L-E144A to R18-Sfi1 was enthalpically driven (Fig. 4). The double mutant C-SdCenF109L-E144A exhibited almost the same thermodynamic values (Ka, ΔH, ΔG, TΔS) as the single mutant C-SdCenF109L (Table 2).
3. Discussion
3.1. In green algae, a single centrin must bind several targets regardless of the orientation of their centrin-binding motif
Although the genome of S. dubia has not been sequenced yet, several other genomes from green algae have been sequenced, including O. lucimarinus and O. tauri. The sequences of the centrin found in these green algae were retrieved from genomes databases using the psiBlast program with the SdCen sequence as the query and aligned with SdCen using the ClustalW program (Fig. 1). Full-length SdCen has 90% identity with OstLu centrin, and the C-terminal domain of SdCen (M93-F168) is identical to the OstLu C-terminal domain (M90-F165). The SdCen sequence was also aligned with the human centrins, HsCen1 and HsCen2. In terms of the C-terminal domain, SdCen has the corresponding K131 residue of HsCen2 but lacks the S158 and N160 residues found in HsCen2. However, the S158 residue is only found in some organisms such as primates, and certain bird and some fish species. This suggests that they appeared through the evolution after the appearance of HsCen1 upon retrotransposition of HsCen2. Thus, sequence comparison suggests that the green algal centrins belong to the HsCen2 family, whereas the yeast centrin Cdc31 belongs to the HsCen3 family. However, in both cases the unique centrin must bind several targets, including XPC, Sac3 and Sfi1, which differ in the orientation of the centrin-binding motif.
We were able to identify Sfi1 and Rad4 homologues in the genomes of the algae O. lucimarinus and O. tauri (Table 1). We further analyzed the evolution of the hydrophobic triad of these targets. The three important residues W1, L4, and L8 that form the triad of found in Sfi1 repeats are more conserved in green algae than in higher eukaryotes. Indeed, in all Sfi1 repeats, there is a W residue in position 1 and an F residue in position 4 of the triad. Such a strict conservation at position 4 is not found in higher organisms although the F residue is common at this position.
The centrin residues involved in target binding have been analyzed using various methods. First, the isolation of yeast temperature sensitive mutants allowed the identification of functionally important residues [11]. Second, the structures of human centrin 2 [24–26] and yeast Cdc31 [29] in complex with targets have shed light on which residues play a role in target binding. For HsCen2, residues E105, A109, F113, and E148 have been identified as target-binding residues. These residues are highly conserved in centrins across species as well as in isoforms from the same organism.
3.2. General properties of C-SdCen binding to the cellular targets XPC and Sfi1 which have opposite centrin-binding motifs
Regardless of the orientation of their centrin-binding motif, the binding of targets to SdCen was exothermic with one binding site in the C-terminal domain of SdCen. For both XPC and Sfi1 targets, the binding reaction was enthalpically driven. Calcium was not required for binding although its presence enhanced the binding. ITC measurements performed at 30 °C in the presence of EDTA showed that C-SdCen had a greater affinity for P17-XPC than for R18-Sfi1, with binding affinities of 30 × 106 M−1 and 4.5 × 106 M−1, respectively.
The temperature sensitive mutations identified in yeast centrin result in the arrest of cell division [11]; however, which centrin-binding interaction was perturbed was not elucidated. We attempted identify which centrin-target interaction is involved in the arrest of cell division by exploring the effect of temperature on centrin binding to XPC and Sfi1. Our ITC experiments showed that a temperature increase had a more pronounced effect on Sfi1 most likely because several centrin contacts are reduced in the case of Sfi1. Indeed the previously published structure of human Cen2 showed that contacts between F113 (corresponding to SdCenF109) and Sfi1 were reduced compare to XPC [30].
3.3. The F109 residue of SdCen is the main centrin residue involved in target binding regardless of centrin-binding motif orientation
Structural analysis of HsCen2 has shown that the F113 residue makes contacts with several target residues [30]. The important role of F113 has already been described in yeast where the corresponding mutation F105L in Cdc31 results in cell cycle arrest when the temperature is increased [11]. Mutating F109 to leucine decreased the binding of C-SdCen to both XPC and Sfi1. However, the F109L mutation had a more negative effect on the affinity constant for P17-XPC than for R18-Sfi1, with a decrease in Ka by a factor of 15 and of 6, respectively. In addition, the change in enthalpy ΔΔH was more pronounced in the case of P17-XPC than R18-Sfi1 (Table 3). These results are consistent with previous observations made when the structures of Cen2/P17-XPC [26] and Cen2/R17-Sfi1 [30] are superimposed. Indeed, the dipole inversion in Sfi1 reduces contacts between the centrin (F113) and the target-triad (L4).
3.4. The E144 residue of SdCen, which corresponds to the E148 residue of HsCen2, discriminates between the DNA repair protein XPC and the centrosomal protein Sfi1
The C-SdCenE144A variant allowed us to confirm our hypothesis concerning the role of the E148 residue of HsCen2 in complex with target-derived peptides [30]. The observed effects of dipole inversion in centrin targets presented here can be considered a general effect because it was first elucidated in the human Cen2 structure and is now demonstrated using mutagenesis and thermodynamic experiments with green alga centrin.
The E148 residue of HsCen2 (or SdCenE144), which discriminates between XPC and Sfi1, is strongly conserved in organisms that possess either a single centrin or several centrin isoforms. The E148 residue is located at the junction of helix 7 and the calcium-binding loop IV. Could it be required for a calcium-induced conformational change? This does not seem to be the case, because addition of calcium enhanced the affinity of XPC for both wild-type SdCen and the SdCenE144A variant. Thus, the question remains why the E148 residue (in human) is conserved among centrin isoforms from the same organism. Does it mean that all isoforms bind two types of targets regardless of the orientation of the centrin-binding motif? All ITC experiments were performed using target-derived peptides, and the interaction with E148 depends on the charge of peptide terminus facing it. In the future, the binding reaction should be characterized using full-length targets to avoid charge effects that may interfere with contacts involving the E148 residue.
3.5. What is the advantage of the centrin-binding motif orientation?
The ITC experiments presented here and in previous work demonstrated that the affinity of centrin is better for XPC than for other targets such as Sfi1 [22,25,30] and transducin β [Grecu and Assairi to be published elsewhere]. This could suggest that the orientation of the centrin-binding motif of XPC is the best orientation for effective centrin binding. However, because calcium is more important for Sfi1 binding [25,30] and absolutely required for binding to transducin β [17, Grecu and Assairi to be published elsewhere], the orientation of their centrin-binding motifs may be optimized for calcium-regulated binding.
4. Material and methods
4.1. Materials
The peptides used in this study were: R18-HsSfi1, 677-SQHNRQLLRGALRRWK-692, and P17-XPC, 846-NWKLLAKGLLIRERLKR-863. These peptides were purchased from GENECUST; the peptides were acetylated at their N-terminus and amidated at their C-terminus for increased stability. Oligonucleotides were purchased from Eurogentec; dNTPs, NdeI and XhoI restriction enzymes, T4 DNA ligase and Finnzymes Phusion DNA polymerase were purchased from New England Biolabs. The pET24a vector and Escherichia coli NM554, BL21(DE3) strains were purchased from Novagen; DEAE-TSK and phenyl-TSK were purchased from Interchim; Sephadex G25 was purchased from GE-Healthcare.
4.2. Methods
4.2.1. Cloning of S. dubia centrin (wild type and variants) and expression in E. coli
The S. dubia integral centrin (SdCen) coding sequence was amplified from a plasmid containing the centrin gene (gift of Michael Melkonian, University of Köln, Germany) by polymerase chain reaction (PCR) using Phusion DNA polymerase, dNTPs, and flanking 5′- and 3′-primers (5′-GGAATTCCATATGAGCTACAGGAAGGCTGCC-3′ and 5′-GGCGCTCGAGTCAGAACAGTGACGTCTTCTTCATG-3′) to create the plasmid pLA17.36.1 encoding the full-length SdCen. The PCR products were digested with the NdeI and XhoI restriction enzymes, and T4 DNA ligase was used to insert them between the NdeI and XhoI restriction sites of the pET24a expression vector. This recombinant plasmid was used as a template for site-directed mutagenesis. Site-directed mutagenesis of SdCen was accomplished via the double PCR method [31], using the full-length wild-type clone as the template, and adding dNTPs, Phusion DNA polymerase, both flanking 5′- and 3′-primers, and mutagenic oligonucleotides, F109L: 5′-CGTCTCGTCGTCGTCTAACAGGCGGAAGGCCTT-3′ to create the plasmid pLA17.36.9; or E144A: 5′-GCCATCGCGGTCCGCCGCGTCAATCATCTCCTG-3′ to create the plasmid pLA17–36–10. The double mutant E144A-F109L was constructed by using the plasmid pLA17–36–10, containing the mutation E144A, as the DNA template and the mutagenic oligonucleotide F109L: 5′-CGTCTCGTCGTCGTCTAACAGGCGGAAGGCCTT-3′ to create the plasmid pLA17.36.11.
The C terminal domain (residues Met93-Phe168) of wild-type SdCen and its variants was constructed by using the plasmids listed above as the DNA template encoding the full-length SdCen and the flanking primers: 5′-GGAATTCCATATGGGCGAGCGCGACTCCC-3′ and 5′-GGCGCTCGAGTCAGAACAGTGACGTCTTCTTCATG-3′ to create the plasmids pLA17.36.5 (wild type), pLA17.36.9 (F109L), pLA17–36–10 (E144A), pLA17.36.11 (E144A-F109L).
The PCR products were digested with the NdeI and XhoI restriction enzymes, and T4 DNA ligase was used to insert them between the NdeI and XhoI restriction sites of the pET24a expression vector. The resulting plasmids were used to transform E. coli NM554 cells, and the transformed clones were sequenced to verify their integrity and incorporation of the desired mutation. DNA from successfully transformed clones was isolated and used to transform the E. coli BL21(DE3)/pDIA17 strain [32], which is chloramphenicol resistant, in order to express the recombinant proteins.
For overproduction of SdCen and its variants, the transformants containing the centrin-encoding plasmid were grown at 37 °C in 2YT medium (Difco) supplemented with chloramphenicol (30 μg/mL) and kanamycin (70 μg/mL). When the absorbance reached 1.5 at 600 nm, the production of the protein was induced by addition of 1 mM isopropyl-β-d-1-thiogalactopyranoside, and the growth was continued for an additional 3 h at 37 °C. The cells were then pelleted by centrifugation and used as a protein purification source.
4.2.2. Purification of the C-terminal domain of SdCen and its variants
Bacterial extracts were heated at 63 °C for 10 min, and the denatured proteins were removed by centrifugation. The wild-type and mutant variants of SdCen C-terminal domain were purified using three successive chromatographic steps, including DEAE-TSK (ion exchange), phenyl-TSK (reversed phase) and G25 (gel filtration). Centrins were then lyophilized and the degree of purity was assessed using SDS–PAGE [33]. The SdCen concentration was determined by UV spectrophotometry using the extinction coefficients (E) of 1490 and 2860 M−1 cm−1 at 280 and 259 nm, respectively, based on the tyrosine and phenylalanine content [34].
4.2.3. Isothermal Titration Calorimetry (ITC)
The thermodynamic parameters of the molecular interactions between SdCen (C-terminal domain of wild type and variants) and the target peptides (P17-XPC, R18-Sfi1) were investigated via ITC using a MicroCal MCS instrument (MicroCal Inc., Northampton, MA). The protein and peptide were equilibrated in the same buffer (50 mM MOPS, 100 mM NaCl pH 7.5) with 1 mM CaCl2 or 2 mM EDTA. The centrins (10 μM) in the calorimeter cell were titrated with the target peptide (100 μM) by automatic injections of 8–10 μL at 30 °C or 37 °C. The first injection of 2 μl was ignored in the final data analysis. Integration of the peaks corresponding to each injection and baseline correction were performed using the Origin-based software provided by the manufacturer. The data were fit to an interaction model to generate the stoichiometry (n), equilibrium binding constant (Ka), and enthalpy of complex formation (ΔH). The free energy (ΔG) was determined using the following equations: ΔG = −RTln (1/Ka), where R (1.987 cal/mol) is the gas constant and T (303 K) is the absolute temperature, and ΔG = ΔH − TΔS. The following units were used: dissociation constant (Ka) M−1; Gibbs energy change (ΔG) kcal/mol; enthalpy change (ΔH) kcal/mol; entropy change (ΔS) kcal/mol.
4.2.4. Identification of Sfi1, Rad4 and centrins
Sfi1, Rad4, and centrins from various eukaryotes were identified via searches in the National Center for Biotechnology Information (NCBI) database, using the Psi-Blast program [35] with human and yeast homologue proteins as a query sequence.
4.2.5. Determination of the Sfi1 repeats and sequence analysis of Sfi1 protein
The Sfi1 sequences were aligned with HsSfi1 and ScSfi1 sequences using the ClustalW program [36]. Centrin-binding motifs are known for both HsSfi1 and ScSfi1, making it possible to determine the centrin-binding motifs of the new Sfi1 sequences from the alignments. Hydrophobic cluster analysis (HCA) [37,38] was performed for the new Sfi1 proteins, allowing a better determination of their respective repeats. The program for detection of hydrophobic amino acids clusters (HCA) is accessible on the Ressource Parisienne en Bioinformatique (RPBS) server. The programs used to perform protein sequence analyses are accessible on the SWISS-PROT web site [39]. We used GOR4 [40], SOPMA [41] for protein secondary structure predictions.
Acknowledgements
This work was supported by the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale and the Institut Curie. Dora Grecu acknowledges a PhD grant from the Université Pierre et Marie Curie.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary material
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fob.2013.11.005.
Appendix. Supplementary materials
This document contains Supplementary Figs. S1 and S2.
References
- 1.Salisbury J.L., Baron A., Surek B., Melkonian M. Striated flagellar roots: Isolation and partial characterization of a calcium-modulated contractile organelle. J. Cell Biol. 1984;99:962–970. doi: 10.1083/jcb.99.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang B., Mengersen A., Lee V.D. Molecular cloning of cDNA for caltractin, a basal body-associated Ca2+-binding protein: homology in its protein sequence with calmodulin and the yeast CDC31 gene product. J. Cell Biol. 1988;107:133–140. doi: 10.1083/jcb.107.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanders M.A., Salisbury J.L. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamymonas reinhardtii. J. Cell Biol. 1994;124:795–805. doi: 10.1083/jcb.124.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J.K., Bressan R., Hasegaw P. An Atriplex nummularia cDNA with sequence related to the algal caltractin gene. Plant Physiol. 1992;99:1734–1735. doi: 10.1104/pp.99.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum P., Furlong C., Bayers B. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc. Natl. Acad. Sci. USA. 1986;33:5512–5516. doi: 10.1073/pnas.83.15.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfrum U. Cytoskeletal elements in arthropod sensilla and mammalian photoreceptors. Biol. Cell. 1992;76:373–381. doi: 10.1016/0248-4900(92)90441-3. [DOI] [PubMed] [Google Scholar]
- 7.Lee V.D., Huang B. Molecular cloning and centrosomal localization of human caltractin. Proc. Natl. Acad. Sci. USA. 1993;90:11039–11043. doi: 10.1073/pnas.90.23.11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Errabolu R., Sanders M.A., Salisbury J.L. Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J. Cell Sci. 1994;107:9–16. doi: 10.1242/jcs.107.1.9. [DOI] [PubMed] [Google Scholar]
- 9.Madeddu L., Klotz C., LeCaer J.P., Beisson J. Characterization of centrin genes in Paramecium. Eur. J. Biochem. 1996;238:121–128. doi: 10.1111/j.1432-1033.1996.0121q.x. [DOI] [PubMed] [Google Scholar]
- 10.Wiech H., Geier B.M., Paschke T., Spang A., Grein K., Steinkötter J., Melkonian M., Schiebel E. Characterization of green alga, yeast, and human centrins, specific subdomain features determine functional diversity. J. Biol. Chem. 1996;271:22453–22461. doi: 10.1074/jbc.271.37.22453. [DOI] [PubMed] [Google Scholar]
- 11.Ivanovska I., Rose M.D. Fine structure analysis of the centrin yeast, Cdc31p, identifies residues specific for cell morphology and spindle pole body duplication. Genetics. 2001;157:503–518. doi: 10.1093/genetics/157.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paoletti A., Bordes N., Haddad R., Schwartz C.L., Chang F., Bornens M. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol. Biol. Cell. 2003;14:2793–2808. doi: 10.1091/mbc.E02-10-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koblenz B., Schoppmeier J., Grunow A., Lechtreck K.F. Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J. Cell Sci. 2003;116:2635–2646. doi: 10.1242/jcs.00497. [DOI] [PubMed] [Google Scholar]
- 14.Molinier J., Ramos C., Fritsch O., Hohn B. Centrin2 modulates homologous recombination and nucleotide excision repair in Arabidopsis. Plant Cell. 2004;16:1633–1643. doi: 10.1105/tpc.021378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Araki M., Masutani C., Takemura M., Uchida A., Sugawasa K., Kondoh J., Ohkuma Y., Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 2001;276:18665–18672. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- 16.Fischer T., Rodriguez-Navarro S., Pereira G., Racz A., Schiebel E., Hurt E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat. Cell Biol. 2004;6:840–848. doi: 10.1038/ncb1163. [DOI] [PubMed] [Google Scholar]
- 17.Puvermüller A., Giessl A., Heck M., Wottrich R., Scmitt A., Ernst O.P., Choe H.W., Hofmann K.P., Wolfrum U. Calcium dependent assembly of centrin/G-protein complex in photoreceptor cells. Mol. Cell Biol. 2002;22:2194–2203. doi: 10.1128/MCB.22.7.2194-2203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giessl A., Pulvermuller A., Trojan P., Park J.H., Choe H.W., Ernst O.P., Hofmann K.P., Wolfrum U. Differential expression and interaction with the visual G-protein transducin of centrin isoforms in mammalian photoreceptor cells. J. Biol. Chem. 2004;279:51472–51481. doi: 10.1074/jbc.M406770200. [DOI] [PubMed] [Google Scholar]
- 19.Kilmartin J.V. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 2003;162(7):1211–1221. doi: 10.1083/jcb.200307064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma P.J., Widerick D., Nauwelares F., Dumortier A., De Doncker J.M., Thevelei J.M., Van Dijck P. Deletion of SFI1, a novel suppressor of partial Ras-cAMP pathway deficiency in the yeast Saccharomyces cerevisiae, causes G2 arrest. Yeast. 1999;15:1097–1109. doi: 10.1002/(SICI)1097-0061(199908)15:11<1097::AID-YEA437>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 21.Sheehan J.H., Bunick C.G., Hu H., Fagan P.A., Meyn S.M., Chazin W.J. Structure of the N-terminal calcium sensor domain of centrin reveals the biochemical basis for domain-specific function. J. Biol. Chem. 2006;281:2876–2881. doi: 10.1074/jbc.M509886200. [DOI] [PubMed] [Google Scholar]
- 22.Radu L., Durussel I., Assairi L., Blouquit Y., Miron S., Cox J.A., Creascu CT. Scherffelia dubia centrin exhibits a specific mechanism for Ca2+-controlled target binding. Biochemistry. 2010;20:4383–4394. doi: 10.1021/bi901764m. [DOI] [PubMed] [Google Scholar]
- 23.Yang A., Miron S., Duchambon P., Assairi L., Blouquit Y., Craescu C.T. The N-terminal domain of human centrin 2 has closed structure, binds calcium with a very low affinity, and plays a role in the self-assembly. Biochemistry. 2006;45:880–889. doi: 10.1021/bi051397s. [DOI] [PubMed] [Google Scholar]
- 24.Thompson J.R., Ryan Z.C., Salisbury J.L., Kumar R. The structure of the human centrin 2-Xeroderma Pigmentosum Group C protein complex. J. Biol. Chem. 2006;281:18746–18752. doi: 10.1074/jbc.M513667200. [DOI] [PubMed] [Google Scholar]
- 25.Charbonnier J.B., Renaud E., Miron S., Le D., MH, Blouquit Y., Duchambon P., Christova P., Shosheva A., Rose T., Angulo J.F., Craescu C.T. Structural, thermodynamic and cellular characterization of human centrin 2 interaction with Xeroderma Pigmentosum Group C protein. J. Mol. Biol. 2007;373:1032–1046. doi: 10.1016/j.jmb.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 26.Yang A., Miron S., Mouwad L., Duchambon P., Blouquit Y., Craescu C.T. Flexibility and plasticity of human centrin 2 binding to the protein XPC from nuclear excision repair. Biochemistry. 2006;45:3653–3663. doi: 10.1021/bi0524868. [DOI] [PubMed] [Google Scholar]
- 27.Lutz W., Lingle W.L., McCormick D., Greenwood T.M., Salisbury J.L. Phosphorylation of centrin during the cell cycle and its role in centriole separation preceding centrosome duplication. J. Biol. Chem. 2001;276:20774–20780. doi: 10.1074/jbc.M101324200. [DOI] [PubMed] [Google Scholar]
- 28.Jani D., Lutz S., Marshall N.J., Fisher T., Köhler A., Ellisdon A.M., Hurt E., Stewart M. Sus1, Cdc31, and the Sac3 CDI region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol. Cell. 2009;33:727–737. doi: 10.1016/j.molcel.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S., Sandercock A., Conduit P., Robinson C.V., Williams R.L., Kilmartin J.V. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 2006;173(6):867–877. doi: 10.1083/jcb.200603153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Sanz J., Kateb F., Assairi L., Blouquit Y., Bodenhausen G., Abergel D., Mouawad L., Craescu C.T. Structure, dynamics and thermodynamics of the human centrin2/hsSfi1 complex. J. Mol. Biol. 2010;395:191–204. doi: 10.1016/j.jmb.2009.10.041. [DOI] [PubMed] [Google Scholar]
- 31.Picard V., Bock S.C. Rapid and efficient one-tube PCR-based mutagenesis method. Methods Mol. Biol. 1997;67:183–188. doi: 10.1385/0-89603-483-6:183. [DOI] [PubMed] [Google Scholar]
- 32.Munier H., Gilles A.M., Glaser P., Krin E., Danchin A., Sarfati R.S., Bârzu O. Isolation of catalytic and calmodulin-binding domains of Bordetella pertussis adenylate cyclase. Eur. J. Biochem. 1991;196:469–474. doi: 10.1111/j.1432-1033.1991.tb15838.x. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Pace C.N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul S.F., Madde T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positision-pecific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaboriaud C., Bissey V., Benchetrit T., Mornon J.P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987;224:149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- 38.Callebaut I., Labesse G., Durand P., Poupon A., Canard L., Chomilier J., Henrissat B., Mornon J.P. Deciphering protein sequence information though hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol. Life Sci. 1997;53(8):621–645. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bairoch A., Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL. Nucleic Acids Res. 1997;25(1):31–36. doi: 10.1093/nar/25.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garnier, J., Gibrat, J.F., Robson, B. (1996) GOR secondary structure prediction method version IV. Methods Enzymol. R.F. Doolittle Ed., 266, 540–553. [DOI] [PubMed]
- 41.Geourjon C., Delage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995;11(6):681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This document contains Supplementary Figs. S1 and S2.


