Summary
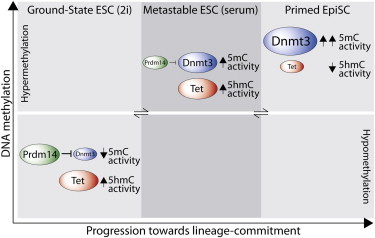
Pluripotent stem cells (PSCs) occupy a spectrum of reversible molecular states ranging from a naive ground-state in 2i, to metastable embryonic stem cells (ESCs) in serum, to lineage-primed epiblast stem cells (EpiSCs). To investigate the role of DNA methylation (5mC) across distinct pluripotent states, we mapped genome-wide 5mC and 5-hydroxymethycytosine (5hmC) in multiple PSCs. Ground-state ESCs exhibit an altered distribution of 5mC and 5hmC at regulatory elements and dramatically lower absolute levels relative to ESCs in serum. By contrast, EpiSCs exhibit increased promoter 5mC coupled with reduced 5hmC, which contributes to their developmental restriction. Switch to 2i triggers rapid onset of both the ground-state gene expression program and global DNA demethylation. Mechanistically, repression of de novo methylases by PRDM14 drives DNA demethylation at slow kinetics, whereas TET1/TET2-mediated 5hmC conversion enhances both the rate and extent of hypomethylation. These processes thus act synergistically during transition to ground-state pluripotency to promote a robust hypomethylated state.
Graphical Abstract
Highlights
-
•
Distinct genome-wide 5mC and 5hmC profiles in diverse pluripotent stem cells
-
•
Poised enhancers and promoters are enriched in 5hmC in ESCs in serum, but not 2i
-
•
Prdm14 overexpression in serum ESCs promotes partial demethylation at slow kinetics
-
•
Mutations in Tet1/Tet2 partially block DNA hypomethylation in ground-state cells
Pluripotent stem cells (PSCs) can give rise to all embryonic lineages. Hackett, Surani, and colleagues analyzed the epigenetic landscape of PSCs in distinct but interchangeable pluripotent “states” and found they are associated with discrete 5mC and 5hmC profiles at regulatory elements and genome wide. Notably, ground-state PSCs are globally hypomethylated via the synergistic effects of PRDM14-dependent repression of Dnmt3 genes and TET-mediated 5hmC conversion.
Introduction
Pluripotent stem cells (PSCs) exhibit the core properties of self-renewal and the capacity to differentiate into all embryonic lineages. These properties are underpinned by expression of a core circuitry of pluripotency transcription factors and reinforced by epigenetic mechanisms (Surani et al., 2007). Mouse embryonic stem cells (ESCs) are classically cultured in serum/leukemia inhibitory factor (LIF) and considered to harbor “naive” pluripotency, which reflects their unrestricted potential to contribute to all somatic lineages and the germline in chimeras (Nichols and Smith, 2009). However, the undefined signaling in serum conditions means that ESCs exhibit heterogeneous gene expression profiles and exist in at least two distinct but interchangeable populations corresponding to a naive and a more developmentally restricted “primed” state and are thus termed metastable (Hayashi et al., 2008; Toyooka et al., 2008).
Recent studies have revealed that inhibition of mitogen-activated protein kinase kinase (MEK) and glycogen synthase kinase-3 (GSK3) by culture in 2i harnesses ESCs in an apparently homogeneous naive “ground-state,” which more closely recapitulates the transcriptional and epigenetic state of the preimplantation epiblast cells from which ESCs are derived (Ying et al., 2008; Marks et al., 2012; Leitch et al., 2013). In contrast, epiblast stem cells (EpiSCs), which are established from the postimplantation epiblast, occupy a pluripotent state “primed” toward lineage differentiation, which is reinforced by culture with FGF and activin (Brons et al., 2007; Tesar et al., 2007). An additional PSC type termed embryonic germ cells (EGCs) can be derived from primordial germ cells (PGCs). Although PGCs are specified from primed postimplantation epiblast, EGCs appear to be transcriptionally and functionally comparable to ESCs and not to EpiSCs, implying that EGCs undergo reprogramming to a more naive state (Matsui et al., 1992; Leitch et al., 2010, 2013). Collectively, these mouse PSCs represent a wide spectrum of pluripotent states that can be used for investigation of developmental processes in vitro. Moreover, understanding the molecular determinants of naive pluripotency in mouse ESCs may yield insights toward derivation and benchmarking of bona fide naive human ESCs (hESCs) (De Los Angeles et al., 2012).
Epigenetic mechanisms including histone modifications and DNA methylation play an important role in stabilizing cell identity and orchestrating many developmental processes. DNA methylation (5mC) is associated with transcriptional silencing and is essential for mouse embryonic development but dispensable for the derivation of ESCs (Okano et al., 1999). Nonetheless, DNA methylation-deficient ESCs are unable to differentiate, indicating that 5mC is required for exit from pluripotency (Jackson et al., 2004). Indeed, de novo promoter methylation occurs relatively early during differentiation—at transition to progenitor cells rather than upon terminal differentiation—which may reflect an early and stable “lock” on lineage commitment (Mohn et al., 2008). Moreover, 5mC at key gatekeeper genes such as Elf5 acts to restrict ESCs to embryonic fates and prevent their contribution to extraembryonic tissues (Ng et al., 2008). Notably, metastable ESCs in serum exhibit relatively high global DNA methylation levels that are comparable with differentiated tissues, whereas naive ground-state ESCs exhibit a globally hypomethylated state (Leitch et al., 2013; Yamaji et al., 2013; Ficz et al., 2013; Habibi et al., 2013). Thus, the genomic landscape of DNA methylation may play an important role in defining the balance between restriction and plasticity in PSCs.
Recent studies have shown that 5mC can be oxidized to 5-hydroxymethycytosine (5hmC) by TET enzymes and that this process may play a role in global DNA demethylation during preimplantation development and in the germline (Tahiliani et al., 2009; Iqbal et al., 2011; Hackett et al., 2013). Conversion of 5mC to 5hmC can potentially engender both replication-dependent “passive” or replication-independent “active” demethylation (Hackett et al., 2012). These pathways could allow turnover or resetting of DNA methylation to promote and maintain pluripotent states. Indeed, both 5hmC and TET enzymes are relatively abundant in PSCs (Ficz et al., 2011; Koh et al., 2011). However, Tet1/Tet2 null ESCs can be efficiently derived and are pluripotent (Dawlaty et al., 2013), and thus, any potential role of 5hmC in promoting dynamic DNA methylation turnover in the context of each pluripotent state remains unclear. Indeed, alternative DNA demethylation mechanisms involving suppression of de novo or maintenance methylation activity have been suggested (Yamaji et al., 2013; Seisenberger et al., 2012), implying that multiple parallel mechanisms may promote DNA demethylation in PSCs (Hackett et al., 2012). Importantly, whereas recent studies have mapped the hypomethylated epigenome of ground-state ESCs at base resolution (Habibi et al., 2013; Ficz et al., 2013), they did not distinguish between 5mC and 5hmC. Moreover, the global profiles of 5mC and 5hmC in the primed state (EpiSC) and in EGCs remain to be determined, as do the precise mechanisms that drive interconversion between distinct DNA methylation landscapes.
Here, we set out to investigate genome-wide 5mC and 5hmC in diverse PSCs and found significant changes associated with distinct pluripotent states. Moreover, we show that dual TET-mediated 5hmC conversion and PRDM14-dependent Dnmt3 repression synergistically drive DNA demethylation during transition to ground-state pluripotency.
Results
Global 5mC and 5hmC in Pluripotent Cells
To investigate the roles and dynamics of 5mC and 5hmC in PSCs derived from distinct developmental origins, or under different culture conditions, we used five genetically matched cell types: male XY and female XX ESCs on feeders in classical serum/LIF media, ground-state ESCs and EGCs under serum-free 2i/LIF conditions (both XY), and primed EpiSCs derived from postimplantation epiblast. We first determined the global levels of 5mC and 5hmC quantitatively by ELISA and found that total 5mC levels were highly comparable among XY ESCs in serum, EpiSCs, and control mouse embryonic fibroblasts (MEFs). However, 5mC levels were reduced slightly in XX ESCs and significantly reduced by 2.2-fold in both ESCs and EGCs maintained in 2i (Figure 1A). The global levels of 5hmC followed a similar pattern, with relatively high levels in ESCs in serum and EpiSCs but reduced levels in cells in 2i. The global reduction of 5hmC in 2i was comparable in extent to 5mC (1.9-fold); however, 5hmC is significantly higher in all PSCs relative to MEFs (Figure 1A). Thus, global 5mC and 5hmC levels are comparable in ESCs in serum and primed EpiSCs but significantly and proportionately reduced in ground-state PSCs cultured in 2i, which is consistent with recent studies (Leitch et al., 2013; Ficz et al., 2013; Habibi et al., 2013).
Figure 1.
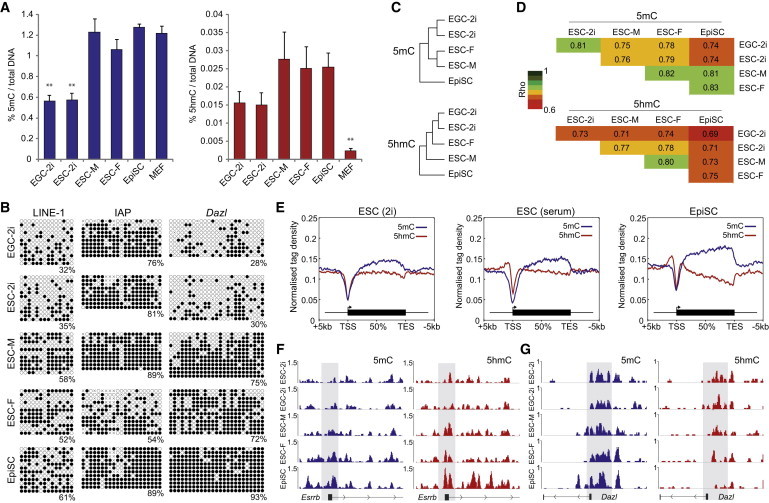
5mC and 5hmC Distributions in Distinct Pluripotent States
(A) Global levels of 5mC (blue) and 5hmC (red) in EGCs in 2i (EGC-2i), ESCs in 2i (ESC-2i), male ESCs in serum (ESC-M), female ESCs in serum (ESC-F), EpiSCs, and MEFs, as a percentage of total nucleotides.
(B) Bisulfite sequencing of repetitive and single-copy loci in distinct PSCs. Open circles represent unmethylated CpGs, and closed circles represent methylated CpGs.
(C) Hierarchical clustering of PSCs based on genome-wide 5mC and 5hmC distribution.
(D) Correlation coefficients of 5mC and 5hmC between PSCs.
(E) Profile shows the distribution of 5mC and 5hmC over an averaged metagene in each pluripotent state.
(F and G) Profiles of 5mC and 5hmC in PSCs at (F) Esrrb and (G) Dazl. Shaded area indicates promoter region.
∗∗p < 0.01 of biological triplicate experiments.
To confirm changes in DNA modification, we used bisulfite sequencing to assay repetitive and single-copy genomic regions. This revealed that LINE-1 elements, CpG-dense promoters at Dazl and Rhox9, CpG-poor promoters at Esrrb and Elf5, an intronic region at Cul1, and exonic sequences of Rex1 are all significantly hypomethylated in ESCs/EGCs in 2i relative to ESCs in serum or EpiSCs, suggesting that hypomethylation occurs at most genomic landmarks in 2i (Figure 1B; Figure S1 available online). A notable exception is IAP elements, which showed no significant difference regardless of the culture condition (Figure 1B), a result consistent with IAPs being resistant to DNA hypomethylation in vivo (Hajkova et al., 2002; Hackett et al., 2013). However, IAPs did exhibit significantly reduced DNA methylation in XX ESCs, which is apparently a specific effect because tested promoter regions and LINE-1 elements were not differentially methylated between our low-passage XX and XY ESCs, and global levels only showed a modest reduction in XX ESCs (Figures 1A and 1B). Interestingly, we observed that XX, but not XY, ESCs exhibited additional global hypomethylation coincident with transition to feeder-free culture, while maintaining XX status, implying that female ESCs may be more susceptible to DNA demethylation as a consequence of in vitro culture parameters than male ESCs (Figure S1). In particular, feeder-free culture may promote global hypomethylation in XX ESCs (Habibi et al., 2013; Zvetkova et al., 2005). Our data suggest that PSCs acquire distinct DNA methylation profiles that reflect their developmental origin and response to culture conditions.
The Distribution of 5mC and 5hmC in Pluripotent Cells
To further investigate the DNA modification landscape in pluripotent cells, we performed genome-wide methylated DNA immunoprecipitation sequencing (meDIP-seq) and hydroxymethylated DNA immunoprecipitation sequencing (hmeDIP-seq). To facilitate comparison with (h)meDIP profiles from low-number PGCs, which share certain attributes of pluripotency and undergo dynamic DNA demethylation, we used a low-input protocol with a high-fragment resolution of ∼150 bp (Hackett et al., 2013). Hierarchical clustering of DNA modification profiles revealed that pluripotent cells cluster into distinct groups that correspond to ground-state, metastable, or primed cells (Figure 1C). Indeed, we observed similar clustering based only on 5mC and 5hmC profiles at binding sites of key chromatin factors (Figure S1). Consistently, the highest correlation coefficients were observed between cells within the same culture condition, regardless of developmental source, such that ESCs and EGCs derived from the ICM and PGCs, respectively, were highly comparable in 2i conditions. In contrast, the greatest difference was seen between ESCs/EGCs in 2i and EpiSCs, which also corresponds to the biggest difference in developmental potential (Figure 1D). Consistent with global hypomethylation under 2i conditions, ESCs in serum were more correlated with EpiSCs than with ESCs in 2i.
Analysis of 5mC enrichment across a metagene revealed a comparable distribution in each pluripotent state, whereby 5mC is enriched within genic regions and strongly depleted from transcriptional start sites (TSSs) (Figure 1E). We observed that 5hmC is also depleted from the TSS but found significant peaks of 5hmC enrichment in the flanking proximal promoter regions in ESCs in serum and EpiSCs, which was absent in ground-state cells (Figure 1E). We did note that 5hmC was enriched at the TSS specifically at low CpG density promoters but only when also bound by TET1 (Figure S1). Notably, some studies previously reported that 5hmC is enriched directly at the TSS in general but used a 300–500 bp hmeDIP resolution rather than the 150 bp used here, which may have enabled some “bridging” of the TSS by fragments that also overlapped flanking promoter/exonic regions enriched in 5hmC (Figure S2). Examination of specific loci such as the Esrrb promoter showed that it is highly enriched with 5hmC in ESCs in serum (XY and XX), as previously reported by Ficz et al. (2011), and that this is lost in 2i, coincident with reduction of 5mC (Figure 1F). In contrast, 5hmC enrichment was maintained at the Dazl promoter in ground-state cells, which may be due to the retention of some 5mC signal under 2i conditions. However, there was a complete loss of 5hmC in primed EpiSCs together with a concomitant increase in 5mC enrichment (Figure 1G).
Differential Promoter Methylation
To investigate the genome-wide changes between PSCs, we examined promoters for differential methylation using XY ESCs in serum as a reference. A previous study has shown that feeder-free XX ESCs are globally hypomethylated (Zvetkova et al., 2005). However, consistent with our observation of only a modest global depletion (∼10%) of 5mC in XX ESCs on feeders, we found only 69 high-confidence (p < 0.01) differential 5mC promoters between XX and XY ESCs. Surprisingly, there were more promoters methylated in XX ESCs (n = 55) relative to XY ESCs (n = 14), albeit there were very few differentially methylated promoters (DMPs) in absolute terms, which suggests that male and female ESCs share a highly similar promoter methylome, at least when cultured on feeders (Figure 2A). Comparison of ESCs in serum with ESCs in 2i revealed 228 high-confidence DMPs, with most being hypomethylated in 2i conditions (n = 179). Gene ontology (GO) analysis indicated that promoters hypomethylated in ground-state 2i relative to serum are not associated with specific processes/pathways after Bonferroni correction, suggesting that promoter hypomethylation is a general rather than a targeted effect in 2i. Similarly, the few promoters that apparently exhibited higher 5mC in 2i were not enriched for GO categories but did apparently include Lin28b, which can promote both self-renewal and differentiation pathways depending on context (Shyh-Chang and Daley, 2013). Relatively elevated DNA methylation at Lin28b is consistent with lower Lin28b expression in 2i (Figure S3) (Marks et al., 2012). Interestingly, rare single-copy loci that escape DNA demethylation in PGCs, such as Sfi1 and Srrm2 (Hackett et al., 2013), are also resistant to demethylation in 2i conditions, although Vmn2r29 is hypomethylated (Figure S3).
Figure 2.
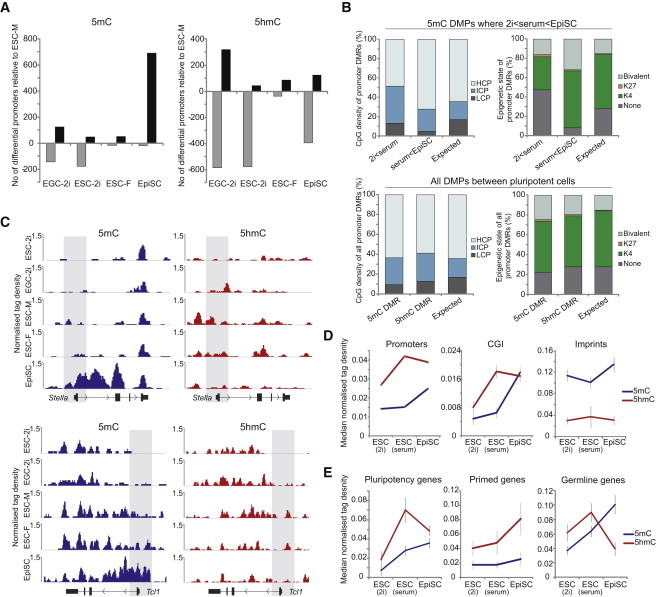
Differentially DNA-Modified Promoters between PSCs
(A) Number of DMPs and DhMPs in PSCs relative to male ESCs in serum. Black bars indicate the number of promoters with higher DNA modification, and gray bars indicate the lower levels in the indicated cell type.
(B) Genetic and epigenetic features of differentially methylated promoters (DMP). Upper panel shows CpG density and chromatin state of DMPs that exhibit gain of 5mC between each successive pluripotent state (2i to serum, serum to EpiSC). Promoters that get methylated between serum ESC and EpiSC states are enriched in being bivalent. Lower panel shows collated CpG density and chromatin state of all promoters that become significantly differentially (hydroxy)methylated between at least one pairwise PSCs comparison.
(C) Profiles of 5mC and 5hmC at Stella and Tcl1 in PSCs. Shaded area indicates promoter region.
(D and E) Enrichment dynamics of 5mC (blue) and 5hmC (red) by (h)meDIP-seq across ground-state ESCs in 2i, metastable ESCs in serum, and primed EpiSCs at (D) distinct genomic landmarks or (E) gene network promoters.
Error bars represent SD of target enrichment(s). CGI, biologically defined CpG island.
Examination of promoter 5mC in EpiSCs revealed a striking number of promoters (n = 668) that become significantly hypermethylated relative to ESCs in serum, with only 19 exhibiting lower 5mC (Figure 2A). Promoters that become methylated in EpiSCs are highly enriched for being bivalently or H3K27me3 marked in ESCs and include many pluripotency and germline-specific genes such as Stella, Rex1, and Tcl1 (Figures 2B and 2C). In addition to acquisition of promoter 5mC, these genes often acquire DNA methylation throughout their entire transcription unit in EpiSCs, which may be related to stable heterochromatinization of these loci (Figure 2C). In general, when collated, the DMPs between all PSCs are enriched for an intermediate CpG density and depleted of CpG-poor promoters (Figure 2B).
Taken together, this analysis suggests that there is a general progressive gain of promoter 5mC coincident with transition from ground state > metastable > primed states. Notably, the most significant acquisition of promoter 5mC occurs in primed EpiSCs, which is in contrast to global 5mC, where significant gain occurs between ground-state ESCs in 2i and ESCs in serum. Thus, in general, promoters in ESCs cultured in serum are resistant to acquiring 5mC despite a high global DNA methylation context. In contrast, a significant number of promoters acquire DNA methylation in EpiSCs, coincident with a constraint on cellular plasticity.
Analysis of differentially hydroxymethylated promoters (DhMPs) showed that both ground-state cells and primed EpiSCs were associated with a significantly reduced number of 5hmC-enriched promoters relative to ESCs in serum (Figure 2A). The observation that the highest relative number of 5hmC-enriched promoters occurs in serum ESCs is in contrast to 5mC, where in general, enrichment of methylated promoters follows a linear pattern that correlates with increasingly restricted cells (ground state > metastable > primed). The enhanced promoter 5hmC in ESCs in serum may contribute to the relative resistance of promoters to acquiring 5mC, despite the high global 5mC levels. Consistent with this, tracking 5mC and 5hmC at all promoters across pluripotent states showed that promoter 5mC enrichment peaks in EpiSCs, whereas 5hmC enrichment peaks in ESCs in serum, with a similar result at biologically defined CpG islands (Figure 2D) (Illingworth et al., 2010). This effect was particularly prominent at pluripotency network genes, which presumably need to remain methylation-free in ESCs to ensure robust expression and therefore may be enriched in 5hmC (Figure 2E). In contrast, “primed” genes continue to acquire 5hmC in EpiSCs and remain depleted of 5mC, which may reflect the requirement to keep these genes competent for expression in EpiSCs. Finally, germline-specific genes become robustly methylated in EpiSCs, which correlates with a dramatic loss of promoter 5hmC and may “lock in” a silenced state (Figure 2E). These data indicate a dynamic interplay between promoter 5mC and 5hmC that may contribute to the maintenance of each pluripotent state.
Imprint Stability in PSCs
We next asked how 2i conditions affect genomic imprints, which are generally resistant to hypomethylation in vivo and only become demethylated in PGCs. We found that enrichment of 5mC at imprints was highly comparable between ESCs in 2i and ESCs in serum, suggesting that they are resistant to the global hypomethylation in ground-state cells (Figure 3A) (Leitch et al., 2013). Moreover, imprints were fully maintained in EpiSCs, with the slight increase in 5mC enrichment possibly a consequence of denser DNA methylation within each methylated allele in EpiSCs rather than an increase in allelic methylation. In contrast, we observed that 5mC was significantly depleted from many imprints in EGCs in 2i, which reflects their origin from PGCs (Figures 3A and 3B). We also found imprints to be almost completely erased in our low-passage female ESCs (Figures 3A and 3B), which we confirmed in two further independent XX ESC lines (Figure 3C). Thus, genomic imprints are resistant to the globally hypomethylated state in 2i conditions, but like EGCs, XX ESCs are highly disposed to imprint erasure. Indeed, EGCs and XX ESCs cluster together according to 5mC enrichment of “imprints,” whereas EpiSCs and XY ESCs in 2i or serum cluster separately (Figure 3D). Because our XX ESCs exhibited no significant loss of promoter 5mC and only a modest reduction in global 5mC, they apparently become hypomethylated preferentially at genomic features that are normally most resistant to DNA demethylation in vivo—IAP and genomic imprints—through an as yet undetermined mechanism. Notably, imprints were highly depleted of 5hmC in all cell types, which contrasts with PGCs, which exhibit highly significant 5hmC enrichment coincident with imprint erasure (Figure 3E) (Hackett et al., 2013). Interestingly, EGCs exhibited the greatest enrichment of 5hmC at imprints despite reduced levels of 5mC substrate, which could be a consequence of their derivation from PGCs (Figure 3A). However, the low absolute levels of 5hmC at genomic imprints in PSCs, in general (Figures 3 and 2D), likely contribute to their relative resistance to demethylation in most contexts, including in the globally hypomethylated state of 2i conditions.
Figure 3.
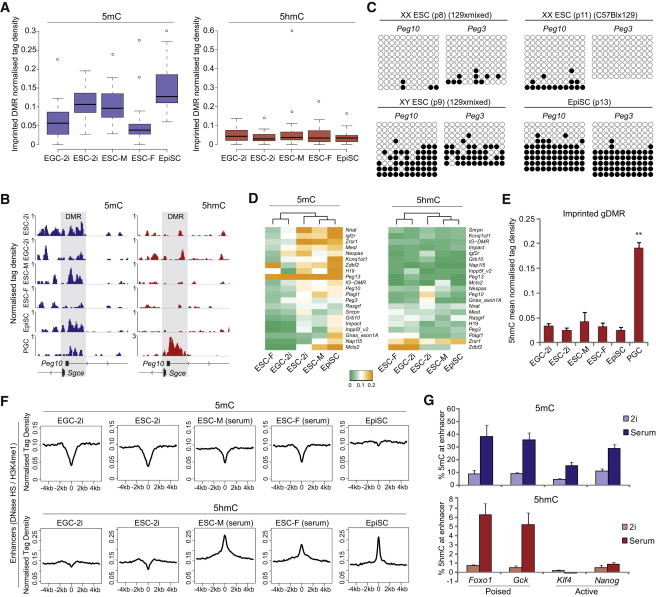
5mC and 5hmC Enrichment at Genomic Imprints and Enhancers
(A) Box plots show enrichment of 5mC and 5hmC over gametic-imprinted DMRs (n = 21) in PSCs.
(B) Distribution of 5mC and 5hmC over the imprinted Peg10 DMR (gray shading) in PSCs and E11.5 PGCs.
(C) Bisulfite sequencing reveals loss of imprints at Peg3 and Peg10 DMRs in independent early-passage XX ESC lines but imprint maintenance in control EpiSCs and XY ESCs.
(D) Clustering of PSCs according to 5mC and 5hmC enrichment at imprinted DMRs. EGCs and XX ESCs (ESC-F) cluster separately from other PSCs due to erasure of imprints.
(E) Average enrichment of 5hmC at all gametic-imprinted DMRs in PSCs and E11.5 PGCs. 5hmC is enriched specifically in PGCs, where imprints are in the process of dynamic erasure.
(F) The profiles of 5mC (upper) and 5hmC (lower) over poised enhancers in EGCs and ESCs in 2i, male and female ESCs in serum, and EpiSCs (G) GluMS-qPCR of quantitative (%) 5mC and 5hmC levels at the Foxo1, Gck, Klf4, and Nanog enhancers in ESCs (XY) cultured in serum or 2i.
Error bars represent SD of biological duplicate experiments. ∗∗p < 0.01.
Differential DNA Modifications at Enhancers
We next tested the enrichment of 5mC and 5hmC at enhancers in PSCs. Poised enhancers, defined by DNase I-hypersensitive sites (DNase HSs) and H3K4me1 enrichment in ESCs, were depleted of 5mC but enriched for 5hmC in ESCs in serum, as expected (Figure 3F) (Yu et al., 2012). In contrast, active enhancers (additionally marked by H3K27ac) exhibited no enrichment for 5hmC and remained depleted of 5mC, suggesting that once activated, enhancers dispense with the 5hmC signature (Figure S3). A similar profile at enhancers was observed for XX ESCs, further underscoring the general similarity between XX and XY ESC methylomes on feeders (excepting imprints and IAP) (Figure 3F). Interestingly, in contrast to 5hmC enrichment in serum conditions, poised enhancers exhibited a significant depletion of 5hmC in both ESCs and EGCs in 2i. Moreover, whereas EpiSCs maintained the 5hmC signature at this enhancer set, it was also associated with acquisition of 5mC (Figure 3F). These data suggest that cells in each pluripotent state may utilize or are premarked for the use of distinct enhancer sets, which may have roles in cellular competence and epigenetic priming (Song et al., 2013; Shen et al., 2013).
To further investigate distal regulatory elements, we used glucosyltransferase methylation-sensitive qPCR (GluMS-qPCR) to examine poised enhancers linked with the Pro-B cell- and liver-specific genes Foxo1 and Gck, respectively, and active enhancers linked to the pluripotency genes Klf4 and Nanog (Whyte et al., 2013). We found 5mC levels to be between 10% and 50% at all these enhancers in ESCs in serum, consistent with them being low-methylated regions (LMRs) that are associated with regulatory elements (Figure 3G) (Stadler et al., 2011). At all enhancers examined, 5mC was reduced 2- to 4-fold in 2i, which is proportionately in line with the global hypomethylation in ground-state cells. Moreover, consistent with hmeDIP-seq, the poised Foxo1 and Gck enhancers were highly enriched for 5hmC (>5%) in ESCs in serum, whereas the active Klf4 and Nanog enhancers were relatively depleted (<1%). However, the enrichment of 5hmC at poised enhancers was dramatically lost in 2i (>10-fold), to a significantly greater extent than global 5hmC depletion (Figure 3G). Thus, 5hmC is apparently a specific marker of poised lineage-specific enhancers in ESCs in serum, but not 2i. The enrichment of 5hmC may be important for resisting the high de novo methyltransferase activity in serum conditions, thereby antagonizing accumulation of repressive DNA methylation at enhancers that need to remain competent for downstream commitment.
Molecular Events during Transition to Ground-State Pluripotency
Because of the dramatic DNA hypomethylation in 2i conditions, we next investigated the kinetics of transition from serum to 2i. We initially shifted ESCs in serum to 2i conditions and tracked gene expression changes. As expected, naive pluripotency markers such as Prdm14, Esrrb, and Stella were upregulated upon transfer to 2i, whereas primed/early differentiation markers including Fgf5, Lefty1, and Bmp4 were strongly downregulated (Figure 4A) (Marks et al., 2012). We also observed that the de novo methylation machinery including Dnmt3a, Dnm3b, and Dnmt3L was rapidly repressed in 2i, whereas maintenance genes such as Dnmt1 and Uhrf1 exhibited no change or were upregulated, respectively. Expression levels of Tet1 were reduced by ∼2-fold in 2i but remained expressed at very high overall levels, whereas Tet2 and Tet3 were not significantly affected, albeit the latter was expressed at very low absolute levels. Notably, changes in gene expression occurred rapidly upon transfer to 2i, with most changes being comparable to ESCs maintained in 2i (more than five passages) within 48 hr (Figure 4A).
Figure 4.
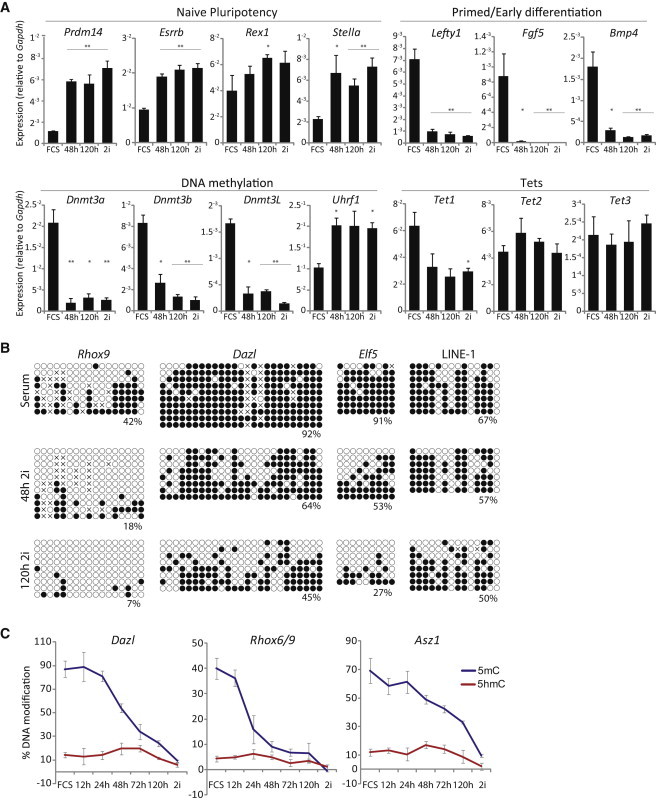
Molecular Dynamics during Transition to Ground-State Pluripotency
(A) Gene expression levels in ESCs in serum (FCS) and at 48 and 120 hr and more than five passages after switch to 2i conditions. Shown are absolute mRNA levels as a proportion of Gapdh.
(B) Bisulfite sequencing shows DNA methylation of ESCs in serum and at 48 and 120 hr after switch to 2i at gene promoters and LINE-1 repeats.
(C) Kinetics of quantitative (%) 5mC and 5hmC levels upon transition of metastable ESCs in serum (FCS) to ground-state pluripotency (2i) assayed by GluMS-qPCR.
∗p < 0.05 and ∗∗p < 0.01, paired Student’s t test. Error bars are SD of independent duplicate experiments performed in technical triplicates.
The rapid transition of a serum ESC gene expression profile to a profile comparable to 2i conditions prompted us to examine the kinetics of DNA methylation changes. We initially used bisulfite sequencing and found that LINE-1 and the promoters at Dazl, Rhox6/9, and Elf5 showed some loss of DNA modification at 48 hr with significant demethylation by 120 hr (Figure 4B). In particular, Rhox6/9 and Elf5, which are representative of relatively CpG-dense and CpG-poor promoters, respectively, were highly hypomethylated after 120 hr in 2i. In contrast, we observed no significant changes at IAP and the imprinted Kcnq1ot1 locus, consistent with our observation that these regions are resistant to demethylation in 2i. We next used GluMS-qPCR to track quantitative changes of 5mC and 5hmC at a high temporal resolution at genes with CCGG motifs. We found that Dazl, Rhox6/9, and Asz1 promoters underwent a slight reduction of 5mC within 24 hr with a progressive loss thereafter. In parallel, the quantitative levels of 5hmC remained at least stable over the first 72 hr, despite the reduction in its obligate 5mC substrate (Figure 4C). Indeed, we observed that 5hmC levels may actually increase modestly at these loci, with a peak between 48 and 72 hr after shift to 2i conditions. Because there is no known 5hmC maintenance mechanism (Ficz et al., 2011) and ESCs transferred to 2i replicate normally, the observation that 5hmC is initially maintained, and possibly enriched, whereas 5mC is lost suggests that 5hmC is being continually targeted to these loci at an increasing rate to account for the decreasing level of the underlying 5mC substrate. Indeed, plotting the ratio of 5hmC:5mC at tested loci demonstrates the relative enrichment of 5hmC at these loci during transition to ground-state pluripotency (Figure S4). These data suggest that 5hmC turnover and retargeting may play a role in mediating demethylation upon transfer to 2i.
Prdm14 Contributes to DNA Demethylation in Ground-State ESCs
To further investigate the mechanism that leads to global hypomethylation in 2i, we thus considered two possibilities, which may operate synergistically. First, that 5hmC is targeted to the genome at an increased rate during transition to ground-state pluripotency, perhaps owing to changes in chromatin accessibility or enhanced TET activity, leading to faster turnover of 5mC and an equilibrium shift toward hypomethylation. Second, the upregulation of Prdm14 observed in 2i (Figure 4A) leads to a rapid repression of the de novo methyltransferase machinery, thereby promoting the passive loss of 5mC owing to inefficient maintenance (Chen et al., 2003). Indeed, we and others have recently reported that Prdm14−/− ESCs exhibit higher steady-state DNA methylation at tested loci than wild-type (Grabole et al., 2013; Yamaji et al., 2013).
To examine whether Prdm14 directly mediates the dynamic DNA demethylation effects of transfer to 2i, we generated ESCs carrying DOX-inducible expression of Prdm14. Induction of Prdm14 (+DOX) in serum resulted in downregulation of primed/early differentiation markers, likely due to repression of FGF signaling (Yamaji et al., 2013; Grabole et al., 2013), and some upregulation of naive pluripotency markers by 48 hr (Figure 5A). Importantly, PRDM14 targets Dnmt3a, Dnmt3b, and Dnmt3L all exhibited significant repression upon DOX treatment, suggesting that the de novo methylation machinery is impaired upon Prdm14 expression, even in serum conditions (Figures 5A and 5B). Nonetheless, GluMS-qPCR revealed that whereas there was some DNA demethylation at Dazl, Asz1, and Rhox6/9 upon Prdm14 overexpression, the rate and extent were significantly lower than seen upon transfer to 2i conditions (compare Figure 5C with Figure 4C); this despite ∼20-fold Prdm14 upregulation compared with ∼6-fold in 2i. This observation was confirmed by bisulfite sequencing (Figure S4). To test whether Prdm14 could promote demethylation over extended time periods, we induced Prdm14 for more than five passages. Under these conditions, we did observe significant demethylation relative to uninduced (−DOX) at tested loci; however, DNA methylation still remained at significantly higher levels than in ESCs shifted to 2i (Figure 5D). This difference between Prdm14 overexpression and 2i may in part reflect the fact that Dnmt3a, Dnmt3b, and Dnmt3L are only partially repressed by Prdm14 overexpression alone but may also be indicative of other Prdm14-independent DNA demethylation pathways being operative during transition to ground-state pluripotency. Because Prdm14−/− ESCs retain DNA methylation in 2i (Leitch et al., 2013; Grabole et al., 2013; Yamaji et al., 2013), but overexpression of PRDM14 does not fully recapitulate global hypomethylation, our results suggest that Prdm14 is necessary but not sufficient to induce a fully hypomethylated epigenome in ground-state pluripotent cells.
Figure 5.
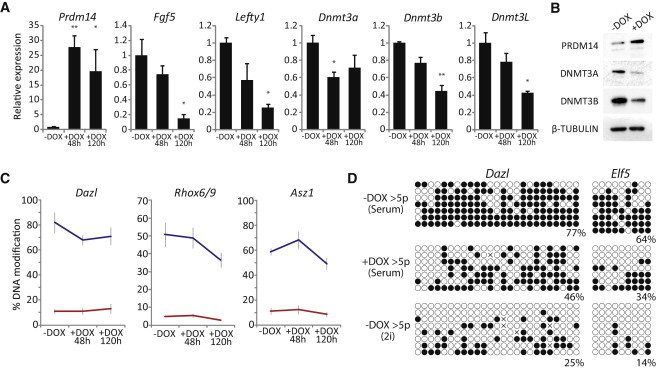
PRDM14 Can Drive Partial DNA Demethylation
(A) Induction of exogenous Prdm14 (+DOX) in ESCs in serum has significant effects of gene expression. Shown are expression levels at 48 and 120 hr relative to uninduced (−DOX) ESCs.
(B) Western blot validates PRDM14 overexpression after DOX treatment and downregulation of DNMT3A and DNMT3B.
(C) Quantitative 5mC and 5hmC levels by GluMS-qPCR during induction of exogenous Prdm14 in ESCs in serum.
(D) Bisulfite sequencing of Dazl and Elf5 from ESCs in serum that have uninduced Prdm14 (−DOX), induced Prdm14 for more than five passages (+DOX), or control uninduced in 2i conditions.
∗p < 0.05 and ∗∗p < 0.01, paired Student’s t test. Error bars represent SD of biological duplicate experiments performed in technical triplicates.
TET-Mediated 5hmC Contributes to Global Hypomethylation
We next turned to the role of 5hmC in promoting DNA demethylation by using Tet1 and Tet2 compound double-knockout (DKO) ESCs (Dawlaty et al., 2013). Expression analysis suggested that upon shift to 2i, naive markers such as Prdm14 and Esrrb showed no significant change in the rate or extent of upregulation in DKO ESCs relative to wild-type (Figure 6A). Moreover, Lefty1 and Fgf5 were both rapidly repressed in control and mutant cells. However, we did observe that methylation-dependent genes such as Dazl and Asz1 were upregulated faster in control cells as compared to DKO, implying an inhibition of DNA demethylation at these loci in DKO ESCs (Figure 6A).
Figure 6.
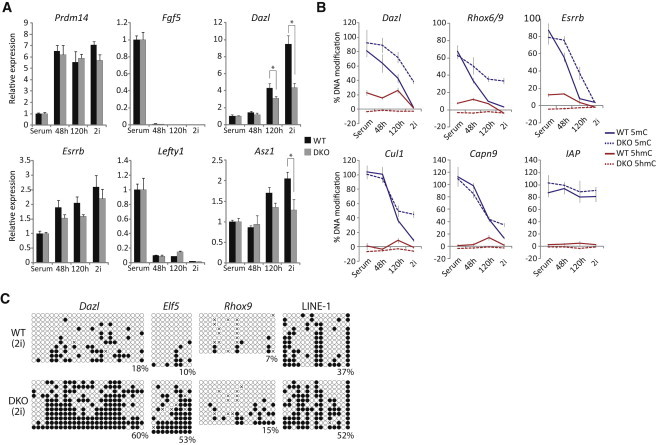
TET-Mediated 5hmC Contributes to DNA Demethylation in 2i
(A) Gene expression dynamics of wild-type and Tet1/Tet2 knockout (DKO) ESCs upon transition to ground-state pluripotency normalized to wild-type and DKO in serum.
(B) Kinetics of 5mC and 5hmC in wild-type and DKO ESCs at 48 and 120 hr and more than five passages after shift from serum to 2i conditions. Shown are CpG-dense promoters (Dazl and Rhox6/9), CpG-poor promoters (Capn9 and Esrrb), introns (Cul1), and repeats (LINE-1).
(C) Bisulfite sequencing of Dazl, Elf5, and Rhox9 promoters and LINE-1 in wild-type and DKO ESCs after five passages in 2i conditions.
∗p < 0.05, Student’s t test. Error bars are SD of independent duplicate experiments performed in technical triplicates.
To further investigate this, we examined the DNA demethylation dynamics in DKO ESCs. In serum conditions, we observed no significant difference in DNA methylation levels between control and DKO ESCs by bisulfite sequencing. However, upon transfer to 2i, the rate of demethylation was impaired specifically in DKO cells (Figure S4). GluMS-qPCR analysis showed that CpG-dense promoters (Dazl and Rhox6/9), CpG-poor promoters (Esrrb), and exons (Cul1) all exhibited a delay in the rate of 5mC depletion in DKO cells, whereas some loci (Capn9) were apparently unaffected (Figure 6B). As expected, 5hmC was undetectable in DKO ESCs at all loci, but a peak of quantitative 5hmC enrichment occurred in wild-type controls at 48 or 120 hr after shift to 2i. The relative delay of 5mC erasure in DKO cells was greater than the reported slight proliferation defect (Dawlaty et al., 2013), suggesting that these results do not only reflect slower DNA replication in DKO ESCs. Indeed, importantly, even after more than five passages in 2i conditions, DKO cells still exhibited significantly elevated levels of 5mC at multiple loci, suggesting that in the absence of TET1 and TET2, ESCs are intrinsically blocked from reaching complete global hypomethylation (Figure 6B). Bisulfite sequencing confirmed that Dazl, Elf5, and Rhox6/9 promoters and LINE-1 repeats are all substantially more methylated in DKO cells relative to wild-type in 2i, albeit these loci have undergone significant demethylation relative to serum conditions (Figure 6C). Collectively, these results support an important role for TET-mediated 5hmC conversion in enhancing complete DNA demethylation in ground-state pluripotency. This likely occurs synergistically with repression of the de novo methyltransferases, in part by PRDM14, to promote a robust hypomethylated state.
Discussion
Our findings document that discrete pluripotent states—naive ground state (2i), metastable (serum), and primed—are each associated with distinct 5mC and 5hmC profiles. Because these states are interchangeable, culture conditions thus dictate the epigenomic landscape of pluripotent cells. Notably, culture in 2i directs a globally hypomethylated state, with our genome-wide analysis indicating that most loci are susceptible to DNA demethylation, as recently suggested by Ficz et al. (2013) and Habibi et al. (2013). Exceptions are imprints and IAPs, which in accordance with their resistance to demethylation during preimplantation reprogramming, remain methylated in 2i. We found that in general, 5mC is acquired during progression from ground-state pluripotency to primed pluripotency. However, whereas the levels of global 5mC dramatically increase between ESCs in 2i and those in serum, promoters are relatively resistant to acquisition of 5mC until progression to the primed state, characterized by EpiSCs. In contrast, 5hmC is particularly enriched at promoters in ESCs cultured in serum, which may function to protect promoter regions from the high global 5mC and de novo methylase activity in this condition, thus maintaining a permissive transcriptional state. Consistently, this observation is particularly prominent at pluripotency-associated genes. Moreover, poised, but not active, enhancers are highly enriched for 5hmC specifically in ESCs in serum, which may function to protect these regions from stable DNA methylation in the absence of functional activation. This could potentially maintain their competence to enable unbiased lineage commitment. Indeed, ESCs depleted of TET proteins exhibit skewed lineage differentiation (Koh et al., 2011; Dawlaty et al., 2013).
The dual presence of high de novo methylase activity and high 5hmC activity in ESCs in serum may represent capture of a unique epigenetic state not present in vivo, whereby the methylome is reflective of a more advanced state of differentiation (hypermethylated) but retains competence for pluripotency through demethylation of key regulatory elements. Such targeted hypomethylation (at some enhancers and promoters) may explain the conundrum that ESCs in serum are associated with high levels of global DNA methylation normally linked with lineage restriction but still exhibit the pluripotent properties of the hypomethylated preimplantation epiblast. In contrast, the striking global hypomethylation in 2i culture parallels the naive epiblast in vivo and supports the paradigm that 2i conditions more faithfully recapitulate the unrestricted state of the inner cell mass from which ESCs are derived.
Notably, ESCs in serum can rapidly initiate DNA demethylation upon shift to 2i. Mechanistically, this is founded upon Prdm14 upregulation driving repression of Dnmt3 genes and TET-mediated conversion to 5hmC. These processes are likely complementary and shift the equilibrium between de novo methylation activity and 5hmC-mediated demethylation toward hypomethylation. Thus, upon transition to 2i, Dnmt3 enzymes are repressed, in part by Prdm14, which suppresses the potential for de novo DNA methylation activity at the same time as 5hmC activity is at least maintained. This promotes greater demethylation both independent of 5hmC, through inefficient maintenance methylation, and via 5hmC, owing to the absence of cyclical remethylation following 5hmC conversion. Indeed, it is possible that TET catalytic activity is actually enhanced in the 2i system either through altered chromatin states, such as reduced H3K27me3/H3K9me3 (Marks et al., 2012), or by a direct effect of culture conditions (Blaschke et al., 2013) because we observed a modest peak of 5hmC at 48–72 hr after transfer to 2i. In contrast, the reciprocal transition toward a primed state is associated with increased de novo activity through upregulation of Dnmt3 genes and parallel downregulation of Tet1 (Figure S4), which shifts the equilibrium toward stable 5mC maintenance.
Knockout of Dnmta3 and Dnmt3b in ESCs leads to progressive hypomethylation, supporting the notion that de novo activity is required to maintain 5mC in ESCs, possibly to offset persistent 5hmC conversion (Chen et al., 2003). However, importantly, Dnmt3a/b null ESCs only exhibited an ∼10%–20% reduction in global 5mC after five passages compared with >2-fold in 2i (Jackson et al., 2004; Chen et al., 2003; Leitch et al., 2013), suggesting that ground-state conditions must activate additional DNA demethylation processes independent of Dnmt3a/b repression. Indeed, Tet1/Tet2 null ESCs used here exhibit a partial block to DNA demethylation in 2i conditions, supporting an important role for 5hmC conversion. Nonetheless, a significant degree of hypomethylation was still observed in DKO ESCs, implying that DNA demethylation is not reliant on 5hmC per se. Thus, a dual model may operate whereby repression of de novo methylation activity is sufficient to drive hypomethylation at slow kinetics, whereas the presence of abundant 5hmC activity may enhance both the rate and the extent of demethylation, particularly at CpG-dense regions that are preferential TET-binding sites, such as the Dazl promoter (Williams et al., 2012). Notably, though, base-resolution data reveal that 5hmC is detectable to some extent throughout the ESC genome (Yu et al., 2012), and thus, 5hmC may play at least a subordinate role in promoting hypomethylation genome wide. Indeed, we observed a partial suppression of demethylation at introns (Cul1) and CpG-poor promoters (Elf5, Essrb, and Capn9) in DKO ESCs in addition to CpG-dense regions (Dazl and Rhox9). This model of synergistic demethylation mechanisms is reminiscent of reprogramming phases in PGCs and the preimplantation embryo, where significant global 5hmC conversion has been reported, and that coincides with periods of suppressed maintenance and/or de novo methylation activity (Hackett et al., 2013; Kagiwada et al., 2013; Iqbal et al., 2011; Pastor et al., 2013).
The functional consequences of global hypomethylation in 2i conditions are difficult to disentangle from the direct effects of ground-state pluripotency. However, one possible outcome is a relaxation on epigenetic barriers imposed by DNA methylation. For example, hypermethylation of the Elf5 promoter has been reported to act as an epigenetic barrier to prevent ESCs in serum from entering into extraembryonic lineages (Ng et al., 2008). Because we observed rapid and dramatic DNA demethylation and activation of Elf5 expression after switching ESCs to 2i (Figure S4), this should enable these ESCs to contribute to extraembryonic tissues and thus be functionally “totipotent.” Indeed, we found that ESCs carrying a reporter for constitutive-Venus expression and cultured in 2i robustly contributed to extraembryonic tissues and that this property occurred soon after the switch in culture conditions (unpublished data), a finding supported by a recent study (Morgani et al., 2013). Thus, global hypomethylation in 2i may contribute to generating a less-restricted in vitro state, which is closer to functional totipotency, possibly through demethylation of Elf5. Additionally, DNA hypomethylation per se appears to promote homogeneity among sister cells after ESC division and may therefore directly promote self-renewal (Jasnos et al., 2013).
In summary, we have established the 5mC and 5hmC profiles of distinct PSCs and revealed that DNA demethylation during transition to ground-state pluripotency is directed by synergistic TET-mediated 5hmC and PRDM14-directed repression of Dnmt3a/Dnmt3b/Dnmt3L. The globally hypomethylated state of cells in 2i may be an in vitro correlate to the preimplantation epiblast or migrating PGCs, and thus, transfer to 2i conditions may provide a tractable system for mechanistic studies into developmental epigenetic transitions.
Experimental Procedures
Cell Lines and Culture Conditions
Animal studies were authorized by a UK Home Office Project License and carried out in a Home Office-designated facility. Cell lines used for (h)meDIP-seq profiling were derived from embryos generated from crossing mixed background Oct4ΔPE-GFP transgenic males with 129SvEv strain females and cultured as follows. For 2i culture, cells were maintained on laminin (10 μg/ml) in N2B27 basal media supplemented with 1 μM PD0325901 (MEK inhibitor), 3 μM CHIR99021 (GSK3 inhibitor), and LIF at 10 μg/ml (prepared in house). For serum conditions, cells were maintained on a MEF feeder layer in DMEM-F12 (GIBCO) supplemented with 15% FCS, 0.1 mM MEM nonessential amino acids, 2 mM L-glutamine, 1 mM sodium pyruvate, 0.1 mM β-mercaptoethanol, and LIF at 10 μg/ml. Cells were passaged by dissociation with trypsin and replating every 2–3 days. Prior to (h)meDIP-seq analysis, cells were purified from feeders by sequential preplating for 2 × 45 min. For primed conditions, EpiSCs were maintained on fibronectin in DMEM-F12 with N2 and B27 supplement (Invitrogen) media with 2 mM L-glutamine, 0.1 mM MEM nonessential amino acids, 0.1 mM β-mercaptoethanol, 50 μg/ml BSA, supplemented with recombinant human activin A (20 ng/ml; PeproTech), and bFGF (12 ng/ml; Invitrogen). EpiSCs were passaged and replated using Accutase (PAA) every 1–2 days. For serum to 2i transfer experiments, feeder-free ESCs were derived on gelatin in GMEM supplemented with 15% FCS, 0.1 mM MEM nonessential amino acids, 2 mM L-glutamine, 1 mM sodium pyruvate, 0,1 mM β-mercaptoethanol, 100 U/ml penicillin/100 μg/ml streptomycin, and LIF at 10 μg/ml and shifted to N2B27 supplemented with 1 μM PD0325901, 3 μM CHIR99021, 1% knockout serum replacement (KSR), and 10 μg/ml LIF on fibronectin. Upon switching to 2i, cells were passaged every 2–3 days by dissociation with TrypLE with no obvious effects on cell viability or the expected proliferation rate.
(h)meDIP-Seq, Bisulfite Sequencing, and GluMS-qPCR
Genome-wide (h)meDIP-seq, bisulfite sequencing, and GluMS-qPCR were performed as described (Hackett et al., 2013). Full experimental procedures are detailed in the Supplemental Experimental Procedures.
Acknowledgments
We thank members of the M.A.S. laboratory for helpful discussions. We thank Professor Rudolf Jaenisch and Dr. Meelad Dawlaty for DKO ESCs. This work was supported by the Wellcome Trust and Human Frontier Science Program.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Jamie A. Hackett, Email: j.hackett@gurdon.cam.ac.uk.
M. Azim Surani, Email: a.surani@gurdon.cam.ac.uk.
Accession Numbers
Sequencing data have been deposited in the Sequence Read Archive under accession number SRA111995.
Supplemental Information
References
- Blaschke K., Ebata K.T., Karimi M.M., Zepeda-Martínez J.A., Goyal P., Mahapatra S., Tam A., Laird D.J., Hirst M., Rao A. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons I.G.M., Smithers L.E., Trotter M.W.B., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A., Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Chen T., Ueda Y., Dodge J.E., Wang Z., Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty M.M., Breiling A., Le T., Raddatz G., Barrasa M.I., Cheng A.W., Gao Q., Powell B.E., Li Z., Xu M. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Angeles A., Loh Y.H., Tesar P.J., Daley G.Q. Accessing naïve human pluripotency. Curr. Opin. Genet. Dev. 2012;22:272–282. doi: 10.1016/j.gde.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G., Branco M.R., Seisenberger S., Santos F., Krueger F., Hore T.A., Marques C.J., Andrews S., Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Ficz G., Hore T.A., Santos F., Lee H.J., Dean W., Arand J., Krueger F., Oxley D., Paul Y.L., Walter J. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabole N., Tischler J., Hackett J.A., Kim S., Tang F., Leitch H.G., Magnúsdóttir E., Surani M.A. Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO Rep. 2013;14:629–637. doi: 10.1038/embor.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E., Brinkman A.B., Arand J., Kroeze L.I., Kerstens H.H., Matarese F., Lepikhov K., Gut M., Brun-Heath I., Hubner N.C. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Hackett J.A., Zylicz J.J., Surani M.A. Parallel mechanisms of epigenetic reprogramming in the germline. Trends Genet. 2012;28:164–174. doi: 10.1016/j.tig.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Hackett J.A., Sengupta R., Zylicz J.J., Murakami K., Lee C., Down T.A., Surani M.A. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P., Erhardt S., Lane N., Haaf T., El-Maarri O., Reik W., Walter J., Surani M.A. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Lopes S.M., Tang F., Surani M.A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth R.S., Gruenewald-Schneider U., Webb S., Kerr A.R.W., James K.D., Turner D.J., Smith C., Harrison D.J., Andrews R., Bird A.P. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001134. e1001134–e1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K., Jin S.G., Pfeifer G.P., Szabó P.E. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M., Krassowska A., Gilbert N., Chevassut T., Forrester L., Ansell J., Ramsahoye B. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol. Cell. Biol. 2004;24:8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasnos L., Aksoy F.B., Hersi H.M., Wantuch S., Sawado T. Identifying division symmetry of mouse embryonic stem cells: negative impact of DNA methyltransferases on symmetric self-renewal. Stem Cell Rep. 2013;1:360–369. doi: 10.1016/j.stemcr.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiwada S., Kurimoto K., Hirota T., Yamaji M., Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J. 2013;32:340–353. doi: 10.1038/emboj.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K.P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., Laiho A., Tahiliani M., Sommer C.A., Mostoslavsky G. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch H.G., Blair K., Mansfield W., Ayetey H., Humphreys P., Nichols J., Surani M.A., Smith A. Embryonic germ cells from mice and rats exhibit properties consistent with a generic pluripotent ground state. Development. 2010;137:2279–2287. doi: 10.1242/dev.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch H.G., McEwen K.R., Turp A., Encheva V., Carroll T., Grabole N., Mansfield W., Nashun B., Knezovich J.G., Smith A. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A., Stunnenberg H.G. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y., Zsebo K., Hogan B.L. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- Mohn F., Weber M., Rebhan M., Roloff T.C., Richter J., Stadler M.B., Bibel M., Schübeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Morgani S.M., Canham M.A., Nichols J., Sharov A.A., Migueles R.P., Ko M.S., Brickman J.M. Totipotent embryonic stem cells arise in ground-state culture conditions. Cell Rep. 2013;3:1945–1957. doi: 10.1016/j.celrep.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R.K., Dean W., Dawson C., Lucifero D., Madeja Z., Reik W., Hemberger M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Pastor W.A., Aravind L., Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S., Andrews S., Krueger F., Arand J., Walter J., Santos F., Popp C., Thienpont B., Dean W., Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Wu H., Diep D., Yamaguchi S., D’Alessio A.C., Fung H.L., Zhang K., Zhang Y. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Daley G.Q. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.X., Szulwach K.E., Dai Q., Fu Y., Mao S.Q., Lin L., Street C., Li Y., Poidevin M., Wu H. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler M.B., Murr R., Burger L., Ivanek R., Lienert F., Schöler A., van Nimwegen E., Wirbelauer C., Oakeley E.J., Gaidatzis D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Surani M.A., Hayashi K., Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K., Christensen J., Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2012;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M., Ueda J., Hayashi K., Ohta H., Yabuta Y., Kurimoto K., Nakato R., Yamada Y., Shirahige K., Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Hon G.C., Szulwach K.E., Song C.X., Zhang L., Kim A., Li X., Dai Q., Shen Y., Park B. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvetkova I., Apedaile A., Ramsahoye B., Mermoud J.E., Crompton L.A., John R., Feil R., Brockdorff N. Global hypomethylation of the genome in XX embryonic stem cells. Nat. Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


