Abstract
RNA is complementary to the DNA sequence from which it is transcribed. Therefore, interactions between DNA and RNA provide a simple mechanism of genetic self-detection within nuclei. Imprinted RNAs could enable alleles of maternal and paternal origin to detect whether they are the same (homozygous) or different (heterozygous), and thereby provide strategic information about expected relatedness to siblings.
Keywords: inclusive fitness, imprinting, siRNA, green beards, relatedness
‘The situations in which a species discriminates in its social behaviour tend to evolve and multiply in such a way that the coefficients of relationship involved in each situation become more nearly determinate’. [1, p. 24]
1. Introduction
The probabilities that factor into calculations of relatedness can be parsed into probabilities, given a genealogy and uncertainties of genealogy. Genealogy may be uncertain, for instance, because a littermate is sometimes a full-sib, sometimes a half-sib, or because a herd contains kin of different degrees, but members cannot distinguish the categories [2]. Hamilton proposed that natural selection favours reduction of genealogical uncertainty. He further proposed that natural selection favours ‘discrimination of those individuals which do carry one or both of the behaviour-causing genes from those which do not’ ([1], pp. 24–25). Here, he considered the possibility of genes ‘recognizing’ their own copies and directing benefits on the basis of this privileged information [1].
Dawkins called gene-based discrimination ‘the Green-Beard Altruism Effect’. He envisaged a gene that encoded both a phenotypic label, the green beard, and the tendency to be nice to green-bearded individuals [3]. Kinship is a major cause of identity by kind but the label is independent of genealogy. Green-beard effects were considered implausible because genetic self-recognition and altruism were viewed as complex behaviours unlikely to be encoded by a single gene or tightly linked cluster of genes. Kinship seemed a much stronger basis for altruism. The situation is reversed at the genic level. It is much simpler to imagine a gene, or its products, preferentially interacting with identical genes, or their products, than to envisage a gene that recognizes half-cousins [4,5].
The interaction between labels of genic identity (green beards) and parental origin (genomic imprinting) makes possible a novel form of discrimination. I first review effects of genomic imprinting on estimates of genic relatedness, then describe the unusual evolutionary properties of imprinted green beards.
2. Parent-specific relatedness
The standard way to calculate the probability that a gene in A has an identical-by-descent copy in B is to multiply by one-half for each generation back from A to a common ancestor C, by one-half for each generation forward from C to B, and then sum these products for all distinct paths linking A to B. Ascending and descending factors of one-half arise from different sources of uncertainty. For each step backward from offspring to parent, a randomly chosen gene in the offspring may come from either mother or father. For each step forward from parent to offspring, the offspring receives one of two alleles in the parent.
Genomic imprinting enables genes to discriminate between matrilineal and patrilineal kin [6]. The factor of one-half for the first backward step resolves into a factor of one or zero when genes have imprinted expression. In the case of sibs, an imprinted gene of maternal origin is definitely present in an offspring's mother and transmitted to littermates with probability one-half, whereas an imprinted gene of paternal origin is definitely present in the offspring's father and transmitted to littermates with probability p/2, where p is the chance of shared paternity. Because relatedness is lower when genes are paternally derived, imprinted genes are predicted to behave more ‘selfishly’ toward littermates when paternally derived and less ‘selfishly’ when maternally derived.
If genes carried imprints of grandparental origin, then factors of one-half for the first and second backward steps would resolve into factors of either one or zero [7]. Thus, half-cousins who share a maternal grandmother are related by one-quarter for genes of maternal grandmaternal origin but are unrelated for all other genes. As yet, there is no clear evidence for second-order imprints.
3. Imprinted green beards
Complementarity between the strands of a double helix, and between DNA and the RNA transcribed from its sequence, allow allele-specific interactions within nuclei between different copies of the same gene. A diverse fauna of small RNAs participate in a wide variety of regulatory processes. Many of these processes depend on selective interaction with complementary DNA or RNA [8]. Because complementary sequences represent each other, their interaction can be viewed as a simple form of genetic self-recognition that makes possible intranuclear green-beard effects.
24-nucleotide small-interfering RNAs (siRNAs) of Arabidopsis cause RNA-directed DNA methylation (RdDM) and transcriptional silencing of DNA sequences with motifs complementary to the siRNA. The template for synthesis of siRNAs is probably the sequence subject to RdDM [8]. Dosage-sensitive responses to an siRNA would allow a sequence to ‘count’ its copies within a nucleus. Imprinted expression of an siRNA would allow alleles of maternal origin to signal their presence to alleles of paternal origin or vice versa. A silent allele can ‘hear’ what the other allele has to say.
An abundant class of maternally expressed siRNAs (mesiRNAs) are expressed from madumnal (maternally derived) chromosomes of endosperm [9]. MesiRNAs target genes that delay onset of endosperm cellularization and prolong endosperm proliferation [10], consistent with theoretical predictions that maternally expressed imprinted genes should inhibit endosperm growth [11].
Consider the introduction of mesiDNA (a motif that encodes mesiRNA) into a previously unimprinted gene encoding a growth enhancer. Transcription of A, the established allele without mesiDNA, is expected to be a compromise between a lower level favoured as a madumnal allele and higher level favoured as a padumnal (paternally derived) allele. By contrast, A′, the initially rare allele with mesiDNA, will be transcribed at the same level as A when it is a padumnal allele because the siRNA is not expressed, but at lower levels than A when it is a madumnal allele because the siRNA is expressed. Thus, A′ behaves as a padumnally expressed, madumnally silent allele when heterozygous. It will increase in frequency at the expense of A because it makes finer discriminations of relatedness.
By contrast to its behaviour in heterozygotes, A′ mRNA is transcribed from neither allele in homozygotes, because both alleles are silenced by the mesiRNA. The siRNA produced by madumnal A′ informs padumnal A′ that the seed contains an A′A′ embryo rather than an AA′ embryo and that the mother carries at least one copy of A′. Therefore, at least half of other embryos will receive A′ from their mother (in addition to those that receive A′ from their father). Thus, mesiRNA signals a doubling of ‘relatedness’ to self and increased ‘relatedness’ to littermates (rL). If rL more than doubles, the balance of benefits to self and costs to littermates for padumnal A′ shifts in favour of production of less growth enhancer, as occurs in the presence of mesiRNA.
Whether rL is doubled will depend on allele frequencies, the frequency of selfing and the number of fathers per brood. A complete analysis of this problem is beyond the scope of this letter, although some insight can be gained by considering effects when A′ is rare. In an outbreeding population, A′ will be transmitted predominantly by AA′ parents, with padumnal A′ expressed when mothers are AA but inhibited by mesiRNA in 50% of seeds when mothers are AA′. MesiRNA signals a doubling of rL for single paternity of a mother's offspring and more than doubling for multiple paternity. Therefore a reduction in seed size, with concomitant increase in seed number, would appear to benefit A′. MesiRNA functions as a ‘secret hand-shake’ that allows padumnal A′ to recognize its allelic partner as self and to reduce its own transcription for the benefit of madumnal A′ in littermates.
MesiRNA could also function as an adaptive signal of self-fertilization. Selfing shifts the optimal trade-off between seed size and number to smaller seeds for genes expressed in filial tissues [12]. If mothers sometimes self, padumnal A′ will be more likely to encounter mesiRNA in selfed seeds than outcrossed seeds, especially when A′ is rare. Therefore, padumnal A′ will promote smaller seeds on selfing and larger seeds when outcrossed.
A′ produces less mRNA in homozygotes than is optimal for an unimprinted allele expressed in offspring. Therefore, near fixation of A′, rare alleles (such as A) that are expressed more than A′ will be favoured by natural selection. This suggests that A and A′ will be maintained at a polymorphic equilibrium.
Green-beard altruism is vulnerable to ‘cheats’ who flaunt the label, receive its benefits, but do not reciprocate [13]. A°, a version of A′ that retains mesiRNA but is insensitive to its inhibitory action, would increase in frequency at the expense of A′ because madumnal A° induces padumnal A′ to reduce demand, benefiting A°, but A° does not reciprocate when madumnal and padumnal roles are reversed. Once A° eliminates A′, the population is primed for the introduction of A*, an allele that possesses a beard of a different colour (new mesiRNA) [14]. Such an iterative process (figure 1), in which successively introduced mesiRNAs are only transiently effective, could explain the diversity of mesiRNAs, their rapid evolutionary turnover and mild effects [9,15].
Figure 1.
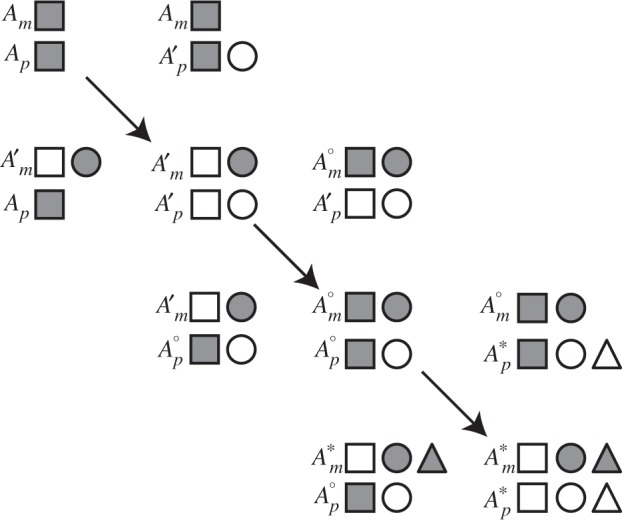
The mesiRNA ratchet. A population initially fixed for allele A (upper left) is successively invaded by an allele A′ that also encodes a mesiRNA; an allele A° that retains the mesiRNA but is insensitive to its effects and an allele A* that encodes a new mesiRNA (lower right). Subscripts m and p indicate madumnal and padumnal alleles. Squares represent the coding sequence of an mRNA. Circles and triangles represent coding sequences of mesiRNAs. Filled symbols are expressed. Unfilled symbols are silent. Homozygotes lie on the main diagonal. Off-diagonal elements are heterozygotes.
Imprinted gene clusters of mammals contain many non-coding RNAs [16]. DNA–DNA associations and RNA–DNA interactions within these clusters may be facilitated by somatic pairing of madumnal and padumnal chromosomes [17,18]. MicroRNAs processed from maternally expressed antiPeg11 cause mRNA degradation of paternally expressed Peg11 [19]. Mutations of madumnal Rasgrf1 silence the padumnal copy of a neighbouring non-coding RNA [20]. Disruption of madumnal Ube3a upregulates padumnal Ube3a-antisense [21]. Such examples suggest the possibility of green-beard effects at mammalian imprinted loci.
References
- 1.Hamilton WD. 1964. The genetical evolution of social behaviour. II. J. Theor. Biol. 7, 17–52 (doi:10.1016/0022-5193(64)90039-6) [DOI] [PubMed] [Google Scholar]
- 2.Haig D. 2011. Genomic imprinting and the evolutionary psychology of human kinship. Proc. Natl. Acad. Sci. USA 108, 10 878–10 885 (doi:10.1073/pnas.1100295108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawkins R. 1976. The selfish gene. Oxford, UK: Oxford University Press [Google Scholar]
- 4.Haig D. 1996. Gestational drive and the green-bearded placenta. Proc. Natl. Acad. Sci. USA 93, 6547–6551 (doi:10.1073/pnas.93.13.6547) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Queller DC, Ponte E, Bozzaro S, Strassmann JE. 2003. Single-gene greenbeard effects in the social amoeba Dictyostelium discoideum. Science 299, 105–106 (doi:10.1126/science.1077742) [DOI] [PubMed] [Google Scholar]
- 6.Haig D. 1997. Parental antagonism, relatedness asymmetries, and genomic imprinting. Proc. R. Soc. Lond. B 264, 1657–1662 (doi:10.1098/rspb.1997.0230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haig D. 2000. Genomic imprinting, sex-biased dispersal, and social behavior. Ann. NY Acad. Sci. 907, 149–163 (doi:10.1111/j.1749-6632.2000.tb06621.x) [DOI] [PubMed] [Google Scholar]
- 8.Castel SE, Martienssen RA. 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14, 100–112 (doi:10.1038/nrg3355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosher RA, Melnyk CW, Kelly KA, Dunn RM, Studholme DJ, Baulcombe DC. 2009. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460, 283–286 (doi:10.1038/nature08084) [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Zhang C, Baulcombe DC, Chen ZJ. 2012. Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 109, 5529–5534 (doi:10.1073/pnas.1203094109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haig D. 2013. Kin conflict in seed development: an interdependent but fractious collective. Annu. Rev. Cell Devel. Biol. 29 (doi:10.1146/annurev-cellbio-101512-122324) [DOI] [PubMed] [Google Scholar]
- 12.de Jong TJ, van Dijk H, Klinkhamer PGL. 2005. Hamilton's rule, imprinting and parent–offspring conflict over seed mass in partially selfing plants. J. Evol. Biol. 18, 676–682 (doi:10.1111/j.1420-9101.2004.00856.x) [DOI] [PubMed] [Google Scholar]
- 13.Gardner A, West SA. 2010. Greenbeards. Evolution 64, 25–38 (doi:10.1111/j.1558-5646.2009.00842.x) [DOI] [PubMed] [Google Scholar]
- 14.Traulsen A, Nowak MA. 2007. Chromodynamics of cooperation in finite populations. PLoS ONE 2, e270 (doi:10.1371/journal.pone.0000270) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Z, Coruh C, Axtell MJ. 2010. Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 22, 1090–1103 (doi:10.1105/tpc.110.073882) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girardot M, Cavaillé J, Feil R. 2012. Small regulatory RNAs controlled by genomic imprinting and their contribution to human disease. Epigenetics 7, 1341–1348 (doi:10.4161/epi.22884) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thatcher KN, Peddada S, Yasui DH, LaSalle JM. 2005. Homologous pairing of 15q11–13 imprinted domains in brain is developmentally regulated but deficient in Rett and autism samples. Hum. Mol. Genet. 14, 785–797 (doi:10.1093/hmg/ddi073) [DOI] [PubMed] [Google Scholar]
- 18.Krueger C, King MR, Krueger F, Branco MR, Osborne CS, Niakan KK, Higgins MJ, Reik W. 2012. Pairing of homologous regions in the mouse genome is associated with transcription but not imprinting status. PLoS ONE 7, e38983 (doi:10.1371/journal.pone.0038983) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis E, Caiment F, Tordoir X, Cavaillé J, Ferguson-Smith AC, Cockett N, Georges M, Charlier C. 2005. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 15, 743–749 (doi:10.1016/j.cub.2005.02.060) [DOI] [PubMed] [Google Scholar]
- 20.Brideau CM, Kauppinen KP, Holmes R, Soloway PD. 2010. A non-coding RNA within the Rasgrf1 locus in mouse is imprinted and regulated by its homologous chromosome in trans. PLoS ONE 5, e13784 (doi:10.1371/journal.pone.0013784) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landers M, Calciano MA, Colosi D, Glatt-Deeley H, Wagstaff J, Lalande M. 2005. Maternal disruption of Ube3a leads to increased expression of Ube3a-ATS in trans. Nucleic Acids Res. 33, 3976–3984 (doi:10.1093/nar/gki705) [DOI] [PMC free article] [PubMed] [Google Scholar]


