Summary
Heterogeneity within pluripotent stem cell (PSC) populations is indicative of dynamic changes that occur when cells drift between different states. Although the role of metastability in PSCs is unclear, it appears to reflect heterogeneity in cell signaling. Using the Fucci cell-cycle indicator system, we show that elevated expression of developmental regulators in G1 is a major determinant of heterogeneity in human embryonic stem cells. Although signaling pathways remain active throughout the cell cycle, their contribution to heterogeneous gene expression is restricted to G1. Surprisingly, we identify dramatic changes in the levels of global 5-hydroxymethylcytosine, an unanticipated source of epigenetic heterogeneity that is tightly linked to cell-cycle progression and the expression of developmental regulators. When we evaluated gene expression in differentiating cells, we found that cell-cycle regulation of developmental regulators was maintained during lineage specification. Cell-cycle regulation of developmentally regulated transcription factors is therefore an inherent feature of the mechanisms underpinning differentiation.
Highlights
-
•
Embryonic stem cells are lineage primed in G1
-
•
Transcription of developmentally regulated genes is cell-cycle regulated
-
•
5hmC is cell-cycle regulated
-
•
Stem cells initiate differentiation from G1
Pluripotent stem cell heterogeneity has been attributed to stochastic variations in signaling pathways across the population. Using Fucci cell-cycle reporters, Dalton and colleagues show that stem cell “lineage priming” in G1 is associated with cell-cycle-dependent changes in the transcription of developmentally regulated genes. Moreover, these changes are paralleled by levels of the epigenetic mark 5-hydroxymethylcytosine. These findings identify the cell cycle as major source of heterogeneity in human pluripotent stem cells.
Introduction
Pluripotent stem cells (PSCs) are heterogeneous under self-renewing conditions in culture (Enver et al., 2009; Graf and Stadtfeld, 2008; Martinez Arias and Brickman, 2011) and during embryonic development (Chazaud et al., 2006). This heterogeneity extends not only to the expression of pluripotency factors such as NANOG, REX1, and STELLA (Chambers et al., 2007; Hayashi et al., 2008; Singh et al., 2007; Toyooka et al., 2008), but also to lineage-specific factors such as HEX, HES1, and GATA6 (Canham et al., 2010; Kobayashi et al., 2009; Singh et al., 2007). Variations in gene expression are transient and reversible, indicating that PSCs alternate between different cell states. Although the function and molecular mechanisms underpinning this heterogeneity are unclear, it appears to be influenced by variations in the activity of signaling pathways at the single-cell level. WNT, BMP, NODAL, and FGF signaling through their downstream effectors has been implicated in contributing to PSC heterogeneity and serves to prime cells for differentiation when transiently activated (Galvin-Burgess et al., 2013; Price et al., 2013). As an example, heterogeneity can be significantly reduced when murine PSCs are cultured in the presence of small-molecule compounds that block ERK and GSK3 signaling (2i media) (Marks et al., 2012; Wray et al., 2011; Ying et al., 2008). In human embryonic stem cells (hESCs), suppression of WNT activity reduces signaling heterogeneities and the sporadic expression of developmental regulators such as BRACHYURY (Blauwkamp et al., 2012; Singh et al., 2012). Together, these observations indicate that signaling heterogeneities reflect alternate cell states that represent different differentiation potentialities.
PSCs exhibit an unusual mode of cell-cycle regulation with a truncated G1 and a large percentage of S phase cells (Singh and Dalton, 2009). As PSCs differentiate, the cell cycle is remodeled, such that G1 is lengthened and the relative amount of time associated with S phase cells is reduced. Recent reports (Calder et al., 2013; Coronado et al., 2013; Pauklin and Vallier, 2013) further documented this using the fluorescent ubiquitination-based cell-cycle indicator (Fucci) system (Sakaue-Sawano et al., 2008). Together, these studies point toward a direct relationship between the cell cycle and differentiation, consistent with earlier reports describing the ability of PSCs to initiate their differentiation program from G1 phase (Chetty et al., 2013; Jonk et al., 1992; Mummery et al., 1987; Sela et al., 2012; Singh and Dalton, 2009). This raises the possibility that heterogeneous gene expression and cell signaling variations in PSCs may also be linked to cell-cycle progression. To address this question, we utilized the Fucci system in hESCs in combination with fluorescence-activated cell sorting (FACS), and performed RNA sequencing (RNA-seq) analysis to establish that heterogeneous expression of developmental regulators is closely coupled to cell-cycle positioning. Our findings provide a rationale for gene-expression heterogeneity in hESCs and a potential mechanism for “lineage priming” in G1 phase. Moreover, we show that transient activation of developmental genes in G1, such as GATA6 and SOX17, is associated with the upregulation of 5-hydroxymethylation (5hmC), an epigenetic mark with proposed roles in gene activation (Szulwach et al., 2011a; Yu et al., 2012b).
Results
Establishment and Validation of Human Fucci ESCs
To construct Fucci hESC lines, fluorescent reporters (Sakaue-Sawano et al., 2008) were introduced into expression vectors under the control of the constitutive CAGi promoter, linked to either a neomycin (neoR) or puromycin (puroR) selectable marker through an internal ribosome entry site (Figure 1A). The principle behind Fucci indicators is that the “destruction box” sequences of the cell-cycle regulators CDT1 and GEMININ are fused to fluorescent proteins and, upon entry into a specific cell-cycle phase, are targeted for degradation. This allows for Fucci indicators to be present only during a specific window of time, with the advantage of reporting cell-cycle position in living cells. The expected pattern of reporter activity is summarized in Figure 1B, showing an initial double-negative (DN) population indicative of early G1 cells, followed by accumulation of CDT1-KO2 (Kusabira orange-2) reporter fluorescence (red) throughout the rest of G1. As the cells enter S phase, KO2 fluorescence is extinguished and GEMININ-Az1 (Azami green-1) fluorescence increases until mitosis is completed. After drug selection and expansion of WA09 hESCs, we confirmed the authentic cell-cycle-regulated expression of Fucci indicators by using several approaches in live cells. Both fluorescent reporters were robustly expressed in a largely nonoverlapping pattern, consistent with their being associated with separate phases of the cell cycle (Figures 1B and 1C). We next validated the phase-specific expression of fluorescent reporters based on DNA content using Vybrant DyeCycle stain. Cells expressing the Kusabira orange-2 (KO2) reporter were exclusively in G1, whereas cells expressing the Azami green-1 (Az1) reporter were composed of S phase and G2/M cells. As the cells transitioned through S phase and G2/M, Az1 reporter intensity increased (Figure 1D).
Figure 1.
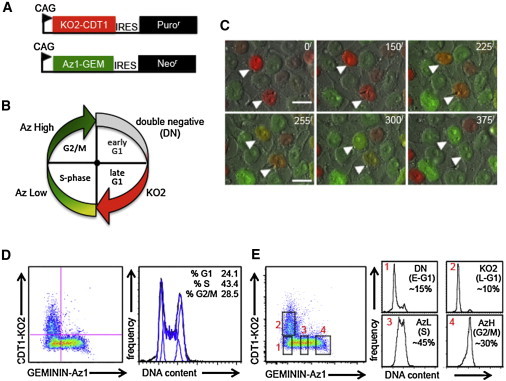
Establishment and Characterization of Fucci hESCs
(A) Diagram of Fucci reporters.
(B) Diagram of cell-cycle phase-specific fluorescent protein expression patterns.
(C) WA09 Fucci hESCs were imaged every 15 min over a 24 hr period. Micron bar: 50 μm. Arrowheads: cell progression from G1 to S to G2.
(D and E) WA09 Fucci hESCs were treated with Violet Vybrant DyeCycle for 2 hr and then analyzed on the MoFlo. The approximate percentage of cells in each cell-cycle phase is shown. Data are representative of biological replicates.
The total cell population stained with DyeCycle stain exhibited a typical hESC DNA profile comparable to that seen with other dyes, such as propidium iodide and Hoechst 33342 (Singh and Dalton, 2009), with a high percentage (∼43%) of S phase cells (Figure 1D). Upon gating of the appropriate fractions, we confirmed the cell-cycle expression of fluorescent reporters. Specifically, the DN fraction represented early G1 cells, the KO2 (red) fraction represented late G1, the Az1-low (AzL, low green) represented S phase, and the Az1-high (AzH, high green) represented G2/M (Figure 1E). Less than 5% of the cells were double positive for KO2 and Az1, indicative of cells in early S phase. These data validate the Fucci reporters as faithful indicators of cell-cycle position, as judged by DNA content.
The Heterogeneity of hESCs Is Dependent on Cell-Cycle Position and Cell Signaling
Although PSCs are known to have an unusual mode of cell-cycle regulation, deep mechanistic insight into this is lacking. Moreover, no global analysis of cell-cycle dependent genes has been performed in pluripotent cells. To examine this issue, we performed RNA-seq analysis on the four FACS-isolated Fucci cell-cycle fractions (DN, KO2, AzL, and AzH). We identified ∼500 transcripts that followed a reproducible pattern of cell-cycle regulation in three biological replicate experiments (Figure 2A; Table S1 available online). Using unsupervised cluster analysis, we identified ten cohorts of transcripts, all of which displayed a similar pattern of periodicity, and then subjected them to Gene Ontology (GO) analysis (Figure 2B; Table S2). Surprisingly, the largest group of cell-cycle-dependent transcripts consisted of developmental regulators (Figure 2C).
Figure 2.
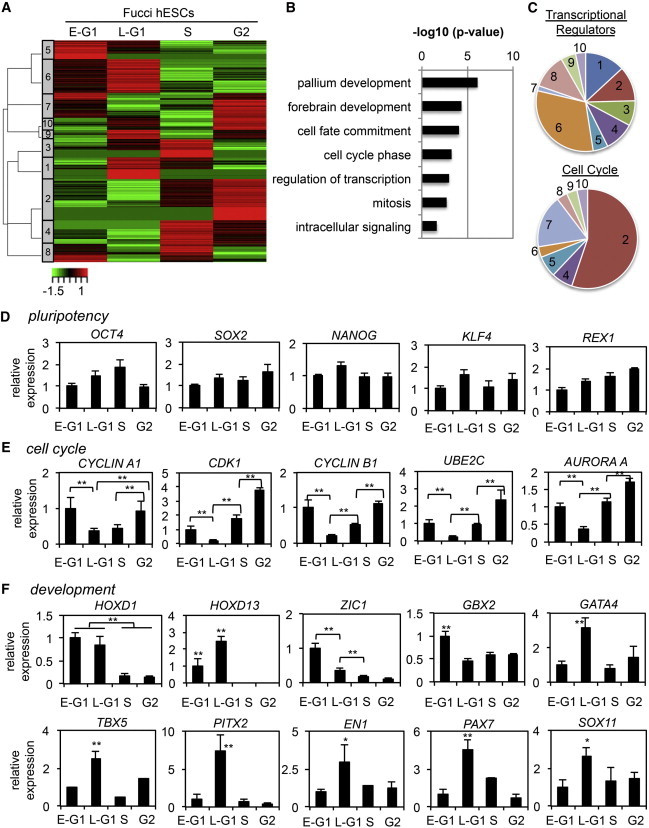
Gene-Expression Profiling of Human Fucci ESCs
(A) Following RNA-seq, cluster analysis was performed on cell-cycle-regulated transcripts in WA09 Fucci hESCs, represented by a heatmap.
(B) GO analysis of cell-cycle-regulated genes from human Fucci RNA-seq.
(C) Percentage of genes by GO analysis in the transcriptional regulation or cell-cycle categories according to RNA-seq cluster analysis. Developmental transcription factors were heavily enriched in cluster 6, and mitotic genes were enriched in clusters 2 and 7.
(D–F) WA09 Fucci hESCs were separated into cell-cycle phases by FACS, and RNA levels were then determined by qRT-PCR for (D) pluripotency genes, (E) cell-cycle mitotic genes, and (F) developmental transcription factors.
Data are representative of biological triplicate experiments displayed as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01. See also Figure S1.
RNA-seq analysis indicated that pluripotency regulators such as OCT4, NANOG, and SOX2 showed no consistent pattern of periodicity in the cell cycle (Figures 2A and 2D). Cell-cycle regulators associated with mitosis, such as CYCLIN B1, CDK1, and AURORA A, showed a consistent pattern of cell-cycle regulation, but regulators of G1 and S phase progression, such as CYCLIN E and E2F1, lacked periodicity at the transcript and protein levels (Figures 2E, S1A, and S1B). To independently validate these findings, we sorted live hESCs based on DNA content using Vybrant DyeCycle (Figures S1C–S1E). Although it is not possible to separate early and late G1 cells using this approach, transcript analysis of cell-cycle genes in G1, S, and G2/M can still be reliably performed. Quantitative RT-PCR (qRT-PCR) analysis of Vybrant DyeCycle FACS-isolated hESCs generated results similar to those obtained with Fucci-sorted cells (Figure S1). This further validates the Fucci indicator hESC line as a reliable tool for studying the cell cycle.
The most pronounced class of transcripts that showed cell-cycle-dependent periodicity was composed of developmentally regulated transcription factors. Transcripts identified by GO analysis as being involved in development, cell-fate commitment, and transcriptional regulation were heavily enriched in cluster 6, coinciding with a cohort of genes with elevated expression in G1 phase (DN and KO2) (Figure 2C). To confirm these findings, we assayed the transcript levels of several randomly selected developmental regulators from cluster 6 (G1 cohort) by qRT-PCR analysis. This analysis confirmed that transcripts important for embryonic patterning (HOXD1 and HOXD13), neural development (ZIC1, GBX2, EN1, PAX7, and SOX11), and mesoderm/definitive endoderm (DE) specification (GATA4, TBX5, and PITX2), were all selectively upregulated in G1 phase (Figure 2F). These data show that hESCs in G1 have higher expression of transcription factors that are important for differentiation and development, indicating that hESCs exist in a “lineage-primed” state during the G1 phase of the cell cycle.
The selective upregulation of transcripts for developmental genes in G1 was surprising, but it raised the possibility that such regulation could be related to previous observations of heterogeneity in PSCs (Enver et al., 2009; Graf and Stadtfeld, 2008; Martinez Arias and Brickman, 2011). To establish the relationship between cell cycle and hESC heterogeneity, we characterized in greater detail the developmental genes that are strongly expressed as part of DE differentiation and weakly expressed during PSC self-renewal (Singh et al., 2012). When we assayed Fucci hESC fractions by qRT-PCR, we found that GATA6, SOX17, FOXA2, and GATA4 transcripts were all upregulated in G1 (Figure 3A). To establish that transcriptional control of developmental regulators is a determinant of their cell-cycle regulation, we pulse labeled Fucci cells with ethynyl uridine (EU) for 1 hr and evaluated the levels of newly synthesized transcripts by qRT-PCR (Figure S2). The levels of nascent transcripts for developmental regulators from cluster 6 were all clearly upregulated in G1, consistent with global RNA-seq and qRT-PCR analyses (Figure 2F; Table S1). This indicates that transcriptional control of developmental genes is a major contributor to cell-cycle-dependent heterogeneity in hESCs. Immunostaining confirmed these results, demonstrating that developmentally regulated transcription factors are expressed during a narrow window of time in the hESC cell cycle (Figure 3B). Analysis at the single-cell level showed that developmental factors are coexpressed with pluripotency factors, indicating that spontaneously differentiating cells are not responsible for this pattern of heterogeneity (Figure S2). Expression of developmental regulators is extinguished as cells transition into S phase, thereby establishing a strong link between cell-cycle position and PSC heterogeneity. To further explore the cell-cycle regulation of developmental genes, we differentiated Fucci hESCs toward neuroectoderm progenitors using dual inhibition of ACTIVIN and BMP signaling (Chambers et al., 2009). After 5 days of differentiation, the expression of neural genes emerged in a cell-cycle-regulated pattern, peaking in early G1 (Figure S2). These data support the general idea that developmental regulators are cell-cycle regulated and contribute to heterogeneity in PSCs, regardless of which germ layer these factors are associated with.
Figure 3.
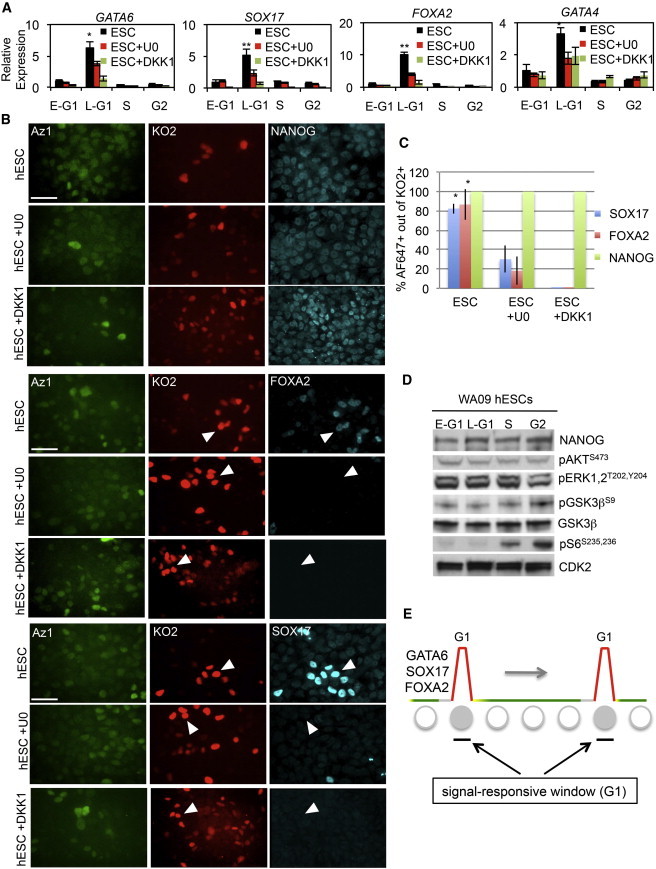
WNT and ERK Signaling Promote Heterogeneity in Late G1 Cells
(A) WA09 Fucci hESCs grown in self-renewal media were treated with U0126 (20 μM) or DKK1 (150 ng/ml) for 3 days and then separated into cell-cycle phases by FACS. RNA expression analysis by qRT-PCR was performed for endoderm markers. All experiments were performed in technical triplicate and are representative of multiple experiments.
(B and C) WA09 Fucci hESCs were treated with U0126 (20 μM) or DKK1 (150 ng/ml) for 3 days and immunostaining was performed for indicated proteins and quantitated in (C) as the ratio of Alexa Fluor 647+ (for indicated proteins) to KO2+ cells. Data represent analyses from five different fields of view, and the total number of cells counted per condition was >350. Arrowheads: KO2+ cells that express endoderm markers in self-renewal media, but not in the presence of the MEK/ERK inhibitor (U0126). Data are representative of multiple experiments. Micron bar: 50 μm.
(D) Immunoblotting (20 μg/lane) for FACS-isolated Fucci fractions for the indicated signaling molecules.
(E) Schematic diagram indicating that cells in G1 are most responsive to differentiation-inducing signals. Colors are representative of Fucci fractions (gray, early G1; red, late G1; yellow, G1/S; green, S-G2/M). Circles depict cells progressing through the cell cycle, with solid circles representing cells that express developmental regulators.
Data are representative of biological triplicate experiments displayed as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01. See also Figure S2.
Heterogeneity in PSCs has been largely attributed to stochastic changes in signaling networks, but up to now, no direct links to the cell cycle have been established. The data we obtained thus far introduced another factor that contributes to PSC heterogeneity: the cell cycle. Next, we sought to determine the relationship between cell signaling and cell-cycle position with respect to heterogeneous expression of developmental genes. We specifically addressed this by examining whether signaling pathways activate developmental genes in manner that depends on the cell-cycle-phase. Fucci hESCs were treated with MEK/ERK signaling inhibitor (U0126) or WNT antagonist (DKK1) for 3 days in self-renewal media, and then the levels of developmental regulators were examined by qRT-PCR and immunostaining (Figures 3A–3C). Inhibiting either ERK or WNT signaling reduced transcript levels for the developmental regulators GATA6, SOX17, FOXA2, and GATA4 in KO2+ (late G1) cells. In addition, treatment with U0126 or DKK1 in self-renewal media also led to a significant reduction in background protein levels for FOXA2 and SOX17, but had no effect on NANOG. These data indicate that WNT and ERK signaling pathways promote heterogeneity in hESCs by activating the transcription of developmental regulators in G1 phase. It should be noted that although 70%–80% of KO2+ cells express SOX17 protein, we observed a small percentage of cells (<2% of total cells) that expressed SOX17 in all other fractions of the cell cycle. We believe these cells to be spontaneously differentiated cells. When we evaluated the activity of several signaling pathways in Fucci fractions, we observed no general pattern of G1-dependent cell signaling (Figure 3D). For example, we found no evidence of phospho-dependent regulation of AKT1, ERK1/2, or GSK3β at different cell-cycle positions. In contrast, phosphorylation of ribosomal S6 protein was upregulated in S and G2/M phases, as described previously (Shah et al., 2003), but not in G1.
The above findings rule out the possibility that G1-associated heterogeneity is driven by cell-cycle-regulated activation of signaling pathways. Instead, G1 appears to represent a brief window of time in which cells are competent to respond to external signals at the level of gene activation (Figure 3E). How cells sense differentiation signals as a function of the cell cycle is presently unclear, but PSCs are known to be particularly susceptible to differentiation cues in G1 (Chetty et al., 2013; Mummery et al., 1987; Sela et al., 2012). Using Fucci hESCs, we then went on to confirm these previous findings (Figure S3). Fucci hESCs were sorted into cell-cycle fractions and replated in serum-containing differentiation media. After 48 hr, qRT-PCR analysis was performed for differentiation markers from all three germ layers. Cells in early and late G1 displayed increased marker expression compared with those from S or G2/M phases. To further investigate whether G1 cells are more sensitive to differentiation signals over other phases of the cell cycle, we treated Fucci hESCs for 3 hr in endoderm differentiation conditions, followed by FACS and qRT-PCR (Figure S3). We found that the 3 hr treatment of Fucci hESCs in differentiation conditions was sufficient to increase the expression levels of endoderm genes in G1 cells, but only weakly increased gene-expression levels in S and G2. These data provide further evidence that cells preferentially initiate their differentiation from the G1 phase of the cell cycle.
Cell-Cycle-Dependent Heterogeneity Is Retained during Early Differentiation
Our data show that many developmental regulators are heterogeneously expressed in hESCs by a cell-cycle-dependent mechanism. It was not clear, however, whether these developmental regulators maintain a heterogeneous pattern of expression linked to the cell cycle during differentiation. To examine this question, we utilized the well-established DE differentiation model, in which phosphatidylinositol 3 kinase (PI3-kinase) signaling is limited and ACTIVIN A/SMAD2,3 signaling is increased (D’Amour et al., 2005; McLean et al., 2007; Singh et al., 2012; Teo et al., 2011). Fucci hESCs were differentiated to DE for 2 or 4 days, and then cell-cycle fractions were isolated by FACS and subjected to transcript analysis by qRT-PCR (Figures 4A and 4B). By day 2 (d2), the DE transcripts GATA6, SOX17, and FOXA2 were significantly elevated in all cell-cycle fractions, but most noticeably in G1 (KO2; Figure 4C). Corresponding increases in cell-cycle-regulated SOX17 and FOXA2 protein were also observed at this time (Figures 4D–4F). The initial stage of DE differentiation is therefore characterized by elevated expression of lineage markers (GATA6, SOX17, and FOXA2) while cell-cycle regulation is retained. BRACHYURY however, does not seem to be under this mode of control and shows no obvious cell-cycle regulation in hESCs (Figures 4C and S2B). By day 4 (d4), the endoderm transcript levels increased further, and although they were higher in G1, they were now also significantly elevated in S and G2/M phases. The transition from d2 to d4 under these conditions was associated with significant remodeling of the cell cycle, where KO2-positive cells increased from <10% to >40% (Figure 4B). To rule out the possibility that late G1 (KO2+) cells are simply a differentiated cell population, we examined the self-renewal and differentiation potential of these cells in greater detail (Figure S4). After sorting late G1 cells and replating them in self-renewal or differentiation media, we found that they reestablished an asynchronous cell-cycle profile within 2 days. From these replated late G1 cells, we sorted the Fucci cell-cycle fractions and analyzed gene-expression markers by qRT-PCR. Importantly, the endoderm markers reestablished their cell-cycle-regulated gene-expression patterns during self-renewal and differentiation. Immunostaining of replated late-G1 cells further confirmed these results and showed clear differences in endoderm marker expression between early- and late-G1 cells. Altogether, our data indicate that although an increased proportion of cells in G1 may contribute to elevated levels of developmental regulators, other factors involved in the amplification mechanism appear to play a role. Nevertheless, cell-cycle-dependent heterogeneity is retained as hESCs differentiate toward DE, indicating that transition through G1 is linked to preferential gene activation.
Figure 4.
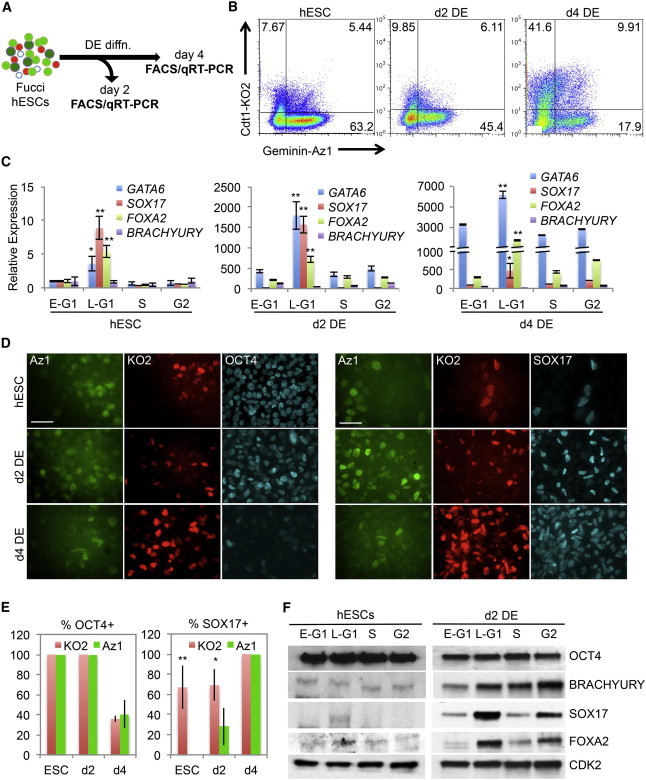
Developmental Regulators Retain Cell-Cycle Regulation during Differentiation
(A) Schematic for examination of DE cell-cycle-regulated transcripts. Fucci hESCs were differentiated to DE for 2 or 4 days, cell-cycle fractions were isolated by FACS, and RNA expression analysis was performed.
(B) Flow-cytometry plots of undifferentiated ESCs, 2-day DE, and 4-day DE.
(C) RNA expression analysis of Fucci cell-cycle fractions after differentiation to DE. All experiments were performed in technical triplicate and are representative of multiple experiments.
(D and E) Immunostaining upon differentiation of Fucci ESCs for 2 or 4 days to DE is quantitated in (E) as the percentage of positive cells in each cell-cycle fraction. Data are representative of multiple independent experiments. Micron bar: 50 μm.
(F) Immunoblotting (20 μg/lane) for the indicated proteins from cell-cycle fractions in undifferentiated ESC or after 2 days of DE differentiation.
Data are representative of biological triplicate experiments displayed as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01. See also Figures S3 and S4.
Epigenetic Changes at Developmentally Regulated Genes in G1 Phase
Although signaling pathways are activated in a cell-cycle-independent manner, cells seem to be responsive only in G1 phase, indicating that uncharacterized mechanisms link signal transduction to target genes required for cell-fate commitment. One mechanism involving signal-regulated transcription factors and cyclin D-associated cyclin-dependent kinase activities was recently described (Pauklin and Vallier, 2013). We hypothesized, however, that factors such as chromatin structure and epigenetic status may also be a corequirement for transcriptional activation by signaling effectors in G1. Because cytosine methylation (5-methylcytosine [5mC]) and 5-hydroxymethylcytosine (5hmC) are epigenetic modifications that are known to be important in modulating transcriptional activity (Szulwach et al., 2011a; Yu et al., 2012b), and little is known about 5mC/5hmC dynamics in the cell cycle, we decided to examine these marks in Fucci cells. To that end, we subjected genomic DNA from the four FACS-isolated Fucci hESC cell-cycle fractions to dot blot analysis and mass spectrometry to evaluate global 5hmC levels. Surprisingly, both analytical approaches revealed a clear upregulation of 5hmC in late G1 followed by a sharp decline during S phase (Figures 5A and 5B). These data indicate that cell-cycle-regulated 5hmC oscillations occur globally during stem cell self-renewal. Furthermore, since 5hmC peaks in late G1 and is almost completely lost in S phase, this indicates that widespread DNA demethylation occurs in S phase.
Figure 5.
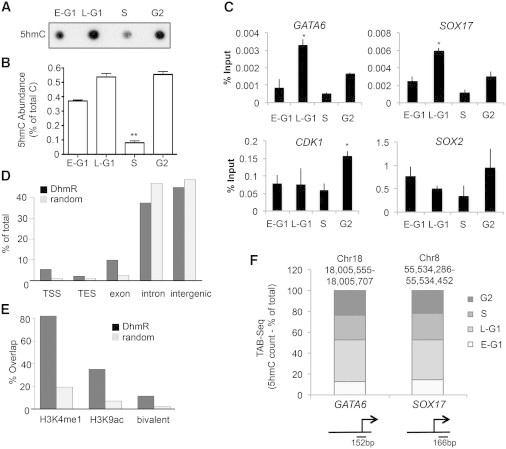
The Epigenetic Mark 5hmC Is Cell-Cycle Regulated
(A) Dot blot analysis reveals a loss of 5hmC in S phase. Representative of assays performed in biological triplicate.
(B) MS analysis for 5hmC performed in technical triplicate and representative of experiments in biological duplicate.
(C) 5hmC-capture qPCR for upstream regions of TSS.
(D) Sequencing of 5hmC-enriched regions reveals enrichment for DhMRs at TSSs and TESs. The experiment was performed in technical triplicate and is representative of duplicate experiments.
(E) DhMRs are colocated with enhancer marks (H3K4me1 and K3K9ac) and bivalent marks (H3K4me3 and H3K27me3).
(F) TAB-seq for promoter regions in Fucci fractions, with diagram indicating primer location.
Data are representative of biological triplicate experiments displayed as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01. See also Figure S5.
To further explore this issue in relation to developmental genes, we performed 5hmC-capture followed by qPCR on genomic regions flanking GATA6, SOX17, CDK1, and SOX2 (Figures 5C and S5). 5hmC was significantly enriched in late G1 (KO2+) cells compared with all other fractions for GATA6 and SOX17, whereas CDK1 showed a slight enrichment in G2/M (AzH+) cells. SOX2, however, did not have any significant enrichment between fractions. Changes in 5hmC at the GATA6 and SOX17 loci therefore parallel those seen in transcriptional activity. To further explore cell-cycle-regulated 5hmC regions globally, we performed 5hmC-capture-seq in biological duplicate. A total of 16,774 differentially 5-hydroxymethylated regions (DhMRs) that changed during the cell cycle were identified (Table S3). Importantly, these 5hmC marks were enriched for regions encompassing transcription start sites (TSSs) and exons (Figure 5D). We further compared how these DhMRs overlapped with epigenetic marks at enhancers (H3K4me1 and H3K9Ac) and bivalent marks (H3K4me3 and H3K27me3) previously identified in hESCs (Figure 5E). Compared with randomly selected regions, the DhMRs were enriched for these epigenetic marks, suggesting that they have broad roles in chromatin organization and links to cell-cycle progression.
To examine 5hmC cell-cycle changes with basepair resolution, we performed Tet-assisted bisulfite sequencing (TAB-seq) (Yu et al., 2012b) at the GATA6 and SOX17 loci (Figure 5F; Table S3) over regions where peaks were identified by 5hmC-capture qPCR (see Figure 5C). Importantly, single-base resolution TAB-seq confirmed our findings from the 5hmC-capture qPCR, i.e., 5hmC was enriched in the KO2 fractions and declined as cells entered S phase. Although the exact function of 5hmC is still unclear, these data demonstrate two important points: (1) epigenetic marks are cell-cycle regulated, and (2) cell-cycle regulation of 5hmC may have important roles in regulating gene expression or chromatin structure.
Discussion
PSCs exist in a dynamic, metastable state characterized by the heterogeneous expression of transcriptional regulators. Although variations in cell signaling contribute to these variations, it was not previously clear how heterogeneity within a cell population was generated. In this report, we show that developmental regulators are transiently expressed by a cell-cycle-dependent mechanism in hESCs and that G1 represents a narrow window of time in which hESCs are responsive to differentiation signals (Figure 6). By contrast, we find no cell-cycle regulation for pluripotency factors in hESCs, indicating that the cell cycle may not regulate the heterogeneity of factors such as NANOG or REX1. We propose that under self-renewing conditions, differentiation signals are not of sufficient magnitude or duration to elevate developmental regulators to a point where their expression can support lineage specification. Instead, low levels of signaling can only transiently activate transcription in G1. It follows, then, that when hESCs are exposed to conditions that efficiently promote differentiation, such as elevated ACTIVIN A and low PI3K signaling, the magnitude and duration of transcriptional activation will be sufficient to activate a sustained developmental program.
Figure 6.
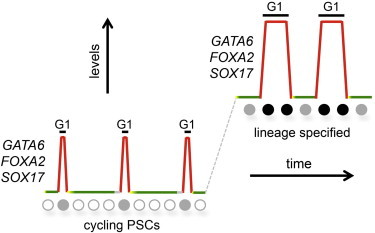
PSC Heterogeneity Is Contingent upon the Cell-Cycle Position
As cells pass through G1 phase, they become sensitized to signaling pathways and exhibit increased levels of developmental regulators (“lineage primed”). During differentiation, the expression of developmentally important transcription factors increases, but they maintain periodicity during the cell cycle. Expansion of the G1 phase is likely to be a factor in this. Colors are representative of Fucci fractions (gray, early G1; red, late G1; yellow, G1/S; green, S-G2/M). Circles depict cells progressing through the cell cycle, with solid gray circles representing cells that weakly express developmental regulators, and solid black circles indicating cells that strongly express developmental regulators.
Why does G1 represent a special window of time that allows for signaling pathways to be sensed and developmental genes to be activated in response? This question is difficult to answer, but one possibility is that changes in chromatin structure in G1 are compatible with the activation of signal-regulated genes (see Dalton 2013; Pauklin and Vallier, 2013). Although this issue requires further attention, in this report we show that one potentially important level of epigenetic modification, 5hmC, is cell-cycle regulated and changes in a cell-cycle-regulated pattern that follows the expression of developmental regulators such as GATA6 and SOX17. The elevated levels of 5hmC combined with increased levels of transcription for developmentally regulated genes suggest that chromatin may be in a permissive state in G1, which allows it to be more responsive to specification cues. This is consistent with the idea that cells initiate programs of differentiation from the G1 phase. Whether 5hmC is a requirement or a consequence of the transcriptional changes seen in G1 will be a subject of continued investigation. Since GATA6 and SOX17 transcripts displayed significant levels of periodicity during the cell cycle, we chose to focus on these genes for further epigenetic analysis. Additional experimentation will be required to determine whether cell-cycle regulation of 5hmC reflects a general occurrence for all cell-cycle-regulated developmental genes.
Another surprising observation described in this report is that, rather than being constitutively expressed with respect to the cell cycle during differentiation, developmental regulators retained a strong expression signature linked to transition through G1. This again supports the idea that G1 represents a temporal window in which cells preferentially respond to signals, allowing them to activate transcription of developmental genes more efficiently. During the first 2 days of DE differentiation, the expression of GATA6 and SOX17 increased >100-fold with respect to hESCs. This preceded the expansion of G1 phase, a characteristic of cell-cycle remodeling during PSC differentiation (Singh and Dalton, 2009; White and Dalton, 2005), and is likely to reflect strong, sustained signaling as a consequence of cells being switched to media optimized for differentiation. The simplest interpretation is that elevated thresholds of SMAD2,3 target gene binding more efficiently and activate developmental genes in G1 phase (Pauklin and Vallier, 2013; Singh et al., 2012). In asynchronous cultures, DE transcripts such as GATA6 and SOX17 begin to increase by d2 and peak by d4. Although significant transcription of these genes already occurs at d2, this is not apparent in typical differentiation experiments because only 10% of cells are in G1.
Between d2 and d4, transcripts continue to increase, and over this period the proportion of cells in G1 increases dramatically, contributing to a spike in DE transcripts. It should not be overlooked that expression of developmental genes increases in all phases during differentiation, indicating that entry and exit from G1 represent a favorable window of time for transcription rather than serving as an on/off switch. In contrast, expression of BRACHYURY is not linked to cell-cycle progression and has a different mode of regulation during differentiation and in hESCs. In addition, we find that some neural genes, such as PAX6 and SOX1, lack cell-cycle regulation in hESCs but impose a cell-cycle-regulated pattern in neural progenitors. Other neural genes, such as OTX2, are only weakly cell-cycle regulated. Although our data show that a large cohort of developmental regulators were cell-cycle regulated, a subset exhibited no obvious periodicity.
In summary, our findings provide a clear explanation for heterogeneity in the expression of developmental regulators in hESCs. Moreover, we show that cells are more responsive to differentiation signals in G1 than in other phases of the cell cycle, and thus this can be considered to be a “lineage-primed” phase in which cells are more susceptible to differentiation signals. This model is consistent with other reports showing that pluripotent cells initiate differentiation from the G1 phase of the cell cycle (Chetty et al., 2013; Jonk et al., 1992; Mummery et al., 1987; Pauklin and Vallier, 2013; Sela et al., 2012).
Experimental Procedures
hESC Culture and Differentiation
WA09 hESCs were grown as previously described in media containing HEREGULIN β1 (10 ng/ml; Peprotech), ACTIVIN A (10 ng/ml; R&D Systems), and human LONGR3 insulin-like growth factor 1 (IGF-1; 200 ng/ml; Sigma; Singh et al., 2012). Human recombinant DKK1 was supplied by R&D Systems and U0126 was supplied by LC Laboratories. WA09 hESCs were differentiated to endoderm by withdrawing HEREGULIN β1 and IGF-1, and increasing the ACTIVIN A concentration to 100 ng/ml. Also, BIO (2 μM; Calbiochem) was used during the first 24 hr. WA09 hESCs were differentiated to all three germ layers by plating in differentiation media consisting of 20% fetal bovine serum (Sigma) in Dulbecco’s modified Eagle’s medium (DMEM)/F-12. Differentiation to neuroectoderm was performed as previously described (Chambers et al., 2009). WA09 Fucci hESCs were plated in 20% KnockOut Serum Replacement (Invitrogen) in DMEM/F-12 (Cellgro) with SB431542 (20 μM; Tocris) and murine Noggin (500 ng/ml; R&D Systems) for 5 days.
RNA-Seq
Human Fucci ESCs were sorted into early-G1 (DN), late-G1 (KO2), S phase (AzL), and G2/M (AzH) fractions by FACS on a MoFlo (Beckman Coulter) in biological triplicate (three independent passages). RNA was extracted using TriZol (Invitrogen) and subjected to sequencing with an Illumina HiSeq instrument (Hudson Alpha). For each biological replicate, ∼50 M reads per sample were obtained by 2 × 50 nucleotide paired-end sequencing. All analyses were performed based on the human reference genome hg19. Alignment of RNA-seq reads against known genomic annotations downloaded from the University of California Santa Cruz site (http://www.genome.ucsc.edu) was performed using Bowtie version 0.12.7 (Langmead et al., 2009) and TopHat version 1.3.3 (Trapnell et al., 2009). Transcript-level expression analysis was performed using Cufflinks version 1.2.1 (Trapnell et al., 2010). Transcripts that showed consistent patterns of change in all three biological replicate experiments were selected for downstream analysis. Transcripts were filtered for Spearman correlation coefficients, with a cutoff of 0.5 between any two of the three replicates.
5hmC Analyses
Dot blot analysis was performed as previously described (Yu et al., 2012b). Briefly, genomic DNA was denatured with NaOH, neutralized with ammonium acetate pH 7.0. and spotted onto a nitrocellulose membrane. Following blocking in 5% nonfat milk, filters were probed with 5hmC antibody (Active Motif). The filters were then probed with horseradish peroxidase-conjugated secondary antibody and detected with enhanced chemiluminescence reagents. Global 5hmC levels were assessed by liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) as previously described (Yu et al., 2012b). The amount of global 5hmC was expressed as the proportion of total cytosine. Briefly, 1 μg of genomic DNA was first denatured by heating at 100°C. Five units of Nuclease P1 (Sigma) was added and the mixture was incubated at 37°C for 2 hr. A 1/10 volume of 1 M ammonium bicarbonate and 0.002 U of venom phosphodiesterase 1 (Sigma) were added to the mixture and the incubation was continued for 2 hr at 37°C. Next, 0.5 U of alkaline phosphatase (Roche) was added and the mixture was incubated for 1 hr at 37°C. Before injection into an Agilent Zorbax 4.6 mm × 50 mm, 3.5 μm particle size -c18 column, the reactions were diluted with water to dilute out the salts and enzymes. LC separation was performed at a flow rate of 220 l/min. Quantification was done using a LC-ESI-MS/MS system in the multiple reaction monitoring (MRM) mode as previously described (Figueroa et al., 2010). 5hmC-capture-seq and TAB-seq were performed exactly as previously described (Szulwach et al., 2011b; Yu et al., 2012a).
qRT-PCR, Immunoblotting, and Immunostaining
All qRT-PCRs were performed using Taqman Assays on Demand (Applied Biosystems) on an iCycler (Bio-Rad). All assays were performed in triplicate, normalized to glyceraldehyde 3-phosphate dehydrogenase, and analyzed using the ΔΔCT method. The data are representative of multiple experiments. Gene expression of newly synthesized transcripts was determined using the Click-iT Nascent RNA Capture Kit (Invitrogen) followed by qRT-PCR with Taqman assays. Fucci hESCs were treated for 1 hr with EU and kept on ice before and after FACS. FACS isolation was performed at 4°C. Immunoblotting and immunostaining were performed as previously described (Singh et al., 2012) with antibodies against NANOG (GTX100863; GeneTex), OCT4 (sc-8628), p107 (sc-318), p130 (sc-317), CYCLIN A (sc-596), CYCLIN B1 (sc-752), CYCLIN E (sc-481), p27KIP1 (sc-527), Mcm (sc-48407), CDK2 (sc-163; Santa Cruz Biotechnology), p-CDK1 (9111S), pS6 (4858S), pAKT1 (4060S), pERK (9101S), pGsk3β (9323S), GSK3β (9315S; Cell Signaling Technology), SOX17 (AF1924; R&D systems), and FOXA2 (07-633; Millipore).
Time-Lapse Microscopy
Fucci hESCs were imaged with an Olympus VivaView fluorescent microscope-incubator at 15 min intervals over a 24 hr period with 200 ms exposure time.
Statistical Analyses
All assays were performed in technical triplicate (unless otherwise indicated) and are representative of biological replicates. All data are displayed as mean ± SEM. Statistical analyses were performed using one-way ANOVA for all assays.
Acknowledgments
We thank Julie Nelson for assistance with FACS and Brittany Matlock and Kevin Weller at Vanderbilt University for their initial characterization of the Fucci hESCs. This work was supported by grants to S.D. from the National Institute of Child Health and Human Development (HD049647) and the National Institute for General Medical Sciences (GM75334).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information
References
- Blauwkamp T.A., Nigam S., Ardehali R., Weissman I.L., Nusse R. Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat Commun. 2012;3:1070. doi: 10.1038/ncomms2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder A., Roth-Albin I., Bhatia S., Pilquil C., Lee J.H., Bhatia M., Levadoux-Martin M., McNicol J., Russell J., Collins T., Draper J.S. Lengthened G1 phase indicates differentiation status in human embryonic stem cells. Stem Cells Dev. 2013;22:279–295. doi: 10.1089/scd.2012.0168. [DOI] [PubMed] [Google Scholar]
- Canham M.A., Sharov A.A., Ko M.S., Brickman J.M. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 2010;8:e1000379. doi: 10.1371/journal.pbio.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud C., Yamanaka Y., Pawson T., Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Chetty S., Pagliuca F.W., Honore C., Kweudjeu A., Rezania A., Melton D.A. A simple tool to improve pluripotent stem cell differentiation. Nat. Methods. 2013;10:553–556. doi: 10.1038/nmeth.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronado D., Godet M., Bourillot P.Y., Tapponnier Y., Bernat A., Petit M., Afanassieff M., Markossian S., Malashicheva A., Iacone R. A short G1 phase is an intrinsic determinant of naïve embryonic stem cell pluripotency. Stem Cell Res. (Amst.) 2013;10:118–131. doi: 10.1016/j.scr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Dalton S. G1 compartmentalization and cell fate coordination. Cell. 2013;155:13–14. doi: 10.1016/j.cell.2013.09.015. [DOI] [PubMed] [Google Scholar]
- D’Amour K.A., Agulnick A.D., Eliazer S., Kelly O.G., Kroon E., Baetge E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Enver T., Pera M., Peterson C., Andrews P.W. Stem cell states, fates, and the rules of attraction. Cell Stem Cell. 2009;4:387–397. doi: 10.1016/j.stem.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Figueroa M.E., Abdel-Wahab O., Lu C., Ward P.S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H.F. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvin-Burgess K.E., Travis E.D., Pierson K.E., Vivian J.L. TGF-β-superfamily signaling regulates embryonic stem cell heterogeneity: self-renewal as a dynamic and regulated equilibrium. Stem Cells. 2013;31:48–58. doi: 10.1002/stem.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T., Stadtfeld M. Heterogeneity of embryonic and adult stem cells. Cell Stem Cell. 2008;3:480–483. doi: 10.1016/j.stem.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Lopes S.M., Tang F., Surani M.A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonk L.J., de Jonge M.E., Kruyt F.A., Mummery C.L., van der Saag P.T., Kruijer W. Aggregation and cell cycle dependent retinoic acid receptor mRNA expression in P19 embryonal carcinoma cells. Mech. Dev. 1992;36:165–172. doi: 10.1016/0925-4773(92)90067-t. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Mizuno H., Imayoshi I., Furusawa C., Shirahige K., Kageyama R. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 2009;23:1870–1875. doi: 10.1101/gad.1823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A., Stunnenberg H.G. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Arias A., Brickman J.M. Gene expression heterogeneities in embryonic stem cell populations: origin and function. Curr. Opin. Cell Biol. 2011;23:650–656. doi: 10.1016/j.ceb.2011.09.007. [DOI] [PubMed] [Google Scholar]
- McLean A.B., D’Amour K.A., Jones K.L., Krishnamoorthy M., Kulik M.J., Reynolds D.M., Sheppard A.M., Liu H., Xu Y., Baetge E.E., Dalton S. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- Mummery C.L., van Rooijen M.A., van den Brink S.E., de Laat S.W. Cell cycle analysis during retinoic acid induced differentiation of a human embryonal carcinoma-derived cell line. Cell Differ. 1987;20:153–160. doi: 10.1016/0045-6039(87)90429-5. [DOI] [PubMed] [Google Scholar]
- Pauklin S., Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price F.D., Yin H., Jones A., van Ijcken W., Grosveld F., Rudnicki M.A. Canonical Wnt signaling induces a primitive endoderm metastable state in mouse embryonic stem cells. Stem Cells. 2013;31:752–764. doi: 10.1002/stem.1321. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Sela Y., Molotski N., Golan S., Itskovitz-Eldor J., Soen Y. Human embryonic stem cells exhibit increased propensity to differentiate during the G1 phase prior to phosphorylation of retinoblastoma protein. Stem Cells. 2012;30:1097–1108. doi: 10.1002/stem.1078. [DOI] [PubMed] [Google Scholar]
- Shah O.J., Ghosh S., Hunter T. Mitotic regulation of ribosomal S6 kinase 1 involves Ser/Thr, Pro phosphorylation of consensus and non-consensus sites by Cdc2. J. Biol. Chem. 2003;278:16433–16442. doi: 10.1074/jbc.M300435200. [DOI] [PubMed] [Google Scholar]
- Singh A.M., Dalton S. The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell. 2009;5:141–149. doi: 10.1016/j.stem.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.M., Hamazaki T., Hankowski K.E., Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- Singh A.M., Reynolds D., Cliff T., Ohtsuka S., Mattheyses A.L., Sun Y., Menendez L., Kulik M., Dalton S. Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell. 2012;10:312–326. doi: 10.1016/j.stem.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach K.E., Li X., Li Y., Song C.X., Han J.W., Kim S., Namburi S., Hermetz K., Kim J.J., Rudd M.K. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach K.E., Li X., Li Y., Song C.X., Wu H., Dai Q., Irier H., Upadhyay A.K., Gearing M., Levey A.I. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo A.K., Arnold S.J., Trotter M.W., Brown S., Ang L.T., Chng Z., Robertson E.J., Dunn N.R., Vallier L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J., Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131–138. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Gomez-Lopez S., Eckardt D., Cook A., Kemler R., Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Hon G.C., Szulwach K.E., Song C.X., Jin P., Ren B., He C. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat. Protoc. 2012;7:2159–2170. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Hon G.C., Szulwach K.E., Song C.X., Zhang L., Kim A., Li X., Dai Q., Shen Y., Park B. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


