Abstract
Purpose of review
The review aims to update the reader on current developments in our understanding of how the gut microbiota impact on inflammatory bowel disease and the irritable bowel syndrome. It will also consider current efforts to modulate the microbiota for therapeutic effect.
Recent findings
Gene polymorphisms associated with inflammatory bowel disease increasingly suggest that interaction with the microbiota drives pathogenesis. This may be through modulation of the immune response, mucosal permeability or the products of microbial metabolism. Similar findings in irritable bowel syndrome have reinforced the role of gut-specific factors in this ‘functional’ disorder. Metagenomic analysis has identified alterations in pathways and interactions with the ecosystem of the microbiome that may not be recognized by taxonomic description alone, particularly in carbohydrate metabolism. Treatments targeted at the microbial stimulus with antibiotics, probiotics or prebiotics have all progressed in the past year. Studies on the long-term effects of treatment on the microbiome suggest that dietary intervention may be needed for prolonged efficacy.
Summary
The microbiome represents ‘the other genome’, and to appreciate its role in health and disease will be as challenging as with our own genome. Intestinal diseases occur at the front line of our interaction with the microbiome and their future treatment will be shaped as we unravel our relationship with it.
Keywords: carbohydrates, inflammatory bowel disease, irritable bowel syndrome, metagenomics, microbiota
INTRODUCTION
Inflammatory bowel disease (IBD) and irritable bowel syndrome (IBS) both present with diarrhoea, abdominal pain, accelerated transit and altered diet. They share a number of common causative features including genetic predispositions, impaired gut barrier function and altered microbiota, which will be the focus of this review.
In the past 10 years, the revolution in the speed and cost of gene sequencing has made it possible to enumerate and quantify the human microbiota: bacteria and other organisms that live in and around the human body over half of which were previously unculturable and hence unknown. The vast majority of these, estimated at 1014 organisms, live in the colon, and in this review will be referred to as the microbiota.
INFLAMMATORY BOWEL DISEASE
Genetic associations have proliferated in the past few years and a recent meta-analysis of genome-wide association studies reported 163 susceptibility loci, including 71 new associations [1▪▪]. That the majority of these loci, such as NOD2, IRGM, ATG16L1 and IL23R, contribute to pathways for detecting, responding to and processing of microbiota, has emphasized the likely importance of host–microbial interactions in causing disease. Of note, Iliev et al.[2▪▪] have recently reported a single-nucleotide polymorphism (SNP) in the CLEC7A gene associated with treatment-resistant ulcerative colitis. This gene codes for dectin-1, a C-type lectin receptor that recognizes commensal gut fungi [2▪▪]. Mice with a CLEC7A gene deletion compared to wild type developed more severe dextran sodium sulphate (DSS) colitis, with increased tissue invasion, an effect prevented by the antifungal fluconazole.
The earlier findings that lower levels of Faecalibacterium prausnitzii on the ileal mucosa of patients with Crohn's disease was associated with recurrent disease, and that F. prausnitzii could induce interleukin (IL)-10 in vitro with reduced inflammation in mouse models of colitis [3] introduced the concept that specific elements of the microbiota might modify inflammation.
Box 1.
no caption available
Recent studies have confirmed lower levels of F. prausnitzii in adults with both Crohn's disease [4▪] and ulcerative colitis [5], but this is not essential and may be just a marker of an abnormal microbiota. A recent study which looked at new cases of paediatric Crohn's disease actually found increased F. prausnitizii in mucosal biopsies associated with reduced bacterial diversity [6▪]. However, relatives of patients with ulcerative colitis without disease also have reduced F. prausnitizii[5], suggesting that this might relate to genetic factors which in animals clearly influence the microbiota (for review, see [7▪▪]). That this is also true in humans is supported by Li et al.[8▪] who demonstrated lower levels of Clostridium group IV in biopsies of resected ileum from ileal Crohn's disease patients without the NOD2 risk allele though interpretation is difficult owing to confounding by treatments including perioperative antibiotics. Clearer results can be seen in mouse models of ileitis (Toxoplasma gondii infection and indomethacin) in which inflammation leads to marked loss in diversity in ileal mucosal biopsies, with reduction in Firmicutes, replaced by Proteobacteria, including the adherent and invasive Escherichia coli (AIEC) [9▪▪]. NOD2 deletion was associated with reduced numbers of Firmicutes in uninfected mice, and after infection a dramatic shift to 99.8% Proteobacteria and loss of diversity, possibly by impairing antibacterial defences. These effects were somewhat reduced by anti-tumour necrosis factor (TNF) antibody therapy. Smoking, the environmental factor most associated with Crohn's disease, has also been linked to a higher proportions of Bacteroides–Prevotella in faeces [4▪]. Oral microbiota in Crohn's disease patients, but not ulcerative colitis patients, show reduced diversity with reduced Firmicutes and Fusobacteria and an increase in Bacteroidetes [10▪]. A recent paediatric study was able to discriminate patients with IBD from other diagnoses on the basis of microbiota testing alone [11▪▪].
Morgan et al.[12▪▪] inferred the microbiome from the microbiota in a sample of 121 Crohn's disease and 75 ulcerative colitis patients, and 27 healthy controls. Ileal Crohn's disease showed the greatest difference from controls with 12% of the metagenomic pathways analysed significantly affected. IBD patients had lower levels of Ruminococcaceae and Roseburia, metabolically interlinked clades both known to produce short-chain fatty acids (SCFAs) (acetate from Ruminococcaceae utilized by Roseburia to produce butyrate), whereas Faecalibacterium, a major butyrate producer, was again reduced in ileal Crohn's disease, compared to an abundance of Escherichia. 5-Aminosalicylate use was associated with reductions in Escherichia, whereas antibiotic use, greatest in ulcerative colitis, was associated with greatly reduced diversity. Metagenomic profiles showed that IBD microbiomes, particularly in ileal Crohn's disease, had reduced capacity for amino acid biosynthesis and carbohydrate metabolism, with increased presence of enzymes to metabolize mucin, and transport carbohydrate and sulphate. Erickson et al.[13▪] used a multi‘omics’ approach in six twin pairs from the Swedish twin cohort, both concordant and discordant for disease. They confirmed reduced levels of Faecalibacterium, as well as reduced richness of the microbiota in Crohn's disease, most markedly with ileal disease. Analysis of the proteome showed reduced abundance of genes for CAZymes, enzymes capable of degrading complex carbohydrates typical of plant cell walls, though since dietary data were unavailable, it is not clear whether this is due to patient selection of a low fibre diet in an attempt to minimize symptoms [13▪]. The increased presence, however, of Omp proteins, found on gram-negative bacteria, does suggest a shift to a microbiota more able to elicit a proinflammatory response through Toll-like receptor (TLR)-4 signalling.
IRRITABLE BOWEL SYNDROME
Some of the most convincing observational data for an interaction between the microbiome and IBS symptoms comes from studying Campylobacter enteritis which markedly increases gut permeability and leads to IBS in around 10%. Swan et al.[14▪] looked at mucosal gene expression both 6 months after C. jejuni infection and in IBS and identified several common alterations compared to healthy controls, including increased TNFSF15 mRNA. They went on to examine candidate genes whose expression was shown to be abnormal in both patient groups and confirmed a recent report [15] of an increase in the major allele of a TNFSF15 SNP associated with diarrhoea-predominant IBS, and also identified a TNF-α polymorphism associated with postinfectious IBS. A HapMap showed that the TNFSF15 SNPs in the two studies are closely linked so they can be regarded as replications. A meta-analysis of studies preceding this report found an increased incidence of polymorphisms associated with lower levels of IL-10 in IBS, though the small numbers even in the meta-analysis mean these studies need replication [16].
Another similarity between IBS and IBD is the association of both with increased mucosal permeability. IL-22 is recognized to regulate mucosal permeability and its secretion is modulated by IL-23, a recognized risk factor for IBD. IL-22-deficient mice have recently been shown to have altered commensal microbiota with an increased presence of Proteobacteria, and reduced Firmicutes [17▪▪]. They developed more severe colitis in a noninfectious model, and were able to transmit this exaggerated response to co-housed wild-type mice, whose microbiota also included the Proteobacteria seen in the knockouts. IL-22 dysfunction has also been linked to increases in circulating antibodies to enteric pathogens in IBD [18▪]. Several studies show IBS is associated with increased permeability (for review, see [19]). Martinez et al.[20▪▪,21▪▪] examined the underlying mechanism showing structural defects by electron microscopy along with down-regulation of the tight junction protein ZO1 at both gene and protein level in jejunal biopsies from IBS patients with diarrhoea. Abnormalities of faecal microbiota in IBS reported in several series are inconsistent probably because confounders such as diet, smoking, age and antibiotic use have not been adequately controlled for. Better controlled intervention studies are needed to overcome this defect, so the report that faecal microbiota transplant from IBS patients into gnotobiotic mice can induce visceral hypersensitivity is a step in the right direction [22▪▪]. The authors transplanted the microbiota of three IBS patients with constipation (IBS-C) and two healthy controls into germ-free mice. They showed a reduction in bifidobacteria and increase in Enterobacteriaceae and sulphate-reducing bacteria with an increase in caecal sulphide levels and hydrogen production with IBS microbiota compared to controls. This profile was similar to these authors’ previous report in 14 IBS-C patients and 12 healthy controls using selective culture techniques to enumerate bacteria divided into anaerobes, hydrolytic (using a range of substrates including cellulose, xylan, spinach, wheat), H2-consuming and lactate-utilizing bacteria [23]. They showed no difference in hydrolytic bacteria, but a decrease in lactate-utilizing bacteria (mainly Bifidobacteria and Lactobacilli), and 100-fold increase in sulphate-reducing bacteria. Ex-vivo fermentation experiments confirmed the IBS microbiota produced less butyrate but strikingly more sulphide. H2S is now recognized to be an important neurotransmitter causing hypersensitivity to colonic distension and might be a mediator of the microbiota influence. Bile acids can also sensitize the gut to distension. Faecal primary bile acids are increased in both colonic IBD [24▪] and IBS [25▪] compared to healthy controls, possibly due to a reduction in metabolism caused by acceleration of colonic transit or by an abnormal microbiota. Interestingly, secondary bile acids exert an anti-inflammatory effect via the bile acid-specific membrane receptor TGR5, and the authors suggest that this loss of anti-inflammatory effects might be important for inflammation in IBD.
Microbiota may exert an influence on motility via activation of TLRs as shown by the finding that mice deficient in TLR4, a pathogen recognition receptor targeted to lipopolysaccharide (LPS), and its downstream signalling molecule MyD88 both show loss of nitrergic neurones and slower intestinal transit [26▪].
Understanding of the role of microbiota in IBS has been summarized recently [7▪▪], but the number of studies is limited. The largest study to date from Finland [27] found that by using 129 genus-like markers within the 16sRNA gene, they could distinguish the microbiota of healthy individuals from IBS patients. Firmicutes dominated, but the ratio of Firmicutes : Bacteroidetes was higher in IBS who had lower levels of Bifidobacteria and a slight increase in Gammaproteobacteria. There was a negative correlation with overall symptom score for some genus-like groups such as Faecalibacterium and Eubacterium rectale, but positive correlation with Gammaproteobacteria. Jeffery et al.[28▪▪] used a smaller Swedish cohort (37 mixed-type IBS patients compared to 20 healthy individuals). They identified three clusters of IBS patients based on their microbiota, two of which had significant increases in Firmicutes/Bacteroidetes ratio and were distinct from controls, whereas the other 15/37 clustered with the microbiota of healthy individuals. The rates of clinically significant depression were 2/22 (9%) in the first two IBS groups, but 6/15 (40%) in those with normal-like microbiota, suggesting that patients without disordered microbiota may have more social and psychological drivers to their condition [28▪▪]. A recent paediatric study found no differences in phyla between 22 children with IBS and diarrhoea, and 22 healthy controls [29], but with these numbers no definitive conclusions are possible.
MANIPULATING THE MICROBIOTA: ANTIBIOTIC, PROBIOTIC, PREBIOTIC
Two large randomized placebo-controlled trials in IBS patients showed a small therapeutic benefit with rifaximin, a nonabsorbable rifamycin antibiotic [30] [number needed to treat (NNT) = 10], suggesting that a disordered microbiota, or ‘dysbiosis’, might be important, although whether that dysbiosis is present in the small bowel or colon is unclear. A recent study examined the effect of two courses of ciprofloxacin in three individuals. Whereas overall numbers of bacteria fell 1–2 logs, some bacteria were suppressed, whereas others increased. The response was mostly short-lived, but some permanent changes were noted which cautions against the widespread use of antibiotics [31]. Antibiotic treatment leads to an increase in antibiotic resistance phages [32▪] and increased susceptibility to pathogens such as Clostridum difficile.
These complications have encouraged interest in alternative methods of manipulation, one of the most extreme being faecal transplantation. A recent systematic review reported 317 cases of recurrent C. difficile infection treated across 27 case series with a success rate of 92% [33]. This has been confirmed by a recent randomized controlled trial (RCT) of nasojejunal faecal infusion against standard antimicrobial therapy [34▪▪], which backed up impressive clinical outcomes (NNT <2) with data showing the recovery of patients’ microbiotal diversity to levels comparable with faeces donors. Transplantation of a normal microbiome to treat C. difficile infection in an otherwise normal bowel aims to re-establish normal colonization resistance with a persistently altered microbiota. IBD and IBS are both, however, chronic diseases with persistent abnormalities of function which may well alter the transplanted microbiota making its establishment more difficult. A systematic review of the evidence base for faecal transplant in IBD found nine reports and no controlled trials [35], but showed some promising data, though much less convincing than for C. difficile, given the expected placebo response of 13–24% [36].
Devkota et al.[37▪▪] elegantly showed that diets containing saturated milk fat, but not polyunsaturated vegetable oil, increased the percentage of total bile acids that were taurine-conjugated. This dietary change promoted the growth of the pathobiont Bilophila wadsworthia, which induced a colitis in mice deficient in IL-10 [37▪▪]. The interaction between diet and microbiome is illustrated in humans by Smith et al.[38▪] who found that Malawian children who developed kwashiorkor on their usual diet had a disordered microbiota compared to their healthy twins, and that a diet with higher energy availability could change the microbiota, preventing as well as resolving disease. Wu et al.[39] showed that dietary change can alter the microbiome within a day, but that enterotype is more stable and correlates better with long-term dietary habits.
Children in rural Africa have a microbiota very significantly different to that of urban European children [40] related to the substantially greater fibre intake (predominantly the nonabsorbable xylans and cellulose) and reduced dairy and meat intake in Africa. The discovery that SCFAs could regulate the enteroendocrine and immune response via G-protein-coupled receptor 43 (GPR43/FFAR2), and that oral acetate could attenuate DSS colitis which was worse in the absence of commensal microbiota [41], indicated that these microbial metabolic products exert an anti-inflammatory effect. This is mediated via the GPR43 receptor as the attenuation was absent in Gpr43 knockout mice. SCFA are also known to affect enteric neurons, increasing cholinergic neurones and increasing motility [42]. An RCT of 30 days of a poorly digested carbohydrate supplement in obese women showed a significant increase in stool Firmicutes, including F. prausnitzii, with a reduction in Propionibacteria and some Bacteroides[43▪]; however, there was no effect on the numerous metabolic parameters measured. There has been a report of the benefit of an elemental diet in nine patients with pouchitis with nonsignificant increase in selected Firmicutes, but this needs repeating with larger numbers [44]. One case report showed normalization of microbiota in a patient with Crohn's disease treated successfully with an elemental diet of protein and fat, encouraging further larger studies [45].
Patients with IBS often report improvement with diets excluding high-fibre foods, and diets reducing intake of fermentable carbohydrates, labelled low FODMAP diets, have proved popular [46]. There may be a role for restricting FODMAPs after gastroenteritis since mice were less able to clear a pathogen and reconstitute their microbiota when polysaccharides were available than when they were replaced with simple sugars, a manoeuvre which may force commensals to compete with pathogens [47▪▪]. Kashyap et al.[48▪▪], using gnotobiotic mice with and without a humanized microbiome, showed not only that microbiota alter transit but that transit alters the microbiota, making it clear that inferring causation from association will be particularly problematic in this field.
CONCLUSION
The microbiome is potentially readily manipulated for therapeutic benefit. However, translating preclinical progress to bedside intervention will need larger studies than are currently available to provide the needed power, given the large number of variables which alter outcome. Since diet seems to be the overriding factor in the long-term stability of the microbiota, methodologically rigorous evaluation of diet will be important. Both IBD and IBS have uncertain cause and the insights gained from this investigative process will undoubtedly shed new light on their genesis and reveal new approaches to prevention, as well as treatment (Fig. 1).
FIGURE 1.
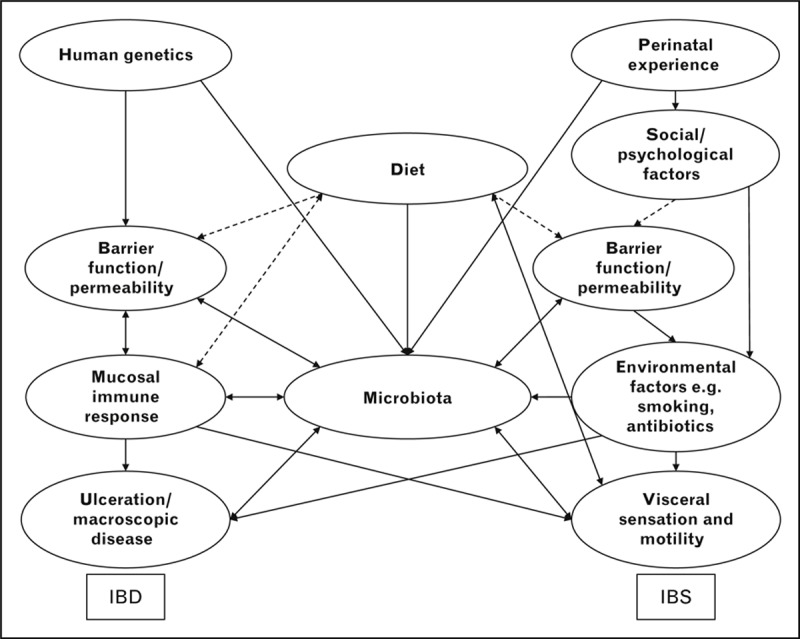
The overlapping factors that influence IBD, IBS and the microbiome. Schematic representation of the factors that contribute to IBD and IBS. Dashed lines represent pathways requiring further evidence. Barrier function and permeability may be the key to pathogenesis in both disorders and be the critical factor in mucosal interaction with the microbiota. Most relationships with the microbiota will be bidirectional, increasing the complexity of understanding its effects. IBD, inflammatory bowel disease; IBS, irritable bowel syndrome.
Acknowledgements
None.
Conflicts of interest
The article presents independent research funded by the National Institute for Health Research (NIHR). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
Footnotes
Correspondence to Professor Robin Spiller, Nottingham Digestive Diseases Centre, University of Nottingham, E Floor West Block, Queen's Medical Centre, Nottingham NG7 2UH, UK. Tel: +44 115 823 1090; e-mail: robin.spiller@nottingham.ac.uk
REFERENCES
- 1▪▪.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012; 491:119–124 [DOI] [PMC free article] [PubMed] [Google Scholar]; Meta-analysis of GWAS studies in IBD with new insights on disease pathogenesis.
- 2▪▪.Iliev ID, Funari VA, Taylor KD, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012; 336:1314–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]; Elegant elucidation of specific polymorphism that may lead to ulcerative colitis through failure to recognize fungi.
- 3.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008; 105:16731–16736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4▪.Benjamin JL, Hedin CR, Koutsoumpas A, et al. Smokers with active Crohn's disease have a clinically relevant dysbiosis of the gastrointestinal microbiota. Inflamm Bowel Dis 2012; 18:1092–1100 [DOI] [PubMed] [Google Scholar]; Observational study of microbiota in smokers and non-smokers, with and without IBD, showing higher Bacteroides–Prevotella group in smokers regardless of disease state. Use of 5-ASA was associated with lower E. coli, but whether any of these associations were causal is unclear.
- 5.Varela E, Manichanh C, Gallart M, et al. Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Aliment Pharmacol Ther 2013; 38:151–161 [DOI] [PubMed] [Google Scholar]
- 6▪.Hansen R, Russell RK, Reiff C, et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am J Gastroenterol 2012; 107:1913–1922 [DOI] [PubMed] [Google Scholar]; Discrepant report on the abundance of F. prausnitzii in Crohn's disease.
- 7▪▪.Simren M, Barbara G, Flint HJ, et al. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut 2013; 62:159–176 [DOI] [PMC free article] [PubMed] [Google Scholar]; Comprehensive review of the role of microbes in functional gastrointestinal disorders with a clinical slant summarizing the many clinical trials aimed at modifying the gut microbiota.
- 8▪.Li E, Hamm CM, Gulati AS, et al. Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS One 2012; 7:e26284. [DOI] [PMC free article] [PubMed] [Google Scholar]; Report of changes in ileal microbiota with comprehensive non-culture characterization demonstrating reduced Clostridia coccoides–E. rectale group in ileal Crohn's disease in those without the NOD2 risk allele.
- 9▪▪.Craven M, Egan CE, Dowd SE, et al. Inflammation drives dysbiosis and bacterial invasion in murine models of ileal Crohn's disease. PLOS One 2012; 7:e41594. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mouse model using Toxoplasma gondii to induce ileal inflammation producing a mucosal microbota dominated by Proteobacteria, particularly the attaching and invasive E. coli (AIEC) seen in Crohn's disease.
- 10▪.Docktor MJ, Paster BJ, Abramowicz S, et al. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm Bowel Dis 2012; 18:935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration of reduced diversity in oral microbiome in children with Crohn's disease but not ulcerative colitis.
- 11▪▪.Papa E, Docktor M, Smillie C, et al. Noninvasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLOS One 2012; 7:e39242. [DOI] [PMC free article] [PubMed] [Google Scholar]; Use of the faecal microbiota to identify paediatric IBD patients with good sensitivity.
- 12▪▪.Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012; 13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]; A large comprehensive study using a metagenomic approach to show important functional changes in IBD microbiome metabolic capabilities not apparent from species profiles. Decreases in SCFA production pathways and increases in cysteine metabolism and other pathways in mucin metabolism suggest a shift towards using mucus as a primary energy source along with adaptation to inflammation associated oxidative stress.
- 13▪.Erickson AR, Cantarel BL, Lamendella R, et al. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn's disease. PLOS One 2012; 7:e49138. [DOI] [PMC free article] [PubMed] [Google Scholar]; Small twin study linking disease to changes in microbiome, metagenome and metabolome confirming reduced diversity and loss of some functions along with increased expression of antigenic bacterial cell surface proteins which may contribute to the host inflammatory response.
- 14▪.Swan C, Duroudier NP, Campbell E, et al. Identifying and testing candidate genetic polymorphisms in the irritable bowel syndrome (IBS): association with TNFSF15 and TNFalpha. Gut 2013; 62:985–994 [DOI] [PubMed] [Google Scholar]; Study of C. jejuni-infected individuals and patients with IBS with both diarrhoea and constipation, demonstrating increased mucosal mRNA for TNFSF15. Susequent genetic analysis confirmed an earlier study [15] of increased major allele of TNFSF15 in IBS similar to the finding in IBD.
- 15.Zucchelli M, Camilleri M, Andreasson A N, et al. Association of TNFSF15 polymorphism with irritable bowel syndrome. Gut 2011; 60:1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bashashati M, Rezaei N, Bashashati H, et al. Cytokine gene polymorphisms are associated with irritable bowel syndrome: a systematic review and meta-analysis. Neurogastroenterol Motil 2012; 24:1102–1566 [DOI] [PubMed] [Google Scholar]
- 17▪▪.Zenewicz LA, Yin X, Wang G, et al. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol 2013; 190:5306–5312 [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration that genetic defects in immune response can alter the microbiota making them colitogenic, a feature which could be transferred to cohoused mice without the immune defect.
- 18▪.Sonnenberg GF, Monticelli LA, Alenghat T, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science 2012; 336:1321–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of bacteria that may proliferate in IL-22 deficiency, with confirmatory findings in patients.
- 19.Camilleri M, Madsen K, Spiller R, et al. Intestinal barrier function in health and gastrointestinal disease. Neurogastroenterol Motil 2012; 24:503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20▪▪.Martinez C, Vicario M, Ramos L, et al. The jejunum of diarrhea-predominant irritable bowel syndrome shows molecular alterations in the tight junction signaling pathway that are associated with mucosal pathobiology and clinical manifestations. Am J Gastroenterol 2012; 107:736–746 [DOI] [PubMed] [Google Scholar]; Companion article to ref. [21]. Detailed mechanistic study of jejunal biopsies in IBS patients with diarrhoea showing abnormal tight junction structure using electron microscopy as well as alteration of tight junction signalling pathways and down-regulation of ZO1 and reduced surface expression of the protein.
- 21▪▪.Martinez C, Lobo B, Pigrau M, et al. Diarrhoea-predominant irritable bowel syndrome: an organic disorder with structural abnormalities in the jejunal epithelial border. Gut 2013; 62:1160–1168 [DOI] [PubMed] [Google Scholar]; Companion article to ref. [20].
- 22▪▪.Crouzet L, Gaultier E, Del’Homme C, et al. The hypersensitivity to colonic distension of IBS patients can be transferred to rats through their fecal microbiota. Neurogastroenterol Motil 2013; 25:e272–e282 [DOI] [PubMed] [Google Scholar]; Elegant animal study showing that transplanting feces from IBS patients can induce visceral hypersensitivity, possibly mediated by increased hydrogen sulphide production.
- 23.Chassard C, Dapoigny M, Scott KP, et al. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment Pharmacol Ther 2012; 35:828–838 [DOI] [PubMed] [Google Scholar]
- 24▪.Duboc H, Rajca S, Rainteau D, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013; 62:531–539 [DOI] [PubMed] [Google Scholar]; Companion article to ref. [25] showing reduced secondary bile acids in IBD associated with impaired deconjugation activities of the microbiota.
- 25▪.Duboc H, Rainteau D, Rajca S, et al. Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil 2012; 24:513–520 [DOI] [PubMed] [Google Scholar]; Companion article to ref. [24]. Small study showing percentage of primary bile acids increased in 14 IBS patients with diarrhoea which correlated with shorter colonic transit and softer stools.
- 26▪.Anitha M, Vijay-Kumar M, Sitaraman SV, et al. Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology 2012; 143:1006–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]; Elegant demonstration using genetically modified mice that LPS acting via TLR-4 or the downstreaming signalling molecule Myd88 have reduced nitrergic neurones and slow gut transit.
- 27.Rajilic-Stojanovic M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 2011; 141:1792–1801 [DOI] [PubMed] [Google Scholar]
- 28▪▪.Jeffery IB, O’Toole PW, Ohman L, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 2012; 61:997–1006 [DOI] [PubMed] [Google Scholar]; Microbiota study showing that IBS patients without abnormal microbiota were more depressed suggesting a subdivision of patients based on microbiota might be valuable.
- 29.Rigsbee L, Agans R, Shankar V, et al. Quantitative profiling of gut microbiota of children with diarrhea-predominant irritable bowel syndrome. Am J Gastroenterol 2012; 107:1740–1751 [DOI] [PubMed] [Google Scholar]
- 30.Pimentel M LA, Chey WD, Zakko S, et al. Rifaximin therapy for patients with irritable bowel syndrome without constipation. N Engl J Med 2011; 364:22–32 [DOI] [PubMed] [Google Scholar]
- 31.Dethlefsen L, Relman D. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. PNAS 2011; 108 Suppl 1:4554–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32▪.Modi SR, Lee HH, Spina CS, Collins JJ. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 2013; 499:219–222 [DOI] [PMC free article] [PubMed] [Google Scholar]; Investigates how antibiotics affects the microbiome and metagenome which may lead to negative outcomes
- 33.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis 2011; 53:994–1002 [DOI] [PubMed] [Google Scholar]
- 34▪▪.van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013; 368:407–415 [DOI] [PubMed] [Google Scholar]; Randomized controlled trial of faecal microbiota transplant with accompanying assessment of the microbiota.
- 35.Anderson JL, Edney RJ, Whelan K. Systematic review: faecal microbiota transplantation in the management of inflammatory bowel disease. Aliment Pharmacol Ther 2012; 36:503–516 [DOI] [PubMed] [Google Scholar]
- 36.D’Haens G, Feagan B, Colombel JF, et al. Challenges to the design, execution, and analysis of randomized controlled trials for inflammatory bowel disease. Gastroenterology 2012; 143:1461–1469 [DOI] [PubMed] [Google Scholar]
- 37▪▪.Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 2012; 487:104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]; Series of experiments showing that, in this mouse model, increased dietary milk fat leads to increased taurocholic acid in bile, promoting growth of Bilophila wadsworthia in the colon which induces inflammation.
- 38▪.Smith MI, Yatsunenko T, Manary MJ, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 2013; 339:548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]; In-depth analysis of the interaction of microbiota and nutrition, and its impact on patient outcomes.
- 39.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011; 334:105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 2010; 107:14691–14696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009; 461:1282–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soret R, Chevalier J, De Coppet P, et al. Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology 2010; 138:1772–1782 [DOI] [PubMed] [Google Scholar]
- 43▪.Dewulf EM, Cani PD, Claus SP, et al. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 2013; 62:1112–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]; How fructans influence the microbiome.
- 44.McLaughlin SD, Culkin A, Cole J, et al. Exclusive elemental diet impacts on the gastrointestinal microbiota and improves symptoms in patients with chronic pouchitis. J Crohn Colitis 2013; 7:460–466 [DOI] [PubMed] [Google Scholar]
- 45.D’Argenio V, Precone V, Casaburi G, et al. An altered gut microbiome profile in a child affected by Crohn's disease normalized after nutritional therapy. Am J Gastroenterol 2013; 108:851–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shepherd SJ, Lomer MC, Gibson PR. Short-chain carbohydrates and functional gastrointestinal disorders. Am J Gastroenterol 2013; 108:707–717 [DOI] [PubMed] [Google Scholar]
- 47▪▪.Kamada N, Kim YG, Sham HP, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 2012; 336:1325–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]; Describes how the microbiome recovers from and expels a pathogen, and the role of dietary carbohydrate.
- 48▪▪.Kashyap PC, Marcobal A, Ursell LK, et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology 2013; 144:967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]; Explores the interactions between gut transit, microbiota and dietary carbohydrates.


