A decrease in Arp2/3 levels results in an NF-κB–dependent increase in the expression of several secreted factors, resulting in nonautonomous effects on chemotaxis.
Abstract
Arp2/3-branched actin is critical for cytoskeletal dynamics and cell migration. However, perturbations and diseases affecting this network have phenotypes that cannot be fully explained by cell-autonomous effects. In this paper, we report nonautonomous effects of Arp2/3 depletion. We show that, upon Arp2/3 depletion, the expression of numerous genes encoding secreted factors, including chemokines, growth factors, and matrix metalloproteases, was increased, a signature resembling the senescence-associated secretory phenotype. These factors affected epidermal growth factor chemotaxis in a nonautonomous way, resolving the recent contradictions about the role of Arp2/3 in chemotaxis. We demonstrate that these genes were activated by nuclear factor κB via a CCM2–MEKK3 pathway that has been implicated in hyperosmotic stress signaling. Consistent with this, Arp2/3-depleted cells showed misregulation of volume control and reduced actin in the submembranous cortex. The defects in osmotic signaling in the Arp2/3-depleted cells can be rescued by hypoosmotic treatment. Thus, perturbations of Arp2/3 have nonautonomous effects that should be considered when evaluating experimental manipulations and diseases affecting the Arp2/3-actin cytoskeleton.
Introduction
The seven-subunit Arp2/3 complex is critical for generating a unique branched actin network underneath the plasma membrane. Arp2/3 complex binds to existing actin filaments and initiates actin daughter filaments as branches off of the mother filaments, creating new actin branches with an angle of ∼70° (Pollard, 2007). The branched actin network generated by Arp2/3 complex is controlled by multiple signaling pathways and is regulated by multiple actin binding proteins (Pollard, 2007; Rotty et al., 2013).
Recent progress has been made in understanding the cellular function of Arp2/3. Using Arp2/3-deficient mammalian cells, two groups showed that Arp2/3-branched actin is essential for forming lamellipodia and maintaining random migration speed (Suraneni et al., 2012; Wu et al., 2012). Because leading edge protrusions have been implicated in directed migration, the effects of Arp2/3 depletion have also been examined in the context of haptotaxis and chemotaxis. With a stable fibroblast cell line depleted of two subunits of the Arp2/3 complex (p34Arc and Arp2, referred to as 2× knockdown [KD] cells throughout), we showed that the branched actin network is essential for sensing and/or responding to changes in extracellular matrix concentration (haptotaxis). Divergent results were reported concerning the role of Arp2/3 in chemotaxis. Using microfluidic devices allowing media exchange, we showed that Arp2/3 complex was not essential for fibroblast chemotaxis up PDGF gradients, suggesting important differences in the molecular machinery of chemotaxis versus haptotaxis (Wu et al., 2012). However, Suraneni et al. (2012) reported that Arp2/3 was required for EGF chemotaxis. Thus, the role of Arp2/3-branched actin in sensing and responding to a soluble gradient remains unresolved.
Cellular senescence is characterized by a state of permanent growth arrest via the up-regulation of p16INK4a and ARF, two linked tumor suppressors encoded by the INK4a/ARF locus (Sharpless, 2004). The surprising viability of 2×KD cells was caused, in part, by the genetic background effects of the loss of Ink4a/Arf tumor suppressors (Wu et al., 2012), suggesting that the loss of Arp2/3 may induce senescence in an Ink4a/Arf-dependent manner. Senescent cells also display altered expression of certain proteins, including the transcription up-regulation of multiple proinflammatory secreted factors, a response known as the senescence-associated secretory phenotype (SASP; Campisi and d’Adda di Fagagna, 2007; Salminen et al., 2012). Emerging data indicate that this SASP response causes nonautonomous effects on disease states such as cancer (Salminen et al., 2012; Lujambio et al., 2013). The nuclear factor κB (NF-κB) and p38 MAPK pathways have been shown to play important roles in regulating SASP (Coppé et al., 2008; Freund et al., 2011; Salminen et al., 2012; Tchkonia et al., 2013).
In the present study, we compared the global transcriptional profiles of cells with and without the Arp2/3 complex and observed an induction of a SASP gene expression response upon Arp2/3 depletion. We also demonstrate that the secreted factors released by Arp2/3-depleted cells affect EGF chemotaxis in a nonautonomous way. Our results resolve the conflicting observations about the role of Arp2/3 in chemotaxis and suggest that experimental manipulations affecting the Arp2/3-branched actin may have both autonomous effects on the cytoskeleton and potential nonautonomous effects, such as confounding inflammatory responses.
Results and discussion
Depletion of Arp2/3 complex induces expression of SASP genes
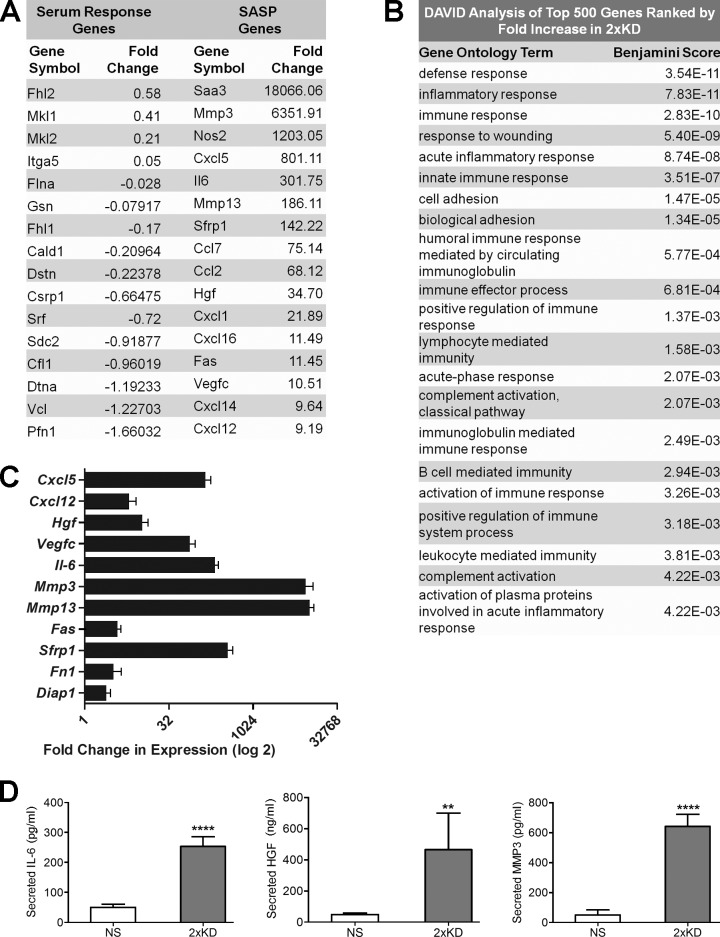
To further understand the role of Arp2/3-branched actin on overall cellular physiology, we performed whole transcriptome RNA-Seq–based expression profiling of the stable Arp2/3-depleted cells we established previously (2×KD cells; Wu et al., 2012). There was no significant pattern of altered expression in genes of the serum response factor pathway that had previously been linked to changes in F-actin content (Posern and Treisman, 2006; Olson and Nordheim, 2010). However, we detected an unexpected increase in the expression of many genes encoding secreted proteins such as chemokines, growth factors and matrix metalloproteinases, a pattern very similar to the SASP gene expression signature (Fig. 1 A and Table S1; Coppé et al., 2008; Kuilman et al., 2008; Salminen et al., 2012). To analyze the altered genes in an unbiased manner, we conducted DAVID (Database for Annotation, Visualization, and Integrated Discovery) analysis of the top 500 genes increased in the 2×KD cells (Huang et al., 2009). Based on this analysis, the list of up-regulated genes contains an overrepresentation of immune and inflammatory genes (Fig. 1 B). The RNA-Seq results were confirmed for a subset of the genes at the transcript level by quantitative RT-PCR (qRT-PCR; Fig. 1 C) and at the protein level by ELISA (Fig. 1 D).
Figure 1.
Arp2/3-depleted cells show increased expression of SASP pathway genes. (A) The transcriptomes of IA32 control fibroblasts and their Arp2/3-depleted derivatives (2×KD) were analyzed by RNA-Seq. Representative genes of the serum response factor and SASP pathways selected from the RNA-Seq data (Table S1) are shown. Fold change represents the fold increase in expression (RPKM) for 2×KD cells from control cells, with negative numbers representing the fold decrease in expression. (B) DAVID analysis of the top 500 genes (>10-fold up-regulated) with the highest fold increase in 2×KD cells, showing overrepresented gene groups as ranked by Benjamini score. (C) qRT-PCR showing fold changes of a subset of cytokines, growth factors, and matrix metalloproteinases up-regulated in 2×KD cells in the RNA-Seq. Two biological replicates (three technical replicates each) for each sample are shown. (D) ELISA showing levels of secreted IL-6, MMP3, and HGF in NS and 2×KD cells. Error bars show 95% confidence interval (CI). ****, P < 0.001; **, P < 0.01 by Student’s t test.
Secreted factors released by Arp2/3-depleted cells cause nonautonomous effects on EGF signaling and chemotaxis
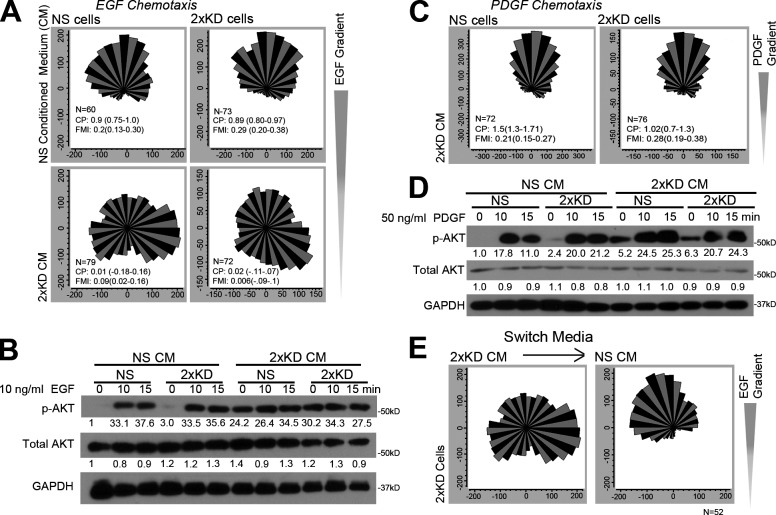
One functional consequence of the change in secreted proteins upon Arp2/3 depletion is that signaling/motility processes, such as chemotaxis, may be affected in a nonautonomous manner depending on the experimental regimen. Given that Arp2/3-deficient cells secrete increased levels of growth factors, such as hepatocyte growth factor (HGF), we reasoned the apparent discrepancy in the need for Arp2/3 in fibroblast chemotaxis could be caused by differences in media exchange in the respective chemotactic chamber designs (closed [Suraneni et al., 2012] vs. continuous flow [Wu et al., 2012]). To mimic the closed chamber design, we used conditioned media (CM) in our continuous flow chambers. CM from the Arp2/3-depleted cells (2×KD) blocked EGF chemotaxis in both control (nonspecific [NS]) and 2×KD cells, but NS CM did not affect this process in either cell line (Fig. 2 A). To test whether CM affected EGF signaling, we compared AKT activation (p-AKT) levels of NS and 2×KD cells after EGF stimulation in the different media. In the presence of 2×KD CM, both cell types had a high basal level of AKT activation that could not be further stimulated by EGF (Fig. 2 B). Together, these results indicate that 2×KD CM disrupts normal EGF signaling sensitivity and perturbs the ability of fibroblasts to migrate up an EGF gradient. Interestingly, 2×KD CM did not show the same effect on PDGF chemotaxis or signaling (Fig. 2, C and D), suggesting that this pathway is relatively insensitive to the factors contained in the 2×KD CM. To establish that the effects of 2×KD CM are reversible, we replaced 2×KD CM with control CM midway through an EGF experiment and observed a restored chemotactic response upon the shift (Fig. 2 E).
Figure 2.
Secreted factors from Arp2/3-depleted cells cause nonautonomous effects on chemotaxis. (A) Wind rose plots showing NS and 2×KD cells in NS CM or 2×KD CM in EGF chemotaxis. (B) Western blots showing phospho-AKT (p-AKT), total AKT, and GAPDH levels in NS and 2×KD cells upon EGF stimulation at the indicated time points. Representative blot from three experiments is shown. Fold changes in band intensity averaged across the three experiments (normalized to loading control and experimental control) are shown below each band. (C) Wind rose plots showing NS and 2×KD cells in NS CM or 2×KD CM during PDGF chemotaxis. (D) Blot showing p-AKT, total AKT, and GAPDH levels in NS and 2×KD cells upon EGF stimulation at the indicated time points. (E) Wind rose plots showing 2×KD cells in EGF chemotaxis, before (right) and after (left) switching from 2×KD CM to NS CM. 2×KD cells were plated in a chemotaxis chamber and serum starved overnight before carrying out the EGF chemotaxis experiment in the presence of 2×KD CM. After 4 h, 2×KD CM was switched to NS CM, and EGF chemotaxis was continued for another 4 h. Compass parameter and forward migration index are indicated on the graphs in A, C, and E; numbers in parentheses show 95% CI.
Depletion or inhibition of Arp2/3 leads to NF-κB activation
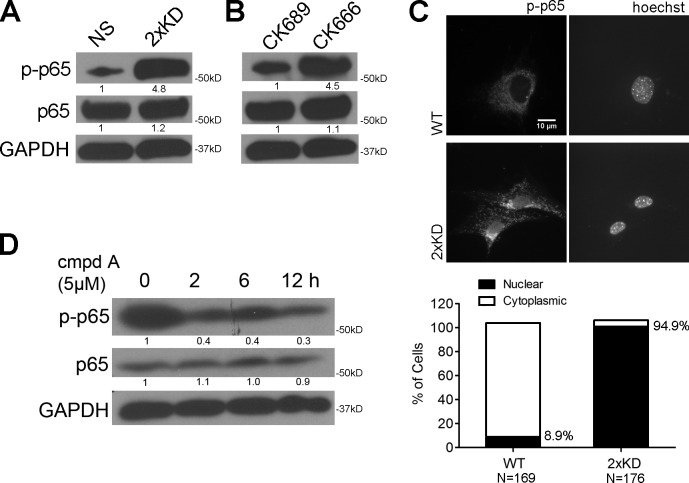
The SASP response has been reported to occur with DNA damage, oncogene activation, and cellular stress and is regulated by the NF-κB pathway (Karin, 2006; Chien et al., 2011; Freund et al., 2011). Thus, we postulated that the loss of Arp2/3 would lead to activation of NF-κB. Using phosphorylated RelA/p65 (p-p65) as a surrogate marker for NF-κB activity, we detected strong activation in Arp2/3-depleted cells (Fig. 3 A) and in multiple cell lines treated with the Arp2/3 inhibitor CK666 but not the inactive CK689 compound (Fig. 3 B and Fig. S1 A). We previously confirmed the specificity of this inhibitor using motility assays (Wu et al., 2012), and in this context, we were able to use it to carefully control the timing of Arp2/3 inhibition. We also observed increased nuclear localization of p-p65 in 2×KD cells, consistent with elevated nuclear NF-κB activity (Fig. 3 C). The cytoplasmic staining observed with this antibody was confirmed to be NS background using p65−/− mouse embryonic fibroblasts (MEFs; Fig. S1 F). Using an IκB kinase (IKK) inhibitor (cmpdA; Stein and Baldwin, 2011), we observed that the elevated p-p65 level in 2×KD cells could be reduced, suggesting that a canonical NF-κB activation pathway was involved (Fig. 3 D). This same treatment decreased the expression of several of the secreted factors by qRT-PCR and ELISA (Fig. S1, B and C). Importantly, this treatment did not affect the reduced migration speed or altered cell morphology that we reported previously with loss of Arp2/3 activity (Fig. S1, D and E; Wu et al., 2012). Together, these results indicate that loss of Arp2/3 activity leads to the activation of a canonical NF-κB pathway.
Figure 3.
Arp2/3 depletion or inhibition activates the canonical NF-κB pathway. (A) Blot showing phospho-p65 (Ser536; p-p65), total p65, and GAPDH levels in NS and 2×KD cells. (B) Blot showing p-p65, total p65, and GAPDH levels in NS cells treated with 100 µM CK666 or its inactive control CK689 for 6 h (right). (C, top) Representative immunofluorescent staining showing p-p65 localization in wild-type (WT) and 2×KD cells. (bottom) Percentages of cells having nuclear p-p65 were quantified and graphed. (D) Blot showing p-p65, p65, and GAPDH levels in 2×KD cells treated with an IKK inhibitor cmpdA for the indicated times. Fold changes in band intensity averaged across the three experiments (normalized to loading control and experimental control) are shown below each band.
The CCM2–MEKK3 pathway is required for the NF-κB activation induced by Arp2/3 inhibition
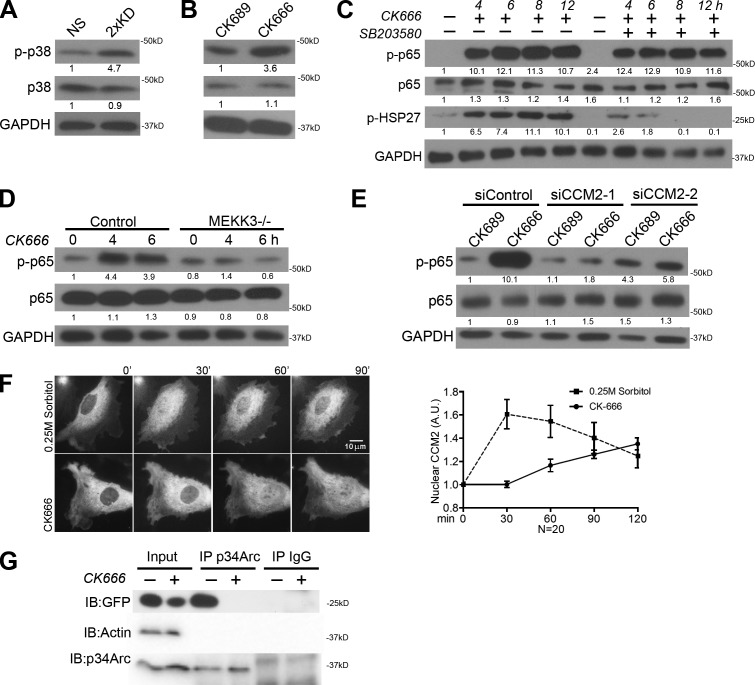
Previous work has implicated p38 MAPK (p38) as a key upstream factor in the activation of the SASP pathway (Freund et al., 2011). To test whether p38 is activated by the loss of Arp2/3, we blotted for p-p38 (active) and observed elevated levels in both Arp2/3-depleted and CK666-treated cells (Fig. 4, A and B). To determine whether p38 is upstream of the observed NF-κB activation in Arp2/3-inhibited cells, we simultaneously treated cells with the p38 inhibitor SB203580 and CK666 and still observed elevated p-p65, suggesting that p38 is not directly upstream in this case (Fig. 4 C). However, consistent with a previous study (Freund et al., 2011), pretreatment with SB203580 for >12 h did inhibit CK666’s effect on p-p65 levels (Fig. S2 A), suggesting that p38 may indirectly affect this response. Moreover, inhibiting p38 activity reduced the expression levels of some but not all of the elevated secreted factors (Fig. S2 B). One upstream candidate molecule that activates both IKK–NF-κB and p38 is the MAPK family member MEKK3 (Schmidt et al., 2003; Uhlik et al., 2003; Sun and Yang, 2010). We tested whether MEKK3 is involved in this pathway by treating MEKK3−/− cells with CK666 and observed no increased activation of NF-κB (Fig. 4 D). To determine how specific MEKK3 is for this pathway, we added CK666 to cells lacking MEKK2 (the closest protein relative of MEKK3; Maruyama et al., 2010) and observed similar NF-κB activation to control cells (Fig. S2 C). MEKK3 participates in multiple signaling cascades, but CCM2 (aka OSM) was originally identified as the scaffold for MEKK3 with F-actin binding capability that is essential for the osmotic stress response (Uhlik et al., 2003). To test whether CCM2 is involved in Arp2/3-based regulation of NF-κB, we depleted CCM2 using two distinct siRNAs and observed no activation of NF-κB in response to CK666 treatment (Fig. 4 E and Fig. S2 D). Previous work has implicated the CCM2–MEKK3 pathway in osmotic stress signaling (Uhlik et al., 2003). Using live-cell imaging of tagged CCM2, we observed a transient nuclear translocation of this protein during hyperosmotic response (Fig. 4 E). Interestingly, CK666 treatment caused a slower, but more persistent, nuclear translocation of CCM2 (Fig. 4 F), with no diminished nuclear signal even after 4 h of treatment (Fig. S2 E). Only a portion of CCM2 translocates to the nucleus in either case, suggesting that it may participate in multiple protein complexes that are under a different regulatory control. To test for an interaction between CCM2 and the Arp2/3 complex, we immunoprecipitated the Arp2/3 complex from cell lysates and blotted for CCM2 (Fig. 4 G). An interaction between these proteins was detected that could be eliminated by pretreating the cells with CK666 before lysis, suggesting that active Arp2/3 is required for this interaction to occur.
Figure 4.
NF-κB pathway activation in Arp2/3-depleted or -inhibited cells requires MEKK3 and CCM2. (A) Blot showing phospho–p38 MAPK (Thr180/Tyr182; p-p38), total p38, and GAPDH levels in NS and 2×KD cells. (B) Blot showing p-p38, total p38, and GAPDH levels in NS cells treated with CK689 or CK666 for 6 h (right). (C) Blot showing p-p65, p65, p-HSP27, and GAPDH levels of IA32 cells with 100 µM CK666 only and simultaneous 10 µM SB203580 + 100 µM CK666 treatments at the indicated time points. Decreased p-HSP27 levels indicate effective inhibition of p-38 activation by SB203580. (D) Blot showing p-p65, p65, and GAPDH levels of control and MEKK3−/− MEFs upon CK666 treatment for the indicated times. (E) Blot showing p-p65, p65, and GAPDH levels of control siRNA and two CCM2 siRNA–treated IA32 cells with CK689 or CK666 treatment for 6 h. (F, left) Time-lapse images showing representative cells expressing YPET-CCM2 treated in 0.25 M sorbitol (top)- or 100 µM CK666 (bottom)-containing media for the indicated times. (right) Quantification of nuclear CCM2 from time-lapse videos. Error bars show 95% CI. (G) Blot showing p34Arc immunoprecipitation (IP) experiments of HEK293 FT cells expressing YPET-CCM2 treated with DMSO or CK666 for 6 h before cell lysis. YPET-CCM2 was detected with an anti-GFP antibody. Fold changes in band intensity averaged across the three experiments (normalized to loading control and experimental control) are shown below each band. A.U., arbitrary unit; IB, immunoblot.
Arp2/3-depleted cells show altered osmotic stress response
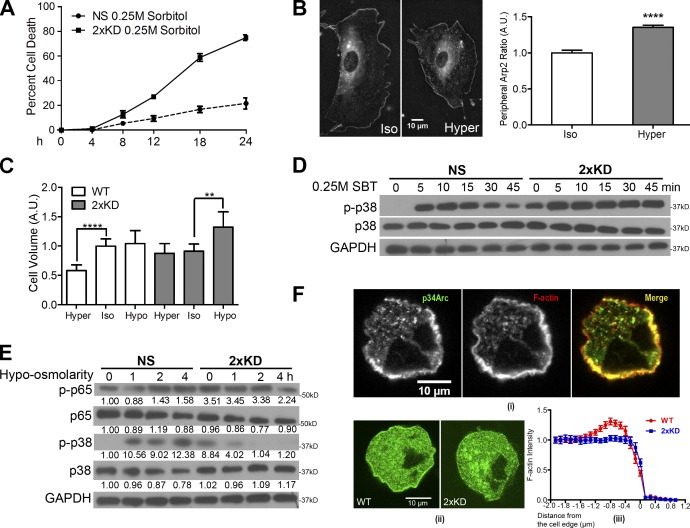
These observations suggest that Arp2/3-branched actin may play a role in osmotic stress responses. Consistent with this idea, cells depleted of Arp2/3 complex show significantly higher cell death under hyperosmotic conditions (Fig. 5 A). Using live-cell imaging, we observed a striking mobilization of the Arp2/3 complex to the cell periphery in response to hyperosmotic stress after 45 min (Fig. 5 B and Video 1). Cells respond to varying osmotic conditions by dynamic volume regulation (Hoffmann and Pedersen, 2006). Whereas control cells decreased their cell volume in response to hyperosmotic conditions, Arp2/3-depleted cells were unable to decrease their cell volume with hyperosmotic stress but did show enhanced swelling under hypoosmotic conditions (Fig. 5 C). A well-known signaling cascade involving p38 is activated in response to osmotic stress (Nielsen et al., 2008). To test the role of Arp2/3 in p38 osmotic signaling, we blotted for p-p38 in control and 2×KD cells treated with sorbitol. Control cells showed the expected transient activation of p38, whereas Arp2/3-depleted cells had high basal p38 activation that increased upon hyperosmotic stress but never decreased, suggesting a defect in adaptation of this response without the Arp2/3 complex (Fig. 5 D). Based on the volume regulation and signaling defects in cells depleted of the Arp2/3 complex, we postulated that reducing osmolarity might rescue the aberrant stress signaling. Indeed, when we treated Arp2/3-depleted cells with hypoosmotic media, we observed decreased p38 activity within 1 h and decreased NF-κB activity within 4 h (Fig. 5 E). Therefore, in the absence of Arp2/3, cells appear to be unable to adapt to hyperosmotic stress with a physiological rapid decrease in volume and exhibit evidence of activated signaling associated with a hyperosmotic state even when in isotonic media.
Figure 5.
Loss of Arp2/3 complex affects volume regulation and osmotic stress signaling. (A) Graph showing cell death rate of NS and 2×KD cells in hyperosmotic conditions. (B, left) Representative images showing Arp2-EGFP–expressing cells under iso- and hyperosmotic conditions. (right) Quantification of peripheral Arp2 under iso- and hyperosmotic conditions. ****, P < 0.001 by Student’s t test. Arp2-EGFP signals were identified manually, and its length versus the whole cell perimeter was defined as the peripheral Arp2 ratio and plotted. (C) Graph showing cell volume of NS and 2×KD cells in hypo (50% mixture of H2O and DMEM media)-, iso-, and hyper (0.25 M sorbitol-containing media)-osmotic conditions. ****, P < 0.001; **, P < 0.01 by Student’s t test. (D) Blot showing p-p38, p38, and GAPDH levels of NS and 2×KD cells under hyperosmotic conditions at different time points. (E) Blot showing p-p65, p65, p-p38, p38, and GAPDH levels of NS and 2×KD cells in hypoosmotic conditions at different time points. Fold changes in band intensity averaged across the three experiments (normalized to loading control and experimental control) are shown below each band. (F, i) Representative images showing endogenous p34Arc and F-actin (phalloidin) localization in the actin cortex. (ii) Representative images showing wild-type (WT) and 2×KD cells expressing Lifeact-EGFP. (iii) Fluorescent intensity profiles of the Lifeact signal near the cell edge were analyzed and plotted. Error bars show 95% CI. A.U., arbitrary unit.
The actin cortex is critical for providing mechanical integrity of the plasma membrane and the homeostasis of cell volume in response to osmotic shifts (Moustakas et al., 1998; Henson, 1999; Pedersen et al., 2001). Although the actin cortex is central to osmotic adaptation, little is known about how cells nucleate, polymerize, and remodel actin networks in the cortex (Salbreux et al., 2012). We tested whether the Arp2/3 complex and branched actin contribute to generating the actin cortex. Because imaging the actin cortex is difficult in flat, spread cells, we synchronized cells and imaged the cortex of cells rounded for mitosis. We detected robust colocalization of the Arp2/3 complex with the cortical actin under the plasma membrane (Fig. 5 F, i). By comparing the fluorescent profile of F-actin in control and 2×KD cells, we observed a significant decrease in F-actin intensity at the plasma membrane in 2×KD cells compared with controls (Fig. 5 F, ii and iii). These data indicate that the Arp2/3 complex plays an important role in generating submembranous cortical actin.
In summary, our findings indicate that loss of the Arp2/3 complex activates the NF-κB pathway, downstream of the CCM2–MEKK3 pathway, which is also critical for osmotic stress signaling. Consistent with this finding, Arp2/3-depleted cells are hypersensitive to osmotic stress and display elevated activation of this pathway under isotonic conditions. Cells dynamically trigger signaling pathways, rearrange cytoskeleton networks, and alter their volume, to adapt to varying osmotic conditions (Lang, 2007; Descot et al., 2009). Whether Arp2/3-generated branched actin affects signaling through a scaffolding function, by providing mechanical rigidity to the cortex or both, remains to be determined. Furthermore, it will be important to determine whether senescence is linked to osmotic stress and regulation of the actin cortex in other cell types.
Our results also indicate that perturbing Arp2/3-branched actin networks leads to the expected cell-autonomous defects in cytoskeletal organization but also nonautonomous effects via the up-regulation of these genes encoding a variety of secreted proteins, such as growth factors and chemokines. We show that fibroblast chemotaxis can be significantly affected by these secreted factors, depending on the precise experimental regimen. This finding resolves the contradictory findings of our group and Suraneni et al. (2012) on the role of the Arp2/3 complex during fibroblast chemotaxis (Wu et al., 2012). In hindsight, each group got the expected result for the experiment performed based on the exchange of fresh media or lack thereof in the chemotaxis chamber. However, this concept has much broader implications than explaining divergent experimental findings. The proinflammatory effects of perturbing Arp2/3 and possibly upstream regulatory pathways such as small GTPases (Rac and Cdc42) and nucleation-promoting factors (e.g., Scar/WAVE) should be taken into consideration when evaluating phenotypes of knockout mice and in other experimental systems. Loss of proper Arp2/3 regulation may explain inflammatory and autoimmune phenotypes in human diseases, such as Wiskott-Aldrich syndrome, in which mutation of the WAS gene (encoding the Arp2/3-activating nucleation-promoting factor Wiskott-Aldrich syndrome protein) changes the transcription of inflammatory genes in addition to altering cell-autonomous cytoskeletal functions (Dupuis-Girod et al., 2003; Prete et al., 2013).
Materials and methods
Reagents and materials
Commercial antibodies were obtained from Cell Signaling Technology (AKT [pan], phospho-AKT [Ser473], p38 MAPK, phospho–p38 MAPK [Thr180/Tyr182], NF-κB p65, phospho–NF-κB p65 [Ser536], and phospho-HSP27 [Ser82]), EMD Millipore (p34Arc), Takara Bio Inc. (GFP), and Ambion/Applied Biosystems (anti-GAPDH). Rhodamine red-X– and HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. Alexa Fluor 647 dye–conjugated phalloidin was obtained from Invitrogen. p38 MAPK inhibitor SB203580, Arp2/3 complex inhibitor CK666, and its inactive control CK689 were obtained from EMD Millipore and used at 100 µM. d-sorbitol is purchased from Sigma-Aldrich. Human plasma fibronectin (FN), PDGF-BB, and EGF were obtained from BD. Western blotting was performed by standard techniques. Propidium iodide was obtained from Sigma-Aldrich. Two siRNAs targeted against siCCM2 were used (SI00943446 and SI00943453; QIAGEN). Protein A–Sepharose beads were obtained from Sigma-Aldrich.
RNA preparation, RNA-Seq, and qRT-PCR
Total RNA was isolated from cells (RNeasy Plus; QIAGEN) and validated to have RNA integrity numbers >8.5 using a bioanalyzer (2100 Bioanalyzer; Agilent Technologies). For RNA-Seq, poly(A)+ RNA was enriched (Oligotex mRNA Mini Kit; QIAGEN) and fragmented (RNA Fragmentation Reagents; Ambion), and cDNA was generated (SuperScript II; Invitrogen) by random priming followed by second strand synthesis (DNA Polymerase I; Enzymatics) and purified (PCR purification kit; QIAGEN). Libraries were prepared according to the manufacturer’s specifications (Illumina). Sequencing was performed using 50-bp single-end reads (HiSeq 2000; Illumina). Reads were aligned to the reference mouse genome (mm9) using TopHat (Trapnell et al., 2009), and gene expression was estimated by calculating RPKM (reads per kilobase per million reads; Mortazavi et al., 2008) for all Ensembl genes, analyzing only exonic reads. For qRT-PCR, cDNA was generated from total RNA (SuperScript II) using a 50:50% mixture of random primers (Invitrogen) and the oligo-dT primer (Promega). This cDNA was used directly with TaqMan Gene Expression Master Mix and TaqMan Gene Expression Assays (Applied Biosystems) to be read on a qRT-PCR machine (ViiA 7; Applied Biosystems). qRT-PCR data were analyzed by the Pfaffl method (Pfaffl, 2001).
Immunoprecipitation
Cell lysates were made in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.5% Na-deoxycholate, 150 mM NaCl, and 1 mM EDTA). An equal amount of each protein lysate was incubated/precleared with protein A–Sepharose beads and anti–rabbit-IgG antibody at 4°C for 1 h. Cleared supernatants were then incubated with anti-p34Arc antibody or anti–rabbit-IgG antibody overnight at 4°C before adding protein A–Sepharose beads for an additional 2-h incubation. After washing the beads 2× with radioimmunoprecipitation assay buffer and 2× with PBS, immunoprecipitates were boiled in SDS sample buffer for immunoblotting as indicated in legend of Fig. 4 G.
Western blotting
Western blotting was performed with standard protocol as previously described (Cai et al., 2007). A representative blot from three experiments was shown for each figure. Fold changes in band intensity averaged across the three experiments (normalized to loading control and experimental control) are shown below each band.
Cell culture and transient transfection
Cells were cultured in regular media (DMEM supplemented with 10% FBS [HyClone], 100 U/ml penicillin, 100 µg/ml streptomycin, and 292 µg/ml l-glutamine). DMEM supplemented with 0.1% fatty acid–free BSA (Equitech-Bio), 100 U/ml penicillin, 100 µg/ml streptomycin, and 292 µg/ml l-glutamine was used to culture cells during serum starvation. Regular media containing 0.25 M sorbitol were used for the hyperosmotic condition. An equal volume of H2O supplemented with 10% FBS (HyClone), 100 U/ml penicillin, 100 µg/ml streptomycin, and 292 µg/ml l-glutamine was added to regular media for the hypoosmotic condition. Transient transfections were performed using X-tremeGENE 9 (Roche) for HEK293 FT cells and NanoJuice (EMD Millipore) for IA32 cells. siRNA transfection was performed with Lipofectamine RNAiMAX (Invitrogen).
Cell synchronization
To synchronize cells and enrich for cells undergoing mitosis, we washed cells twice with PBS at 30–40% cell confluency and added DMEM without serum. After 48 h, we stimulated cells with 10% serum-containing media to induce cell division.
Cell volume measurement
Cells were trypsinized for 3 min until rounded and lifted from the culture dish. A gentle wash with sterile Dulbecco’s PBS was performed before resuspending cells in the indicated osmotic solutions for 20 min at room temperature. Cells in suspension were imaged by differential interference contrast microscopy, and cell diameter was measured with ImageJ (National Institutes of Health) to calculate cell volume.
Cell viability assay
Cells were plated on 10 µg/ml FN-coated glass-bottom culture dishes (MatTek Corporation) for 8–12 h before imaging. Propidium iodide was added to imaging dishes to a final concentration of 1 µg/ml. Time-lapse microscopy was performed on an incubator fluorescent microscope (20× objective; VivaView FL; Olympus) with a camera (ORCA-ER/AG type C4742-80-12AG; Hamamatsu Photonics). Fluorescent cells (indicating loss of membrane integrity and cell death) were counted at the indicated time points. The curve of the percentage of cell death was generated with Prism (GraphPad Software).
ELISAs and CM
ELISA kits were as follows: IL-6 (eBioscience), MMP3 (RayBio), and HGF (Abcam). Cells were washed with serum-free DMEM and incubated in serum-free DMEM for 24 h. Media were collected and filtered before performing ELISAs. All ELISA data are presented as absolute values based on standard curves from the kits.
Light microscopy and image analysis
Immunofluorescence.
For immunofluorescent staining, the cells were fixed, stained, and mounted as described previously (Bear et al., 2002). Cells were plated on acid-washed coverslips coated with 10 µg/ml FN overnight before fixing with 4% PFA and permeabilized in 0.1% Triton X-100 in PBS for 5 min. To stain for p-p65, cells were prepermeabilized with 0.05% saponin for 45 s before fixation. Cells were stained with Alexa Fluor 647 phalloidin for F-actin (1:400 dilution), p-p65 (1:100; Cell Signaling Technology), and Hoechst (1:10,000). After three washes in PBS, the coverslips were mounted onto slides with Fluoromount-G (Electron Microscopy Sciences). Images were captured using an inverted microscope (IX81; Olympus) with a 60×, 1.42 NA objective, a charge-coupled device camera (C4742-80-12AG), and an automated X-Y stage, with MetaMorph imaging software (Molecular Devices). Images were combined and annotated in Photoshop (Adobe) for presentation. To plot the distribution of a protein around the cell edge, a custom ImageJ macro was used to extract pixel intensity as a function of distance from the leading edge (Cai et al., 2007).
Nuclear CCM2 and p-p65 quantification.
For CCM2, fluorescent time-lapse images were taken with cells expressing YPET-CCM2 (Uhlik et al., 2003) in cell culture media supplemented with trolox, ascorbic acid, and catalase under indicated conditions. Images were taken using an inverted microscope (IX81) with a 60×, 1.42 NA objective, a charge-coupled device camera (C4742-80-12AG), and an automated X-Y stage, operated by MetaMorph imaging software. Images at different time points were selected, and the cell nucleus was outlined. Mean intensity of CCM2 signal within the outlined region was measured. Intensity was then normalized to time 0 and plotted using Prism. For p-p65 imaging, 20× fluorescent images were taken with fixed cells stained with p-p65 and Hoechst, cells with colocalization of p-p65 and Hoechst were counted, and fractions of cells with nuclear or cytoplasmic localization were plotted with Prism.
Peripheral Arp2 quantification.
Fluorescent images were taken with the same imaging setup. Images were then processed with ImageJ software. Arp2-EGFP signals near the cell edge were identified and drawn out with the freehand line function. The length l of the line was then measured and recorded. The perimeter of the same cell was measured the same way and recorded as L. The ratio of l/L was defined as the peripheral Arp2 ratio for each cell.
Single-cell tracking.
Cells were plated on 10 µg/ml FN-coated glass-bottom culture dishes for 8–12 h before imaging. Time-lapse microscopy was performed on an incubator fluorescent microscope (20× objective; VivaView FL) with a camera (Orca ER/AG type c4742-80-12AG). Cell speed was measured with ImageJ using the Manual Tracking plug-in.
Directional migration assays
Microfluidic device preparation.
Transparency masks were printed using a high-resolution printer (FineLine Imaging). The pattern for the chemotaxis chamber was fabricated on 4-inch silicon wafers using a two-step photolithography process. Polydimethylsiloxane (PDMS) was then poured on the wafer and cured overnight at 70°C. Individual PDMS devices were cut out from the wafer and placed in a clean dish until use. Ports were then punched out in the devices. The devices were washed with water and ethanol and blow dried. The PDMS device was placed into contact with a glass dish bottom immediately after plasma treatment of both pieces, ensuring that an irreversible seal was formed. The cell culture chamber was then filled with 10 µg/ml FN for 1 h at 37°C followed by flushing with sterile PBS. Cells were loaded into the cell culture chamber using a gel-loading pipette tip. The cell chamber ports were plugged with short pieces of tubing (0.0025 × 0.01325 inch; Upchurch Scientific).
Chemotactic gradients.
The exit ports of the sink and source channels were connected to waste using tubing of an internal diameter of 0.015 inches. Gas tight 100-µl glass syringes (100-µl syringe; 81020 1710TLL; Hamilton) were connected to 27.5-gauge needles connected to tubing. The source syringe and tubing were filled with the indicated CM containing the indicated chemoattractant and 10 µg/ml TRITC-dextran to visualize the gradient. The sink syringe and tubing were filled with the indicated CM only. The tubing was then inserted into the source and sink channels, respectively, and the syringe pump was operated at a flow rate of 20 nl/min. A stable gradient was then established in the cell culture chamber within 30 min, and typically remained stable for 18 h as monitored by TRITC-dextran fluorescent intensity. In PDGF chemotaxis experiments, the source concentration of PDGF was 120 ng/ml, whereas in EGF chemotaxis experiments, 100 ng/ml EGF was in the source chamber.
Directional migration image acquisition and analysis.
Chemotaxis assays were performed on an inverted microscope (IX81) with a camera (Orca-ER) operated by MetaMorph imaging software. A 20× objective was used. Images were collected every 10 min for over 12 h. Individual cells were manually tracked using ImageJ software Manual Tracking plug-in. The tracks obtained were further analyzed using the chemotaxis tool developed by ibidi. This analysis tool was used to extract the forward migration index and histogram of angular direction of tracks from the manual tracking results. To obtain the compass parameter, the histograms obtained were further analyzed in Prism software by performing a nonlinear curve fit to the chemotaxis equation.
Online supplemental material
Fig. S1 shows that NF-κB is activated in Arp2/3-inhibited cells and regulates the secretory phenotype in Arp2/3-deficient or -inhibited cells. Fig. S2 show that p38 MAPK and the MEKK3–OSM complex are involved in the secretory phenotype in Arp2/3-deficient or -inhibited cells. Video 1 shows a representative cell expressing Arp2-GFP (with Arp2 depletion) treated with sorbitol to induce hyperosmotic stress. Table S1 is provided as an Excel (Microsoft) file and shows the RNA-Seq results of this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201306032/DC1.
Supplementary Material
Acknowledgments
We thank Aaron Cook for help with the Arp2-GFP/osmotic stress quantification and Robert Currin of the University of North Carolina–Olympus Imaging Research Center for assistance with imaging.
This work was supported by National Institutes of Health grants to N.E. Sharpless (AG024379), A.S. Baldwin (CA75080 and CA73756), I.J. Davis (CA166447), G.L. Johnson (GM68820), and J.E. Bear (GM083035). J.E. Bear is also supported by the Howard Hughes Medical Institute.
Footnotes
Abbreviations used in this paper:
- CI
- confidence interval
- CM
- conditioned media
- FN
- fibronectin
- HGF
- hepatocyte growth factor
- IKK
- IκB kinase
- KD
- knockdown
- MEF
- mouse embryonic fibroblast
- NF-κB
- nuclear factor κB
- NS
- nonspecific
- PDMS
- polydimethylsiloxane
- qRT-PCR
- quantitative RT-PCR
- SASP
- senescence-associated secretory phenotype
References
- Bear J.E., Svitkina T.M., Krause M., Schafer D.A., Loureiro J.J., Strasser G.A., Maly I.V., Chaga O.Y., Cooper J.A., Borisy G.G., Gertler F.B. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 109:509–521 10.1016/S0092-8674(02)00731-6 [DOI] [PubMed] [Google Scholar]
- Cai L., Marshall T.W., Uetrecht A.C., Schafer D.A., Bear J.E. 2007. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 128:915–929 10.1016/j.cell.2007.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J., d’Adda di Fagagna F. 2007. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8:729–740 10.1038/nrm2233 [DOI] [PubMed] [Google Scholar]
- Chien Y., Scuoppo C., Wang X., Fang X., Balgley B., Bolden J.E., Premsrirut P., Luo W., Chicas A., Lee C.S., et al. 2011. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 25:2125–2136 10.1101/gad.17276711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé J.P., Patil C.K., Rodier F., Sun Y., Muñoz D.P., Goldstein J., Nelson P.S., Desprez P.Y., Campisi J. 2008. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6:2853–2868 10.1371/journal.pbio.0060301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descot A., Hoffmann R., Shaposhnikov D., Reschke M., Ullrich A., Posern G. 2009. Negative regulation of the EGFR-MAPK cascade by actin-MAL-mediated Mig6/Errfi-1 induction. Mol. Cell. 35:291–304 10.1016/j.molcel.2009.07.015 [DOI] [PubMed] [Google Scholar]
- Dupuis-Girod S., Medioni J., Haddad E., Quartier P., Cavazzana-Calvo M., Le Deist F., de Saint Basile G., Delaunay J., Schwarz K., Casanova J.L., et al. 2003. Autoimmunity in Wiskott-Aldrich syndrome: risk factors, clinical features, and outcome in a single-center cohort of 55 patients. Pediatrics. 111:e622–e627 10.1542/peds.111.5.e622 [DOI] [PubMed] [Google Scholar]
- Freund A., Patil C.K., Campisi J. 2011. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 30:1536–1548 10.1038/emboj.2011.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J.H. 1999. Relationships between the actin cytoskeleton and cell volume regulation. Microsc. Res. Tech. 47:155–162 [DOI] [PubMed] [Google Scholar]
- Hoffmann E.K., Pedersen S.F. 2006. Sensors and signal transduction pathways in vertebrate cell volume regulation. Contrib. Nephrol. 152:54–104 10.1159/000096318 [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Karin M. 2006. Nuclear factor-kappaB in cancer development and progression. Nature. 441:431–436 10.1038/nature04870 [DOI] [PubMed] [Google Scholar]
- Kuilman T., Michaloglou C., Vredeveld L.C., Douma S., van Doorn R., Desmet C.J., Aarden L.A., Mooi W.J., Peeper D.S. 2008. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 133:1019–1031 10.1016/j.cell.2008.03.039 [DOI] [PubMed] [Google Scholar]
- Lang F. 2007. Mechanisms and significance of cell volume regulation. J. Am. Coll. Nutr. 26(5 Suppl.):613S–623S [DOI] [PubMed] [Google Scholar]
- Lujambio A., Akkari L., Simon J., Grace D., Tschaharganeh D.F., Bolden J.E., Zhao Z., Thapar V., Joyce J.A., Krizhanovsky V., Lowe S.W. 2013. Non-cell-autonomous tumor suppression by p53. Cell. 153:449–460 10.1016/j.cell.2013.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Kadowaki H., Okamoto N., Nagai A., Naguro I., Matsuzawa A., Shibuya H., Tanaka K., Murata S., Takeda K., et al. 2010. CHIP-dependent termination of MEKK2 regulates temporal ERK activation required for proper hyperosmotic response. EMBO J. 29:2501–2514 10.1038/emboj.2010.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 5:621–628 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Moustakas A., Theodoropoulos P.A., Gravanis A., Häussinger D., Stournaras C. 1998. The cytoskeleton in cell volume regulation. Contrib. Nephrol. 123:121–134 10.1159/000059925 [DOI] [PubMed] [Google Scholar]
- Nielsen M.B., Christensen S.T., Hoffmann E.K. 2008. Effects of osmotic stress on the activity of MAPKs and PDGFR-beta-mediated signal transduction in NIH-3T3 fibroblasts. Am. J. Physiol. Cell Physiol. 294:C1046–C1055 10.1152/ajpcell.00134.2007 [DOI] [PubMed] [Google Scholar]
- Olson E.N., Nordheim A. 2010. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11:353–365 10.1038/nrm2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S.F., Hoffmann E.K., Mills J.W. 2001. The cytoskeleton and cell volume regulation. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130:385–399 10.1016/S1095-6433(01)00429-9 [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. 2007. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36:451–477 10.1146/annurev.biophys.35.040405.101936 [DOI] [PubMed] [Google Scholar]
- Posern G., Treisman R. 2006. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 16:588–596 10.1016/j.tcb.2006.09.008 [DOI] [PubMed] [Google Scholar]
- Prete F., Catucci M., Labrada M., Gobessi S., Castiello M.C., Bonomi E., Aiuti A., Vermi W., Cancrini C., Metin A., et al. 2013. Wiskott-Aldrich syndrome protein–mediated actin dynamics control type-I interferon production in plasmacytoid dendritic cells. J. Exp. Med. 210:355–374 10.1084/jem.20120363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty J.D., Wu C., Bear J.E. 2013. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Biol. 14:7–12 10.1038/nrm3492 [DOI] [PubMed] [Google Scholar]
- Salbreux G., Charras G., Paluch E. 2012. Actin cortex mechanics and cellular morphogenesis. Trends Cell Biol. 22:536–545 10.1016/j.tcb.2012.07.001 [DOI] [PubMed] [Google Scholar]
- Salminen A., Kauppinen A., Kaarniranta K. 2012. Emerging role of NF-κB signaling in the induction of senescence-associated secretory phenotype (SASP). Cell. Signal. 24:835–845 10.1016/j.cellsig.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Schmidt C., Peng B., Li Z., Sclabas G.M., Fujioka S., Niu J., Schmidt-Supprian M., Evans D.B., Abbruzzese J.L., Chiao P.J. 2003. Mechanisms of proinflammatory cytokine-induced biphasic NF-kappaB activation. Mol. Cell. 12:1287–1300 10.1016/S1097-2765(03)00390-3 [DOI] [PubMed] [Google Scholar]
- Sharpless N.E. 2004. Ink4a/Arf links senescence and aging. Exp. Gerontol. 39:1751–1759 10.1016/j.exger.2004.06.025 [DOI] [PubMed] [Google Scholar]
- Stein S.J., Baldwin A.S. 2011. NF-κB suppresses ROS levels in BCR-ABL(+) cells to prevent activation of JNK and cell death. Oncogene. 30:4557–4566 10.1038/onc.2011.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Yang J. 2010. Molecular basis of lysophosphatidic acid-induced NF-κB activation. Cell. Signal. 22:1799–1803 10.1016/j.cellsig.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P., Rubinstein B., Unruh J.R., Durnin M., Hanein D., Li R. 2012. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 197:239–251 10.1083/jcb.201112113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchkonia T., Zhu Y., van Deursen J., Campisi J., Kirkland J.L. 2013. Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J. Clin. Invest. 123:966–972 10.1172/JCI64098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlik M.T., Abell A.N., Johnson N.L., Sun W., Cuevas B.D., Lobel-Rice K.E., Horne E.A., Dell’Acqua M.L., Johnson G.L. 2003. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat. Cell Biol. 5:1104–1110 10.1038/ncb1071 [DOI] [PubMed] [Google Scholar]
- Wu C., Asokan S.B., Berginski M.E., Haynes E.M., Sharpless N.E., Griffith J.D., Gomez S.M., Bear J.E. 2012. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 148:973–987 10.1016/j.cell.2011.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.