Abstract
Porites corals are foundation species on Pacific reefs but a confused taxonomy hinders understanding of their ecosystem function and responses to climate change. Here, we show that what has been considered a single species in the eastern tropical Pacific, Porites lobata, includes a morphologically similar yet ecologically distinct species, Porites evermanni. While P. lobata reproduces mainly sexually, P. evermanni dominates in areas where triggerfish prey on bioeroding mussels living within the coral skeleton, thereby generating asexual coral fragments. These fragments proliferate in marginal habitat not colonized by P. lobata. The two Porites species also show a differential bleaching response despite hosting the same dominant symbiont subclade. Thus, hidden diversity within these reef-builders has until now obscured differences in trophic interactions, reproductive dynamics and bleaching susceptibility, indicative of differential responses when confronted with future climate change.
Keywords: cryptic species, zooxanthellae, coral-bleaching susceptibility, asexual coral propagation, trophic interactions, microsatellites
1. Introduction
Unrecognized species diversity, especially in foundation species, can impede our understanding of major features of ecosystem functioning and the resilience of communities [1,2], thereby complicating projections of the ecological dynamics and future of imperilled coral reefs. Corals are placed in functional groups based on their structural growth forms (e.g. tabular, bushy or massive) [3]. These functional groups show striking differences in life histories and susceptibility to threats [4], and promote species diversity by providing different habitats for reef dwellers [5]. In addition to these readily apparent morphological differences, marine communities harbour many species not easily resolvable without extensive molecular genetic characterization, but which provide additional diversity [1] that is typically not perceived by scientists. Here, we concentrate on two coral species with nearly indistinguishable morphologies and test whether unresolved species diversity masks functional diversity [1], specifically with respect to trophic interactions and stress resistance. We concentrate on the eastern tropical Pacific (ETP), where relatively low species diversity makes the system more tractable compared with richer reefs in the Indo-West Pacific [6,7].
Environmental conditions for reef growth are suboptimal in the ETP, owing mainly to seasonal cold-water upwelling, low aragonite saturation state and recurrent warm-water events associated with the El Niño Southern Oscillation (ENSO) [6,8]. Together, these factors reduce coral growth [9] and reef species diversity [6,7]. Reefs in the ETP are built largely by branching Pocillopora spp. and the massive Porites lobata. However, species identification in these genera is notoriously challenging because colony morphology is plastic [10,11]. Based on genetic data, the number of Pocillopora species described from the ETP has recently been revised [10]. Less is known about the taxonomic status of P. lobata. Seven Porites species are currently recognized in the eastern Pacific [12]. Porites lobata and Porites panamensis are thought to be widespread in the region while much of the remaining diversity in this genus (Porites arnaudi, Porites australiensis, Porites lichen and Porites lutea) is restricted to the higher latitudes [12]. In lower latitudes, P. lobata dominates the reef-building coral community. A survey of genetic variation at the multi-copy internal transcribed spacer (ITS) marker at the genus level found that some colonies from Panama, diagnosed morphologically as P. lobata, clustered genetically with Porites evermanni [11] currently thought to be restricted to Hawaii and the Indo-Pacific [12]. Combined with previous observations of local variation in reproduction [13], this suggests the possibility of unresolved species within P. lobata in the ETP.
Functional differences between morphologically similar coral species have yet to be demonstrated. Although corals in the Orbicella (nee Montastraea) annularis species complex in the Caribbean differ in their reproductive timing and hybridization potential [14], no study, to our knowledge, has shown differences in the way these species interact with other species as we demonstrate here for Porites spp. The ecology and evolution of reproductive mutualisms has received much interest in the terrestrial literature [15,16] but similar studies are rare in corals partly because corals do not rely on pollinators for sexual reproduction. We show here that asexual reproduction of corals might indeed be dependent on other members of the reef community. Elegant work on Orbicella spp. demonstrated that partial colony bleaching can be attributed to symbiotic algae with varying heat tolerances that occupy different niches within a colony [17,18]. Conversely, we present evidence that coral host species harbouring the same dominant algal subclade differ in bleaching response, pointing to the role of the host in temperature tolerance.
2. Results and discussion
Samples of massive Porites were collected from 17 locations in the ETP (see map in figure 3; see the electronic supplementary material, table S1). A principal coordinate analysis (PCoA) based on genetic distance grouped the 448 unique multi-locus genotypes (MLGs) into two clusters along the first principle coordinate (PCo1), which explained 67% of the variation (figure 1a). No geographical structure was evident. Rather, sympatric MLGs assigned with high probability (99.5 ± 2.2% s.d.) to one of two clusters (figure 1b; see the electronic supplementary material for additional clustering and genotyping results). A network analysis of allelic sequences from five single-copy nuclear genes similarly reveals two clusters in our eastern Pacific collections (n = 14): one associated with P. lobata samples from across the Pacific and the other with Hawaiian P. evermanni (M. E. Hellberg 2013, unpublished data).
Figure 3.
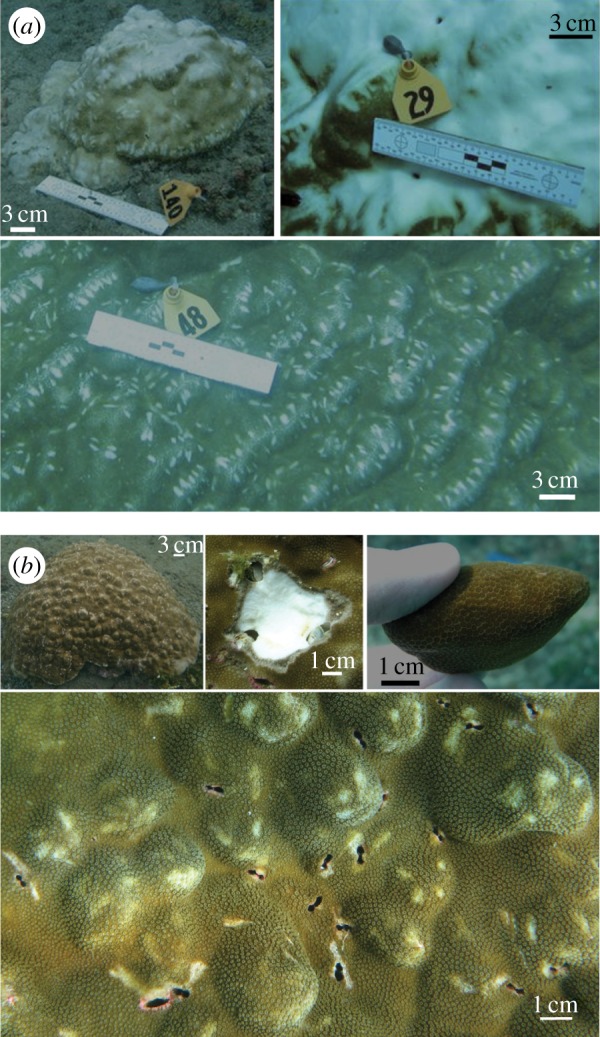
Colony growth forms of Porites spp. in the eastern Pacific. (a) Photographs of P. lobata top row: whole colony; bleached colony; bottom: typical ridge-like morphology. (b) Photographs of P. evermanni top row: whole colony, endolithic Lithophagea mussels exposed during sampling, ‘rolling stone’ fragment; bottom: typical peak-like nodules with Lithophagea boreholes at the base of or between peaks.
Figure 1.
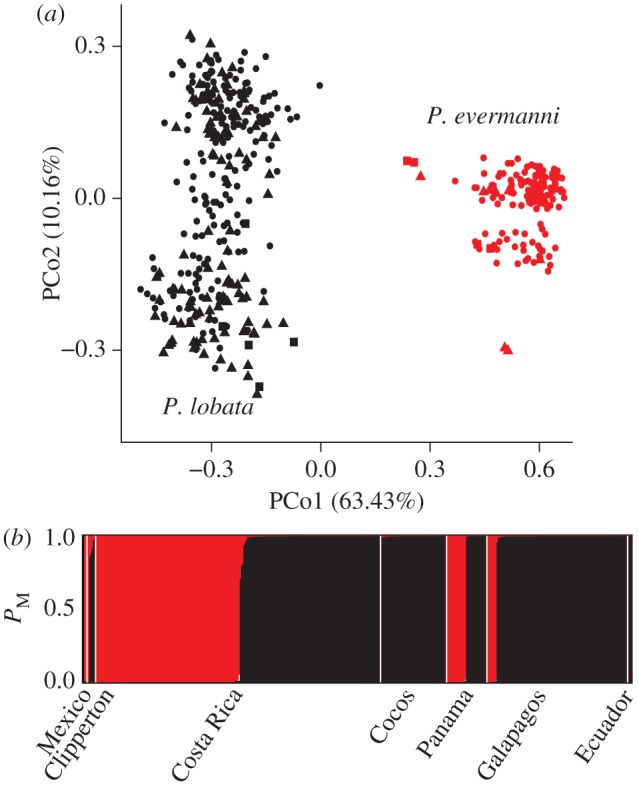
Cluster analysis of 11 microsatellite loci amplified in ETP Porites samples. Strong probability of membership to either the P. lobata or the P. evermanni cluster is demonstrated by individuals in all locations. (a) Principal coordinates (PCo) analysis on genetic identity by region. North (squares): Mexico and Clipperton Island; central (triangles): Costa Rica, Cocos Island and Panama; south (circles): Galápagos and Ecuador. (b) Structure plot with probability of membership (PM) to a cluster given on y-axis, samples are on x-axis. (Online version in colour.)
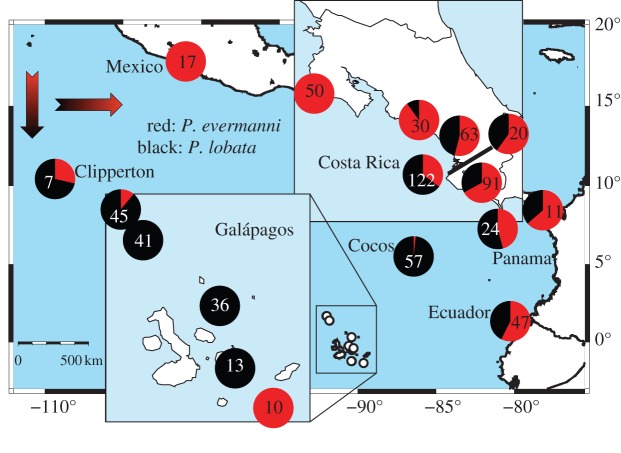
These two species show a strong geographical distribution gradient between inshore and offshore sites (figure 2; analysis of variance (ANOVA); F2 = 73.383, p < 0.001). Insular collections are composed almost entirely of P. lobata (figure 3a) [19]. Porites evermanni (figure 3b) constitutes roughly half of the collections at southern sites and gradually increases along a northern gradient. The two most marginal coastal sites in the north comprised entirely P. evermanni (see the electronic supplementary material for additional species distribution results). Thus, the two related species occupy different environmental niches.
Figure 2.
Distribution of P. evermanni and P. lobata across the eastern Pacific. Only P. evermanni was found in Mexico or the northern-most site in Costa Rica (upper right insert). Porites evermanni was rare offshore (lower left insert). Numbers indicate ramets sampled.
On a local scale, we found substantial differences between species in the relative importance of asexual reproduction. Colonies sampled from 10 sites (four coastal and six insular) using a spatially explicit random method [20] were genotyped as before, revealing that each species was found in seven plots (co-occurring in four). However, standard genotypic diversity estimates (e.g. number of MLGs/number of samples (NG/N); number of observed MLGs/number of expected MLGs (GO/GE); see the electronic supplementary material, table S3) showed that rates of asexual reproduction differed (t-test; nPlots_Pe = 7, nPlots_Pl = 7; NG/N, p = 0.01; GO/GE, p < 0.005) between the species. Porites evermanni often reproduces asexually (NG/N = 0.47 ± 0.20 s.d., GO/GE = 0.30 ± 0.20 s.d.), whereas P. lobata does so rarely (NG/N = 0.81 ± 0.22 s.d., GO/GE = 0.75 ± 0.27 s.d.; figure 4). The mode of asexual reproduction here is probably fragmentation of adult colonies rather than asexual larval production because ramets occur close together and asexually produced larvae have not been reported in gonochoric broadcast spawners as these Porites spp. [21].
Figure 4.
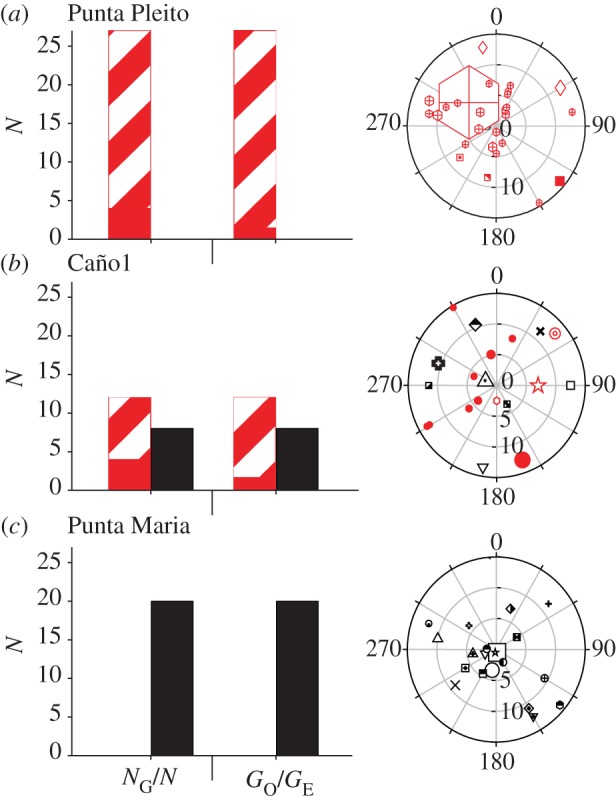
Analysis of clonal structure in Costa Rica revealed greater clonal diversity in P. lobata (black) than P. evermanni (grey; red online). Panels show average genotypic diversity indices (NG/N and GO/GE) and representative polar plots (three out of 10) along the Costa Rican coast: Punta Pleito (a), Caño Island 1 (b), and Cocos Island: Punta Maria (c). In bar charts: striped bars, clones; solid bars, unique multi-locus genotypes (MLGs). In polar plots: unique MLGs are represented by unique symbols. Ramets of the same genet share symbols. Radial axis shows distance in 5 m increments and the angular axis is given in 30° increments. Colony symbols were scaled by colony area (range: 35 cm2–20.4 m2). (Online version in colour.)
Differences in asexual reproduction appear to be driven by interactions between corals, bivalves and triggerfish. Endolithic bivalves (Lithophaga spp.) bore into the carbonate skeleton of corals (figure 3b) and result in colony fragmentation [7,22–25]. Coral skeletal strength is reduced in colonies containing Lithophaga spp. and colonies fracture along boreholes if present [24] (figure 3b). Two endemic triggerfish (Pseudobalistes naufragium and Sufflamen verres) common in the ETP prey upon bioeroding Lithophaga spp., which constitute 83% of the diet of P. naufragium at Caño Island [23]. Where the Porites species occur together, mussel density (N cm−2) is higher in P. evermanni than in P. lobata based on direct counts (t-test; p < 0.001, nPe = 22, nPl = 21) and photographic evidence (t-test; p = 0.011, nPe = 46, nPl = 61; figure 5a).
Figure 5.
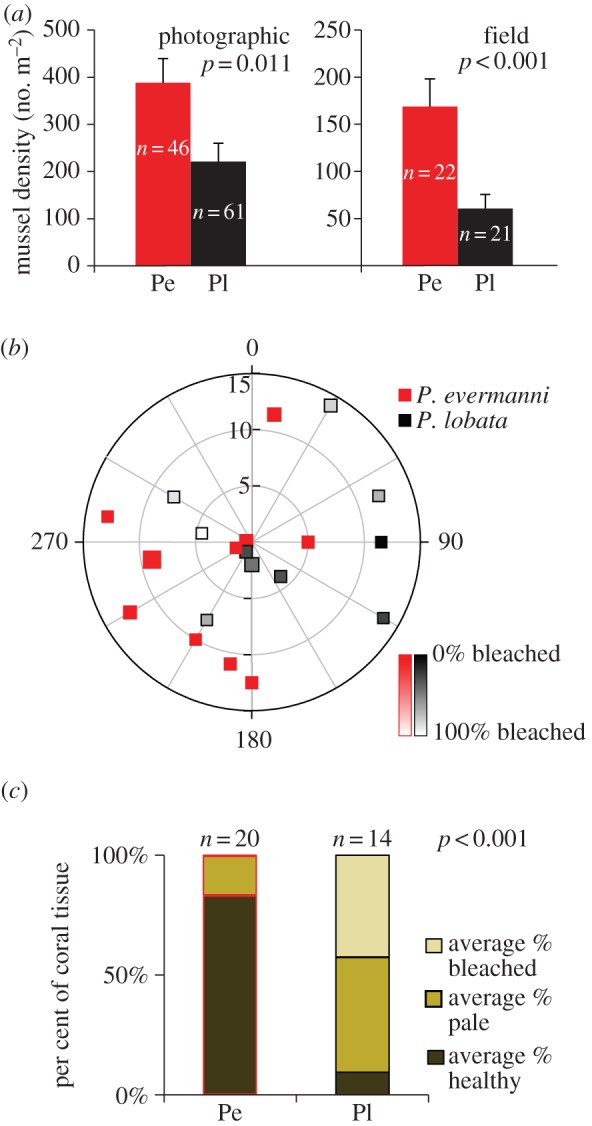
Species-level differences were evident in mussel density and amount of bleaching in each colony. (a) Mean mussel density (number of mussels boreholes per m2 of tissue) ± s.e.m. in P. evermanni and P. lobata based on photographic and field analyses. The mean mussel density was higher (t-test, log transformed) in P. evermanni than in P. lobata. (b) Polar plot depicting location, species and bleaching status for each colony sampled at Caño 2. Radial axes (in 30° increments) show distance (m) in 5 m increments. Symbol indicates species. Amount of tissue bleached (%) is represented by fill. (c) Proportions of bleached (white), healthy (green/brown) and pale (yellow) tissue in photographs of P. evermanni (Pe) and P. lobata (Pl) based on visual inspections of tissue underneath 36 uniformly spaced points on each image. The mean per cent of bleached tissue was higher (t-test, arcsine square root transformed) in P. lobata than P. evermanni.
To extract their prey, the triggerfish break off coral fragments, resulting in ‘rolling stones’ known as coralliths [22] that can reattach to the substrate (figure 3b). A single fish can produce three to 75 fragments in a few minutes from a single colony, with apparently few negative consequences for the coral, as evinced by rapid wound healing of the donor colony (they regenerate in less than four weeks) and high establishment rates of fragments (30–50% of fragments survive) [25]. All coralliths sampled randomly in polar plots were P. evermanni (n = 6, mean maximum dimension = 10 ± 4.1 cm s.d.). Four out of six were fragments from a larger colony sampled in the plot (mean maximum dimension = 56.5 ± 32.6 cm s.d.). The fragments were located at a mean distance of 3.67 ± 1.69 m s.d. from the likely ‘parent’ colony. The differences between the two species in mode of reproduction may play a large part in their disparate geographical distributions because larger colony fragments might be better able to establish in less hospitable settings (continental margins; northerly latitudes) than sexually produced larvae, and fragment production does not require the presence of a sexual partner.
Whereas biting by triggerfish is relatively benign, thermal stress during ENSO events causes coral bleaching and is a major factor in ETP coral mortality [9,26]. Photographic evidence (t-test; p < 0.001) revealed that P. lobata bleached more readily (nPl = 14; mean per cent tissue bleached per colony = 42.5 ± 35.5% s.d.) than P. evermanni (nPe = 20; mean per cent tissue bleached per colony = 0.4 ± 1.9% s.d.) at two sites with sympatric P. lobata and P. evermanni (less than 5 m apart; figure 5c). Such differential bleaching has previously been ascribed to differences in the subclade of algal symbionts hosted in co-occurring coral species [17]. However, denaturing gradient gel electrophoresis (DGGE) fingerprinting and sequencing of the ITS2 region of the Symbiodinium algae present in P. lobata (n = 18) and P. evermanni (n = 19) show that both coral species associate primarily with Symbiodinium subclade C15 (see the electronic supplementary material, figure S2). Therefore, the differential bleaching susceptibility of these coral species points to differences in host physiology, although symbiont density [27], background (minor) symbionts [28] or differences among closely related strains of the ITS2-C15 subclade might play an additional role.
Knowlton [1] proposed that unrecognized species diversity in the sea hinders our understanding of marine ecosystem function and limits our ability to predict how reefs will respond to climate change, but few data have supported this claim. Here, we demonstrate that unresolved species diversity has obscured differences in reproduction, ecological dynamics, trophic interactions and stress resistance between two reef-building coral species.
We propose that a three-way interaction between corals, mussels and triggerfish alters the local distribution of the foundation fauna. The species involved have different geographical distributions, varying from the triggerfish restricted to the ETP to the trans-Pacific range of P. lobata. It follows that variation in their co-distribution could alter community dynamics across the Pacific [29]. An understanding of such geographical shifts in trophic interactions is important because trophic complexity is one driver of ecosystem diversity [30] and marine ecosystem diversity is thought to be linked to increased function [31].
The maintenance and function of ecosystems built by few foundation species rely on the persistence of those species. In P. evermanni, frequent asexual fragmentation allows for local patch reef formation when sexual partners are unavailable [32,33] and the persistence and spread of potentially locally well-adapted genotypes. However, genetically depauperate populations are more susceptible to non-random (with respect to genotype) stressors than diverse populations [34], an alarming prospect given the increasing frequency of disturbance events affecting ETP coral reefs [35,36].
The adaptability of corals in the face of elevated sea surface temperatures and the consequences of more frequent bleaching conditions are not fully understood. Here, however, we have shown that morphological similarity can mask variation in responses to thermal stressors. Furthermore, ecosystem resilience improves when critical species are functionally redundant in some respects (e.g. the form of the colonies they build) but show differential stress responses [4], as observed here. This argues against recent trends of using morphological groups to project the future of reefs [37]. It follows that as long as coral taxonomy remains unresolved, unrecognized species will continue to obscure our understanding of reef ecosystem function and resilience and our ability to predict the fate of the coral reef ecosystem. The functional differences within morphologically similar species observed in this study lead us to predict that there will be a differential response to climate change among the massive Porites species that compose the foundation of ETP reefs.
3. Material and methods
(a). Study sites and sampling
Corals were sampled under randomly generated coordinates in 15 m radius polar plots [20] or haphazardly (more than 5 m separating sampled colonies) at sites where random sampling was not feasible owing to low colony density (see the electronic supplementary material, table S1). Random coordinates were generated using the random number generation function in Microsoft Excel 2007 (Microsoft, WA, USA) with a precision of 5° of arc and of 0.5 m along strike. Coordinates were located by a team of SCUBA divers using a compass and a measuring tape secured to the centre point of a circle. If present, the colony underneath a coordinate was sampled by using a hammer and chisel to break off a small fragment of coral tissue (maximum size 1 × 1 cm). No colony was sampled twice. An underwater photograph was taken of each colony sampled, and colony size was measured as the maximum length, width and height to the nearest 10 cm. Sampling of a polar plot ceased when 20 colonies had been collected. All colonies within each 15 m radius circle were counted. Fragments were placed in individual zip-lock bags underwater, and then transferred to vials containing 95% ethanol on shore. Fragments were stored in a −20°C freezer prior to genotyping.
(b). Genotyping
DNA from coral tissue was extracted from each sample following the manufacturer's instructions in the DNeasy 96 Blood and Tissue Kit (Qiagen, CA, USA). A total of 11 microsatellite loci were used in this study ([38,39]; see the electronic supplementary material, table S2). One additional marker (pl0072) was developed for this study following Polato et al. [38]. DNA was amplified with fluorescently labelled primers in one singleplex and four multiplex reactions consisting of two to three primer pairs each (see the electronic supplementary material, table S2). Thermal cycling was performed in an MJ Research PT200 (Bio-Rad, CA, USA) or an Eppendorf Mastercycler Gradient (Eppendorf, Germany) with an initial denaturation step of 95°C for 5 min followed by 35 cycles of 95°C for 20 s; 52–56°C (see the electronic supplementary material, table S2) for 20 s and 72°C for 30 s. A final extension of 30 min at 72°C ensured the addition of a terminal adenine to all products [40]. Fragments were analysed by using an ABI 3730 with an internal size standard (Genescan LIZ-500, Applied Biosystems, CA, USA; Penn State Genomics Core Facility, University Park, PA, USA). Electropherograms were visualized and allele sizes were called using Genemapper v. 4.0 (Applied Biosystems, CA, USA). Samples that failed to amplify more than two of 11 loci were excluded from the analysis. This resulted in an overall average failure of 5 ± 4% s.d. and a per locus failure rate of less than 12% for each locus in the included samples (n = 684).
(c). Analysis of multi-locus genotype data
MLGs were grouped into genets in GenAlEx v. 6.4 [41] by requiring complete matches at all loci ignoring missing data. Only unique MLGs (n = 448) were used in subsequent analyses. Potential genotyping errors were detected with GenClone v. 2.0 [42] and flagged allele calls were corrected when appropriate by re-examining electropherograms. Despite a lower number of alleles in P. evermanni, the power to distinguish unique MLGs was high in both species as calculated by the combined probability of identity (PID) [41] for each species (PIDlobata = 6.8 × 10–10; PIDevermanni = 1.7 × 10–5). Given the sample sizes of 377 and 307 and the respective PIDs, we do not expect to have mistakenly identified colonies as clonemates of another colony when they were in fact the result of independent sexual reproductive events in P. lobata (estimated number of misidentified colonies = 2.5 × 10–7) or in P. evermanni (estimated number of misidentified colonies = 5.1 × 10–3).
(d). Clustering analysis
A PCoA on standardized codominant genotypic distance was conducted in GenAlEx v. 6.4 [41] (n = 448). We also clustered MLGs using Bayesian assignment methods implemented in Structure v. 2.3.3 [43]. The software uses a Bayesian clustering algorithm to assign genotypes to clusters that minimize deviation from Hardy–Weinberg expectations. Values of K = 1 to 6 were tested, where K = 1 represents a single panmictic population and K = 6 equals the number of subregions (see the electronic supplementary material, table S1) by running three replicate simulations with 106 Markov Chain Monte Carlo repetitions each and a burn-in of 100 000 iterations with an admixture model assuming independent allele frequencies among clusters. Under the null hypothesis that P. lobata formed a cohesive genetic unit, we chose the admixture model to prevent masking a signal of gene flow. However, admixture of individuals was rare (figure 1b). Further, we assumed independent allele frequencies because we were looking for strong signals of differentiation. Altering the assumptions (non-admixture, independent allele frequencies) did not change the optimal number of clusters or the outcome of species assignment for any individual. In fact, under a non-admixture model at K = 2, each MLG assigned to its respective cluster with 100% probability of membership giving no evidence of hybridization between species. The Evanno et al. [44] method implemented in Structure Harvester [45] was used to select the most likely number of clusters. The assignment of individual MLGs to species was congruent between PCoA and Structure in all cases.
Because the markers were developed for P. lobata, we investigated whether the loci showed differential failure rates between species and whether these failures were driving the clustering results. Overall failure rates were low and similar between species (P. lobata: 5% and P. evermanni 4% of samples failed to amplify at any locus). On the locus level, four out of 11 of the microsatellite loci in this analysis (pl1370, pl2258, pl0072 and pl905) showed differential failure rates between species (Fisher's exact test, nPe = 149, nPl = 299, d.f. = 1, p < 0.05) after Bonferonni correction for multiple testing. Including only the seven non-significant loci does not change the outcome of clustering analyses or the membership of any of the samples in the species clusters identified with all loci (data not shown).
(e). Clonal structure analysis
Clonal diversity measures (genotypic richness, genotypic diversity and genotypic evenness) were calculated using Genodive [46] for all randomly sampled polar plots (see the electronic supplementary material for definitions of indices and table S3 for results). The probability that repeated MLGs were the product of different sexual events (psex) was calculated for each repeated genotype included in the clonal structure analysis using MLGsim [47]. The values of psex for each MLG repeated within a sampling plot were significantly low (meanPe = 4.8 × 10–8 ± 7.1 × 10–8 s.d; meanPl = 1.1 × 10–15 ± 1.7 × 10–15 s.d) as to reject origin by sexual reproduction and instead point to an asexual origin (104 simulations, p < 0.05). Colonies were mapped in SigmaPlot v. 10.0 (Systat, IL, USA) by using polar coordinates from the field with each symbol scaled for colony size (figure 4).
(f). Geographical distribution of species
The regional difference in the relative numbers of P. evermanni and P. lobata sampled was analysed with an ANOVA using each polar plot as a sampling point and a Fisher's exact test on the proportions of each species found in a region performed in Systat v. 13.1 (Systat, IL, USA). The regions considered for this analysis were: Oceanic Island (Cocos), Coastal Island (Caño) and Mainland (Mainland Costa Rica). Only the polar plot data (where sampling effort was standardized) were appropriate for this analysis.
(g). Photographic analysis of bleaching
Bleaching was observed primarily at two sampling sites, Caño 2 and Tres Hermanas, where P. lobata and P. evermanni coexist. Caño 2 and Tres Hermanas plots have little topography (depth range = 2.4–5.5 m and 4.6–6.1 m, respectively) over the scale sampled. The average distance between a sampled colony and its nearest sampled congener at Caño 2 was 4.44 ± 2.44 m s.d. and at Tres Hermanas was 4.76 ± 3.07 m s.d., thus P. evermanni and P. lobata colonies were exposed to the same water temperatures and UV light conditions.
Photographs were taken of each colony sampled in the field as described above (nPl = 14, nPe = 20; nphotos = 81). Thirty-six uniformly spaced points were overlain on each image in Photoshop v. CS5.1 (Adobe, CA, USA). A categorical score was given for each point based on visual inspections of the health of the tissue underneath. Categories included substrate, healthy coral, bleached coral, pale coral, flag and photo scale. Flags were temporary, numbered markers of standard size (4 × 5 cm; Allflex USA, Inc., TX, USA) to identify each colony. Healthy coral tissue was green to brown. Bleached tissue was white. Pale tissue was yellow when compared with a white standard included in each photograph (the photo scale). The per cent of tissue of each colony that was bleached, healthy and pale was estimated by dividing the number of points in that category over the total number of points in all three categories. The mean per cent of bleached tissue was compared between species using a t-test in Systat v. 13.1 (Systat, IL, USA).
(h). Lithophagea surveys
The density of Lithophagea bore holes in coral colonies was surveyed both in the field and photographically at sampling sites where P. evermanni and P. lobata co-occur. At Caño 2 and Drake Bay, colonies of similar size (estimated as the surface area of a five-sided rectangular prism using max length, width and height as measured in the field) were selected for each species (nPe = 22, nPl = 21; colony surface area, t-test, t42 = 0.759, p = 0.45) and the number of mussel holes was counted in situ. At sympatric sampling sites, photographs (n = 202) of Porites colonies were scaled using a photo scale included in each photograph in the program AxioVision 4.8 (Zeiss, Germany). Visible surface area in each photo was measured using the Trace tool in AxioVision. There was no difference in the average surface area measured between species (nPe = 46, nPl = 61; t-test, t105 = −0.328, p = 0.743). Discernible mussel boreholes (figure 2b) were counted using the event tool in AxioVision without knowledge of molecular species identification (file names did not reveal species identity). The mean density of mussels (number of mussels per cm2 of tissue) was compared between species using t-tests in Systat v. 13.1 (Systat, IL, USA).
(i). Denaturing gradient gel electrophoresis
To determine symbiont diversity, Symbiodinium internal transcribed spacer 2 (ITS2) sequences were fingerprinted with DGGE. By using standards of known unique fingerprints and direct sequencing of dominant bands from those fingerprints the symbiont diversity can be resolved to the subclade level [48]. We amplified the Symbiodinium ITS2 region in three to five samples of each species from the polar plots where the species coexist (Caño 1, Caño 2, Caño 5 and Tres Hermanas; nPl = 18, nPe = 19) using primers and amplification protocol previously described [48]. The single P. evermanni sample collected at Cocos Island and one P. evermanni sample from the Galápagos were also run. Of the analysed colonies, eight were bleached or partially bleached (nPl = 6, nPe = 2); nine were pale (nPl = 5, nPe = 4); 12 were healthy (nPl = 3, nPe = 9); four were healthy with some pale spots (nPl = 2, nPe = 2) and four had unrecorded tissue health status (nPl = 2, nPe = 2). Reaction products and a diagnostic subclade C15 standard [49] were separated by electrophoresis for 19 h at 120 V at a constant temperature of 60°C on an 8% polyacrylamide gradient gel containing a gradient of 3.15 M urea/18% deionized formamide to 5.6 M urea/37% deionized formamide. Gels were visualized after staining with SYBRGreen (Molecular Probes, OR, USA) using manufacturer's specifications and photographed (see the electronic supplementary material, figure S2 for example). DGGE profiles were characterized by their prominent bands. Two representative bands from each host species and a C15 positive control were excised, re-amplifed, Sanger sequenced (Penn State Genomics Core Facility, University Park, PA, USA) and aligned against a published C15 ITS2 sequence (GenBank AY239369.1) [50] using Geneious 5.5.6 (Biomatters, New Zealand).
Acknowledgements
J. Nivia-Ruiz, S. Luna, D. Wham and T. LaJeunesse provided assistance. A. Lopez, H. Reyes-Bonilla, A. Baker and Z. Forsman provided samples. C. Fisher and J. Marden commented on drafts. J.N.B., J.C., M.E.H. and I.B.B. collected data. J.N.B. performed molecular and statistical analysis. J.N.B. and I.B.B. wrote the paper. All authors edited the paper.
Data accessibility
Coral genotyping data and photographs are available at Dryad (doi:10.5061/dryad.d6108). WGS84 coordinates of sampling locations and primer sequences are uploaded as the electronic supplemental material. The data reported in this paper are tabulated in the electronic supplementary material.
Funding statement
Work was supported by NSF grant no. OCE–0550294 to I.B.B. and M.E.H.
References
- 1.Knowlton N. 1993. Sibling species in the sea. Annu. Rev. Ecol. Syst. 24, 189–216 (doi:10.1146/annurev.es.24.110193.001201) [Google Scholar]
- 2.Hughes AR, Stachowicz JJ. 2004. Genetic diversity enhances the resistance of a seagrass ecosystem to disturbance. Proc. Natl Acad. Sci. USA 101, 8998–9002 (doi:10.1073/pnas.0402642101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes TP, Connell JH. 1999. Multiple stressors on coral reefs: a long-term perspective. Limnol. Oceanogr. 44, 932–940 (doi:10.4319/lo.1999.44.3_part_2.0932) [Google Scholar]
- 4.Bellwood DR, Hughes TP, Folke C, Nystrom M. 2004. Confronting the coral reef crisis. Nature 429, 827–833 (doi:10.1038/nature02691) [DOI] [PubMed] [Google Scholar]
- 5.Idjadi JA, Edmunds PJ. 2006. Scleractinian corals as facilitators for other invertebrates on a Caribbean reef. Mar. Ecol. Prog. Ser. 319, 117–127 (doi:10.3354/meps319117) [Google Scholar]
- 6.Glynn PW, Ault JS. 2000. A biogeographic analysis and review of the far eastern Pacific coral reef region. Coral Reefs 19, 1–23 (doi:10.1007/s003380050220) [Google Scholar]
- 7.Glynn PW, Wellington GM. 1983. Corals and coral reefs of the Galápagos Islands. Berkeley, CA: University of California Press [Google Scholar]
- 8.Cortés J. 1997. Biology and geology of eastern Pacific coral reefs. Coral Reefs 16, S39–S46 (doi:10.1007/s003380050240) [Google Scholar]
- 9.Toth LT, et al. 2012. ENSO drove 2500-year collapse of eastern Pacific coral reefs. Science 337, 81–84 (doi:10.1126/science.1221168) [DOI] [PubMed] [Google Scholar]
- 10.Pinzón JH, LaJeunesse TC. 2011. Species delimitation of common reef corals in the genus Pocillopora using nucleotide sequence phylogenies, population genetics and symbiosis ecology. Mol. Ecol. 20, 311–325 (doi:10.1111/j.1365-294X.2010.04939.x) [DOI] [PubMed] [Google Scholar]
- 11.Forsman ZH, Barshis DJ, Hunter CL, Toonen RJ. 2009. Shape-shifting corals: molecular markers show morphology is evolutionarily plastic in Porites. BMC Evol. Biol. 9, 45 (doi:10.1186/1471-2148-9-45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veron J. 2000. Corals of the World, p. 490 Townsville, Australia: Australian Institue of Marine Science [Google Scholar]
- 13.Glynn PW, Colley SB, Eakin CM, Smith DB, Cortes J, Gassman NJ, Guzman HM, Delrosario JB, Feingold JS. 1994. Reef coral reproduction in the eastern Pacific: Costa Rica, Panama, and Galápagos Islands (Ecuador) II. Poritidae. Mar. Biol. 118, 191–208 (doi:10.1007/BF00349785) [Google Scholar]
- 14.Levitan DR, Fogarty ND, Jara J, Lotterhos KE, Knowlton N. 2011. Genetic, spatial and temporal components of precise spawning synchrony in reef building corals of the Montastraea annularis species complex. Evolution 65, 1254–1270 (doi:10.1111/j.1558-5646.2011.01235.x) [DOI] [PubMed] [Google Scholar]
- 15.Ollerton J, Coulthard E. 2009. Evolution of animal pollination. Science 326, 808–809 (doi:10.1126/science.1181154) [DOI] [PubMed] [Google Scholar]
- 16.Burkle LA, Marlin JC, Knight TM. 2013. Plant-pollinator interactions over 120 years: loss of species, co-occurrence, and function. Science 339, 1611–1615 (doi:10.1126/science.1232728) [DOI] [PubMed] [Google Scholar]
- 17.Rowan R, Knowlton N, Baker A, Jara J. 1997. Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature 388, 265–269 (doi:10.1038/40843) [DOI] [PubMed] [Google Scholar]
- 18.Baker AC, Starger CJ, McClanahan TR, Glynn PW. 2004. Corals’ adaptive response to climate change. Nature 430, 741 (doi:10.1038/430741a) [DOI] [PubMed] [Google Scholar]
- 19.Boulay JN, Cortés J, Nivia-Ruiz J, Baums IB. 2012. High genotypic diversity of the reef-building coral Porites lobata (Scleractinia: Poritidae) in Isla del Coco National Park, Costa Rica. Rev. Biol. Trop. 60, 279–292 [Google Scholar]
- 20.Baums IB, Miller MW, Hellberg ME. 2006. Geographic variation in clonal structure in a reef building Caribbean coral, Acropora palmata. Ecol. Monogr. 76, 503–519 (doi:10.1890/0012-9615(2006)076[0503:GVICSI]2.0.CO;2) [Google Scholar]
- 21.Baird AH, Guest JR, Willis BL. 2009. Systematic and biogeographical patterns in the reproductive biology of scleractinian corals. Annu. Rev. Ecol. Evol. Syst. 40, 551–571 (doi:10.1146/annurev.ecolsys.110308.120220) [Google Scholar]
- 22.Glynn PW. 1974. Rolling stones among the scleractinian: mobile corallith communities in the Gulf of Panama. Proc. Second Int. Coral Reef Symp. 2, 183–188 [Google Scholar]
- 23.Guzmán HM. 1988. Distribución y abundancia de organismos coralívoros en los arrecifes coralinos de la Isla del Caño, Costa Rica. Rev. Biol. Trop. 36, 191–207 [Google Scholar]
- 24.Scott PJB, Risk MJ. 1988. The effect of Lithophaga (Bivalvia, Mytilidae) boreholes on the strength of the coral Porites lobata. Coral Reefs 7, 145–151 (doi:10.1007/bf00300974) [Google Scholar]
- 25.Guzmán HM, Cortés J. 1989. Coral reef community structure at Caño Island, Pacific Costa Rica. Mar. Ecol. 10, 23–41 (doi:10.1111/j.1439-0485.1989.tb00064.x) [Google Scholar]
- 26.Glynn PW. 1988. El-Niño Southern Oscillation 1982–1983: nearshore population, community, and ecoystem responses. Annu. Rev. Ecol. Syst. 19, 309–346 (doi:10.1146/annurev.ecolsys.19.1.309) [Google Scholar]
- 27.Cunning R, Baker AC. 2013. Excess algal symbionts increase the susceptibility of reef corals to bleaching. Nat. Clim. Change 3, 259–262 (doi:10.1038/nclimate1711) [Google Scholar]
- 28.Baker AC. 2003. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 34, 661–689 (doi:10.1146/annurev.ecolsys.34.011802.132417) [Google Scholar]
- 29.Thompson JN. 2005. The geographic mosaic of coevolution. Chicago, IL: The University of Chicago Press [Google Scholar]
- 30.Paine RT. 1966. Food web complexity and species diversity. Am. Nat. 100, 65–75 (doi:10.1086/282400) [Google Scholar]
- 31.Duarte CM. 2000. Marine biodiversity and ecosystem services: an elusive link. J. Exp. Mar. Biol. Ecol. 250, 117–131 (doi:10.1016/s0022-0981(00)00194-5) [DOI] [PubMed] [Google Scholar]
- 32.Highsmith RC. 1980. Passive colonization and asexual colony multiplication in the massive coral Porites lutea (Milne Edwards and Haime). J. Exp. Mar. Biol. Ecol. 47, 55–67 (doi:10.1016/0022-0981(80)90137-9) [Google Scholar]
- 33.Lasker HR, Coffroth MA. 1999. Responses of clonal reef taxa to environmental change. Am. Zool. 39, 92–103 [Google Scholar]
- 34.Reusch TBH, Ehlers A, Hammerli A, Worm B. 2005. Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc. Natl Acad. Sci. USA 102, 2826–2831 (doi:10.1073/pnas.0500008102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins M, et al. 2010. The impact of global warming on the tropical Pacific ocean and El Niño. Nat. Geosci. 3, 391–397 (doi:10.1038/ngeo868) [Google Scholar]
- 36.Glynn PW. 1990. Coral mortality and disturbance to coral reefs in the tropical eastern Pacific. In Global ecological consequences of the 1982–1983 El Nino-Southern Oscillation. (ed. Glynn PW.), pp. 55–126 Amsterdam, The Netherlands: Elsevier [Google Scholar]
- 37.Darling ES, Alvarez-Filip L, Oliver TA, McClanahan TR, Côté IM. 2012. Evaluating life-history strategies of reef corals from species traits. Ecol. Lett. 15, 1378–1386 (doi:10.1111/j.1461-0248.2012.01861.x) [DOI] [PubMed] [Google Scholar]
- 38.Polato NR, Concepcion GT, Toonen RJ, Baums IB. 2010. Isolation by distance across the Hawaiian Archipelago in the reef-building coral Porites lobata. Mol. Ecol. 19, 4661–4677 (doi:10.1111/j.1365-294X.2010.04836.x) [DOI] [PubMed] [Google Scholar]
- 39.Baums IB, Boulay JN, Polato NR, Hellberg M. 2012. No gene flow across the eastern Pacific barrier in the reef-building coral Porites lobata. Mol. Ecol. 21, 5418–5433 (doi:10.1111/j.1365-294X.2012.05733.x) [DOI] [PubMed] [Google Scholar]
- 40.Brownstein MJ, Carpten JD, Smith JR. 1996. Modulation of non-templated nucleotide addition by tag DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20, 1004–1010 [DOI] [PubMed] [Google Scholar]
- 41.Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295 (doi:10.1111/j.1471-8286.2005.01155.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnaud-Haond S, Belkhir K. 2007. Genclone: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol. Ecol. Notes 7, 15–17 (doi:10.1111/j.1471-8286.2006.01522.x) [Google Scholar]
- 43.Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol. Ecol. 14, 2611–2620 (doi:10.1111/j.1365-294X.2005.02553.x) [DOI] [PubMed] [Google Scholar]
- 45.Earl DA, vonHoldt BM. 2012. Structure Harvester: a website and program for visualizing Structure output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361 (doi:10.1007/s12686-011-9548-7) [Google Scholar]
- 46.Meirmans PG, Van Tienderen PH. 2004. Genotype and Genodive: two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 4, 792–794 (doi:10.1111/j.1471-8286.2004.00770.x) [Google Scholar]
- 47.Stenberg P, Lundmark M, Saura A. 2003. MLGsim: a program for detecting clones using a simulation approach. Mol. Ecol. Notes 3, 329–331 (doi:10.1046/j.1471-8286.2003.00408.x) [Google Scholar]
- 48.LaJeunesse TC. 2002. Diversity and community structure of symbiotic dinoflagellates from Caribbean coral reefs. Mar. Biol. 141, 387–400 (doi:10.1007/s00227-002-0829-2) [Google Scholar]
- 49.LaJeunesse TC, Thornhill DJ, Cox EF, Stanton FG, Fitt WK, Schmidt GW. 2004. High diversity and host specificity observed among symbiotic dinoflagellates in reef coral communities from Hawaii. Coral Reefs 23, 596–603 (doi:10.1007/s00338-004-0428-4) [Google Scholar]
- 50.LaJeunesse TC, Loh WKW, van Woesik R, Hoegh-Guldberg O, Schmidt GW, Fitt WK. 2003. Low symbiont diversity in southern Great Barrier Reef corals, relative to those of the Caribbean. Limnol. Oceanogr. 48, 2046–2054 (doi:10.4319/lo.2003.48.5.2046) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Coral genotyping data and photographs are available at Dryad (doi:10.5061/dryad.d6108). WGS84 coordinates of sampling locations and primer sequences are uploaded as the electronic supplemental material. The data reported in this paper are tabulated in the electronic supplementary material.