Abstract
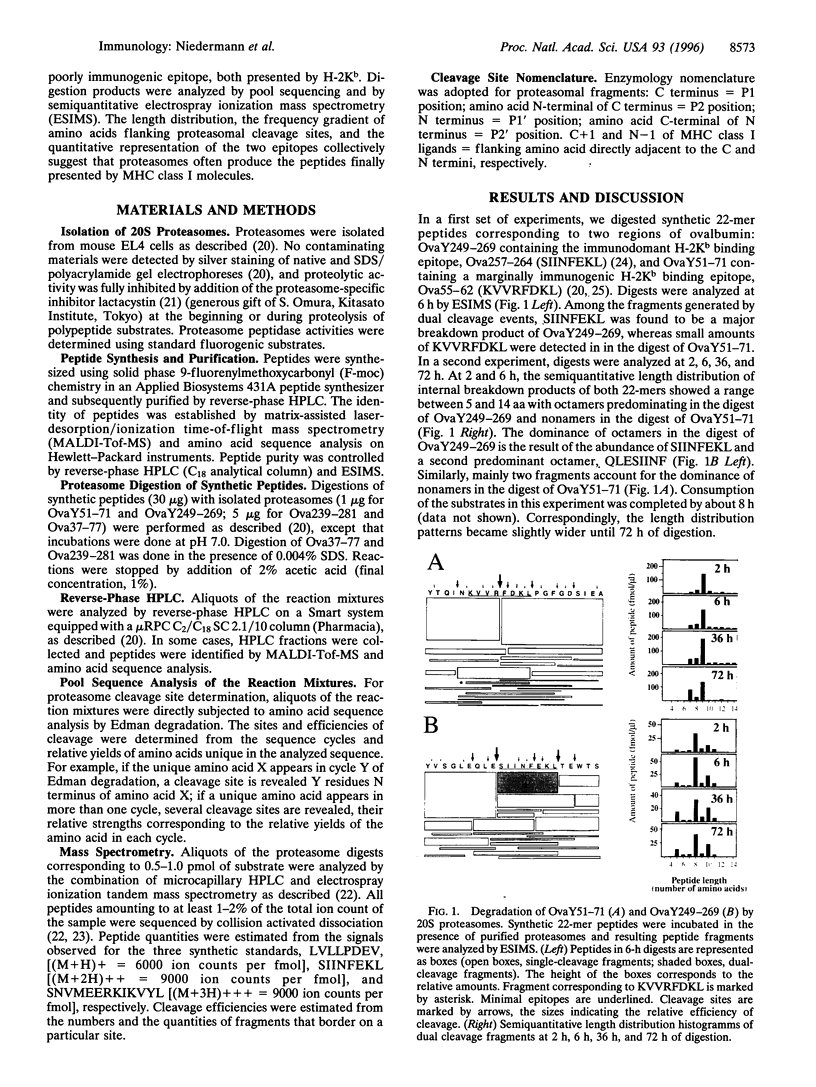
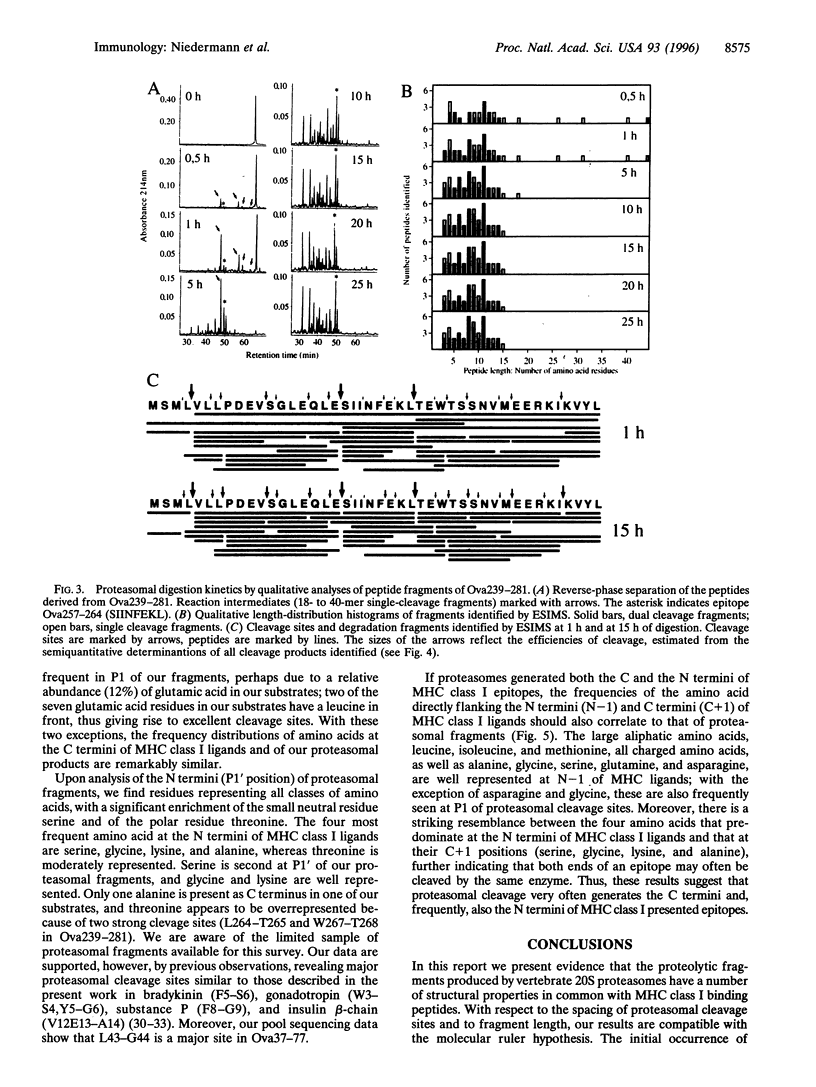
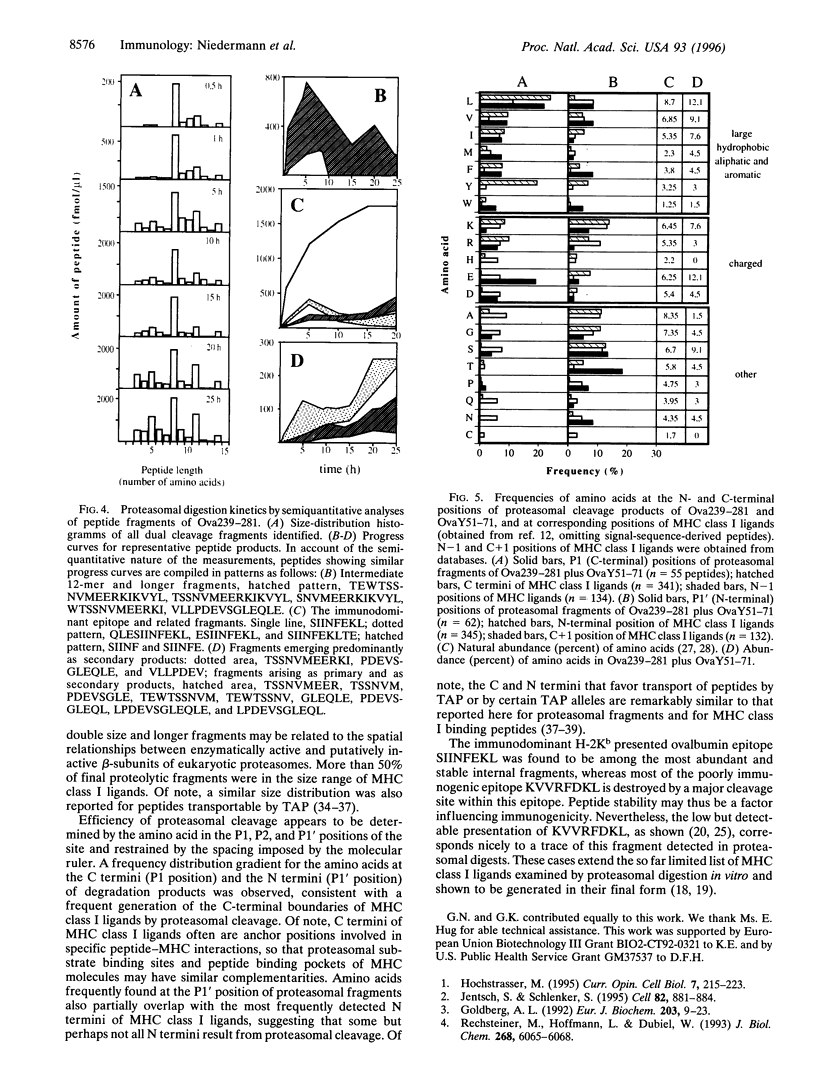
Proteasomes are involved in the proteolytic generation of major histocompatibility complex (MHC) class I epitopes but their exact role has not been elucidated. We used highly purified murine 20S proteasomes for digestion of synthetic 22-mer and 41/44-mer ovalbumin partial sequences encompassing either an immunodominant or a marginally immunogenic epitope. At various times, digests were analyzed by pool sequencing and by semiquantitative electrospray ionization mass spectrometry. Most dual cleavage fragments derived from 22-mer peptides were 7-10 amino acids long, with octa- and nonamers predominating. Digestion of 41/44-mer peptides initially revealed major cleavage sites spaced by two size ranges, 8 or 9 amino acids and 14 or 15 amino acids, followed by further degradation of the latter as well as of larger single cleavage fragments. The final size distribution was slightly broader than that of fragments derived from 22-mer peptides. The majority of peptide bonds were cleaved, albeit with vastly different efficiencies. This resulted in multiple overlapping proteolytic fragments including a limited number of abundant peptides. The immunodominant epitope was generated abundantly whereas only small amounts of the marginally immunogenic epitope were detected. The frequency distributions of amino acids flanking proteasomal cleavage sites are correlated to that reported for corresponding positions of MHC class I binding peptides. The results suggest that proteasomal degradation products may include fragments with structural properties similar to MHC class I binding peptides. Proteasomes may thus be involved in the final stages of proteolytic epitope generation, often without the need for downstream proteolytic events.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boes B., Hengel H., Ruppert T., Multhaup G., Koszinowski U. H., Kloetzel P. M. Interferon gamma stimulation modulates the proteolytic activity and cleavage site preference of 20S mouse proteasomes. J Exp Med. 1994 Mar 1;179(3):901–909. doi: 10.1084/jem.179.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Khilko S., Fecondo J., Margulies D. H., McCluskey J. Determinant selection of major histocompatibility complex class I-restricted antigenic peptides is explained by class I-peptide affinity and is strongly influenced by nondominant anchor residues. J Exp Med. 1994 Oct 1;180(4):1471–1483. doi: 10.1084/jem.180.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick L. R., Aldrich C., Jameson S. C., Moomaw C. R., Pramanik B. C., Doyle C. K., DeMartino G. N., Bevan M. J., Forman J. M., Slaughter C. A. Proteolytic processing of ovalbumin and beta-galactosidase by the proteasome to a yield antigenic peptides. J Immunol. 1994 Apr 15;152(8):3884–3894. [PMC free article] [PubMed] [Google Scholar]
- Dick L. R., Moomaw C. R., DeMartino G. N., Slaughter C. A. Degradation of oxidized insulin B chain by the multiproteinase complex macropain (proteasome). Biochemistry. 1991 Mar 12;30(10):2725–2734. doi: 10.1021/bi00224a022. [DOI] [PubMed] [Google Scholar]
- Ehring B., Meyer T. H., Eckerskorn C., Lottspeich F., Tampé R. Effects of major-histocompatibility-complex-encoded subunits on the peptidase and proteolytic activities of human 20S proteasomes. Cleavage of proteins and antigenic peptides. Eur J Biochem. 1996 Jan 15;235(1-2):404–415. doi: 10.1111/j.1432-1033.1996.00404.x. [DOI] [PubMed] [Google Scholar]
- Engelhard V. H. Structure of peptides associated with MHC class I molecules. Curr Opin Immunol. 1994 Feb;6(1):13–23. doi: 10.1016/0952-7915(94)90028-0. [DOI] [PubMed] [Google Scholar]
- Fenteany G., Standaert R. F., Lane W. S., Choi S., Corey E. J., Schreiber S. L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995 May 5;268(5211):726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L., Rock K. L. Proteolysis, proteasomes and antigen presentation. Nature. 1992 Jun 4;357(6377):375–379. doi: 10.1038/357375a0. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L. The mechanism and functions of ATP-dependent proteases in bacterial and animal cells. Eur J Biochem. 1992 Jan 15;203(1-2):9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- Groettrup M., Ruppert T., Kuehn L., Seeger M., Standera S., Koszinowski U., Kloetzel P. M. The interferon-gamma-inducible 11 S regulator (PA28) and the LMP2/LMP7 subunits govern the peptide production by the 20 S proteasome in vitro. J Biol Chem. 1995 Oct 6;270(40):23808–23815. doi: 10.1074/jbc.270.40.23808. [DOI] [PubMed] [Google Scholar]
- Heemels M. T., Ploegh H. L. Substrate specificity of allelic variants of the TAP peptide transporter. Immunity. 1994 Dec;1(9):775–784. doi: 10.1016/s1074-7613(94)80019-7. [DOI] [PubMed] [Google Scholar]
- Heemels M. T., Ploegh H. Generation, translocation, and presentation of MHC class I-restricted peptides. Annu Rev Biochem. 1995;64:463–491. doi: 10.1146/annurev.bi.64.070195.002335. [DOI] [PubMed] [Google Scholar]
- Heemels M. T., Schumacher T. N., Wonigeit K., Ploegh H. L. Peptide translocation by variants of the transporter associated with antigen processing. Science. 1993 Dec 24;262(5142):2059–2063. doi: 10.1126/science.8266106. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995 Apr;7(2):215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- Howard J. C. Supply and transport of peptides presented by class I MHC molecules. Curr Opin Immunol. 1995 Feb;7(1):69–76. doi: 10.1016/0952-7915(95)80031-x. [DOI] [PubMed] [Google Scholar]
- Hunt D. F., Henderson R. A., Shabanowitz J., Sakaguchi K., Michel H., Sevilir N., Cox A. L., Appella E., Engelhard V. H. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992 Mar 6;255(5049):1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- Hunt D. F., Yates J. R., 3rd, Shabanowitz J., Winston S., Hauer C. R. Protein sequencing by tandem mass spectrometry. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6233–6237. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., Schlenker S. Selective protein degradation: a journey's end within the proteasome. Cell. 1995 Sep 22;82(6):881–884. doi: 10.1016/0092-8674(95)90021-7. [DOI] [PubMed] [Google Scholar]
- Lupas A., Koster A. J., Baumeister W. Structural features of 26S and 20S proteasomes. Enzyme Protein. 1993;47(4-6):252–273. doi: 10.1159/000468684. [DOI] [PubMed] [Google Scholar]
- Löwe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995 Apr 28;268(5210):533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- McDermott J. R., Gibson A. M., Oakley A. E., Biggins J. A. Multicatalytic, high-Mr endopeptidase from postmortem human brain. J Neurochem. 1991 May;56(5):1509–1517. doi: 10.1111/j.1471-4159.1991.tb02045.x. [DOI] [PubMed] [Google Scholar]
- Momburg F., Roelse J., Howard J. C., Butcher G. W., Hämmerling G. J., Neefjes J. J. Selectivity of MHC-encoded peptide transporters from human, mouse and rat. Nature. 1994 Feb 17;367(6464):648–651. doi: 10.1038/367648a0. [DOI] [PubMed] [Google Scholar]
- Momburg F., Roelse J., Hämmerling G. J., Neefjes J. J. Peptide size selection by the major histocompatibility complex-encoded peptide transporter. J Exp Med. 1994 May 1;179(5):1613–1623. doi: 10.1084/jem.179.5.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco J. J. A molecular model of MHC class-I-restricted antigen processing. Immunol Today. 1992 May;13(5):173–179. doi: 10.1016/0167-5699(92)90122-N. [DOI] [PubMed] [Google Scholar]
- Nakashima H., Nishikawa K., Ooi T. The folding type of a protein is relevant to the amino acid composition. J Biochem. 1986 Jan;99(1):153–162. doi: 10.1093/oxfordjournals.jbchem.a135454. [DOI] [PubMed] [Google Scholar]
- Niedermann G., Butz S., Ihlenfeldt H. G., Grimm R., Lucchiari M., Hoschützky H., Jung G., Maier B., Eichmann K. Contribution of proteasome-mediated proteolysis to the hierarchy of epitopes presented by major histocompatibility complex class I molecules. Immunity. 1995 Mar;2(3):289–299. doi: 10.1016/1074-7613(95)90053-5. [DOI] [PubMed] [Google Scholar]
- Orlowski M., Cardozo C., Michaud C. Evidence for the presence of five distinct proteolytic components in the pituitary multicatalytic proteinase complex. Properties of two components cleaving bonds on the carboxyl side of branched chain and small neutral amino acids. Biochemistry. 1993 Feb 16;32(6):1563–1572. doi: 10.1021/bi00057a022. [DOI] [PubMed] [Google Scholar]
- Rammensee H. G., Friede T., Stevanoviíc S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41(4):178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M., Hoffman L., Dubiel W. The multicatalytic and 26 S proteases. J Biol Chem. 1993 Mar 25;268(9):6065–6068. [PubMed] [Google Scholar]
- Rivett A. J. Purification of a liver alkaline protease which degrades oxidatively modified glutamine synthetase. Characterization as a high molecular weight cysteine proteinase. J Biol Chem. 1985 Oct 15;260(23):12600–12606. [PubMed] [Google Scholar]
- Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A. L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994 Sep 9;78(5):761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Rötzschke O., Falk K., Stevanović S., Jung G., Walden P., Rammensee H. G. Exact prediction of a natural T cell epitope. Eur J Immunol. 1991 Nov;21(11):2891–2894. doi: 10.1002/eji.1830211136. [DOI] [PubMed] [Google Scholar]
- Schumacher T. N., Kantesaria D. V., Heemels M. T., Ashton-Rickardt P. G., Shepherd J. C., Fruh K., Yang Y., Peterson P. A., Tonegawa S., Ploegh H. L. Peptide length and sequence specificity of the mouse TAP1/TAP2 translocator. J Exp Med. 1994 Feb 1;179(2):533–540. doi: 10.1084/jem.179.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemüller E., Lupas A., Stock D., Löwe J., Huber R., Baumeister W. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995 Apr 28;268(5210):579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- Wenzel T., Baumeister W. Conformational constraints in protein degradation by the 20S proteasome. Nat Struct Biol. 1995 Mar;2(3):199–204. doi: 10.1038/nsb0395-199. [DOI] [PubMed] [Google Scholar]
- Wenzel T., Eckerskorn C., Lottspeich F., Baumeister W. Existence of a molecular ruler in proteasomes suggested by analysis of degradation products. FEBS Lett. 1994 Aug 1;349(2):205–209. doi: 10.1016/0014-5793(94)00665-2. [DOI] [PubMed] [Google Scholar]
- Wilk S., Orlowski M. Cation-sensitive neutral endopeptidase: isolation and specificity of the bovine pituitary enzyme. J Neurochem. 1980 Nov;35(5):1172–1182. doi: 10.1111/j.1471-4159.1980.tb07873.x. [DOI] [PubMed] [Google Scholar]
- van Endert P. M., Tampé R., Meyer T. H., Tisch R., Bach J. F., McDevitt H. O. A sequential model for peptide binding and transport by the transporters associated with antigen processing. Immunity. 1994 Sep;1(6):491–500. doi: 10.1016/1074-7613(94)90091-4. [DOI] [PubMed] [Google Scholar]