Metacaspase-dependent autophagy in plants promotes cell disassembly during vacuolar cell death and inhibits necrosis.
Abstract
Although animals eliminate apoptotic cells using macrophages, plants use cell corpses throughout development and disassemble cells in a cell-autonomous manner by vacuolar cell death. During vacuolar cell death, lytic vacuoles gradually engulf and digest the cytoplasmic content. On the other hand, acute stress triggers an alternative cell death, necrosis, which is characterized by mitochondrial dysfunction, early rupture of the plasma membrane, and disordered cell disassembly. How both types of cell death are regulated remains obscure. In this paper, we show that vacuolar death in the embryo suspensor of Norway spruce requires autophagy. In turn, activation of autophagy lies downstream of metacaspase mcII-Pa, a key protease essential for suspensor cell death. Genetic suppression of the metacaspase–autophagy pathway induced a switch from vacuolar to necrotic death, resulting in failure of suspensor differentiation and embryonic arrest. Our results establish metacaspase-dependent autophagy as a bona fide mechanism that is responsible for cell disassembly during vacuolar cell death and for inhibition of necrosis.
Introduction
Programmed cell death (PCD) is indispensable for animal and plant development, but the mechanisms of PCD differ between the two kingdoms. Plants lack apoptosis that involves cell fragmentation into discrete bodies and their heterophagic removal, owing to the presence of cell walls and lack of phagocytosis (Beers, 1997; Jones, 2001; Lam, 2004). Furthermore, plant genomes lack the core apoptotic regulators, such as Bcl-2 family proteins and caspases (Koonin and Aravind, 2002).
Although molecular regulation of plant PCD remains poorly understood, most cases of plant cell death can be divided into two classes with distinct kinetics and morphology: vacuolar cell death and necrosis (van Doorn et al., 2011). Vacuolar cell death is a slow process whereby growing lytic vacuoles gradually digest entire or most of the contents of terminally differentiated cells excluding cell walls. This cell death is indispensable for plant development, playing an instrumental role in the formation of conduits of water, nutrients, and hormones (the embryo suspensor and the vascular system) and secretory structures (e.g., laticifers; Beers and McDowell, 2001; Bozhkov et al., 2005a; van Doorn and Woltering, 2005; Bollhöner et al., 2012). We have shown that execution of vacuolar cell death in Norway spruce (Picea abies) embryo suspensor requires type II metacaspase mcII-Pa, a cysteine protease distantly related to caspases (Suarez et al., 2004; Bozhkov et al., 2005b; Tsiatsiani et al., 2011). However, mcII-Pa is a nucleocytoplasmic protein, and its proteolytic action alone is not sufficient to account for progressive growth of lytic vacuoles in the course of execution of PCD. Identification of such a mechanism would significantly advance our understanding of the vacuolar cell death process.
In contrast to vacuolar cell death, necrosis is a rapid mode of plant cell death and, like in animal systems, involves mitochondrial dysfunction and early loss of plasma membrane integrity (van Doorn et al., 2011; Galluzzi et al., 2012). However, although animal necrotic cells swell and burst, their plant counterparts retain undigested cytoplasmic materials surrounded by cell walls. Necrosis is a typical response of plants to abiotic stress and pathogen attack (Heath, 2000; Coll et al., 2011; van Doorn et al., 2011). Thus, vacuolar and necrotic modes of cell death not only differ morphologically but are also implicated in distinct aspects of plant biology: development and stress response, respectively. An intriguing question is what determines the mode of cell death in plants.
Here, we show that autophagy is responsible for cell self-disassembly during vacuolar cell death in P. abies embryos. Activation of autophagy requires metacaspase mcII-Pa and deficiency of either component switches the mode of cell death from vacuolar to necrotic. These findings provide a mechanistic explanation for morphological differences between two major classes of cell death in plants.
Results and discussion
Vacuolar cell death in the embryo suspensor is associated with enhanced autophagy
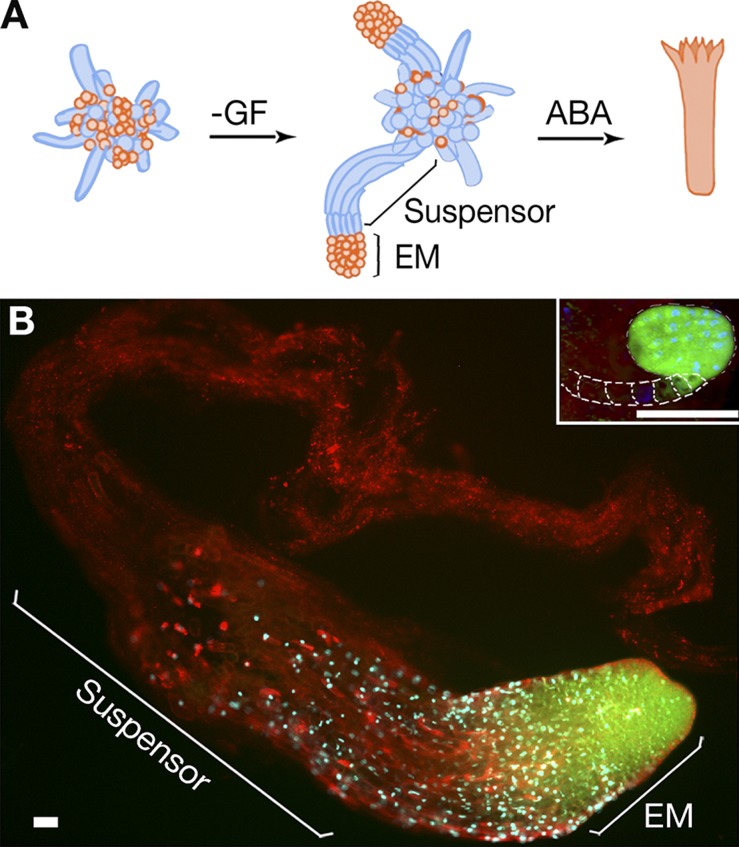
In somatic embryogenesis of P. abies, the formation of early embryos is induced by withdrawal of growth factors (GFs), auxin and cytokinin, whereas further development to the cotyledonary stage requires exogenous abscisic acid (ABA; Fig. 1 A; Bozhkov et al., 2005a). An early P. abies embryo is composed of a proliferating embryonal mass (EM) that will eventually form a cotyledonary embryo and terminally differentiated suspensor, which is gradually eliminated before the cotyledonary stage. Although Arabidopsis thaliana embryos have minute suspensors of seven cells, the suspensors in P. abies and most other gymnosperms are several millimeters long and composed of many cells (Fig. 1 B; Singh, 1978). In addition, suspensors of P. abies consist of several tiers of elongated cells at successive stages of cell disassembly, providing an excellent paradigm for studying vacuolar PCD (Bozhkov et al., 2005a; van Doorn et al., 2011).
Figure 1.
Embryo development in P. abies. (A) A schematic model of somatic embryogenesis. Cell proliferation is stimulated by GF, auxin, and cytokinin. Early embryogenesis is induced by withdrawal of GF. An early embryo is composed of embryonal mass (EM) and a suspensor. Development to the cotyledonary stage is promoted by abscisic acid (ABA). Suspensors are eliminated before the cotyledonary stage. Dying or dead cells are colored in blue. (B) Embryos of P. abies and A. thaliana (inset; dashed lines indicate contour of suspensor) at the developmental stage before formation of cotyledons stained with fluorescein diacetate (FDA; green), DAPI (blue), and FM4-64 (red). The lack of FDA staining in the suspensor denotes the loss of cell viability. Note the giant size, as well as the higher suspensor-to-EM size ratio, for P. abies embryo as compared with the A. thaliana embryo. Bars, 50 µm.
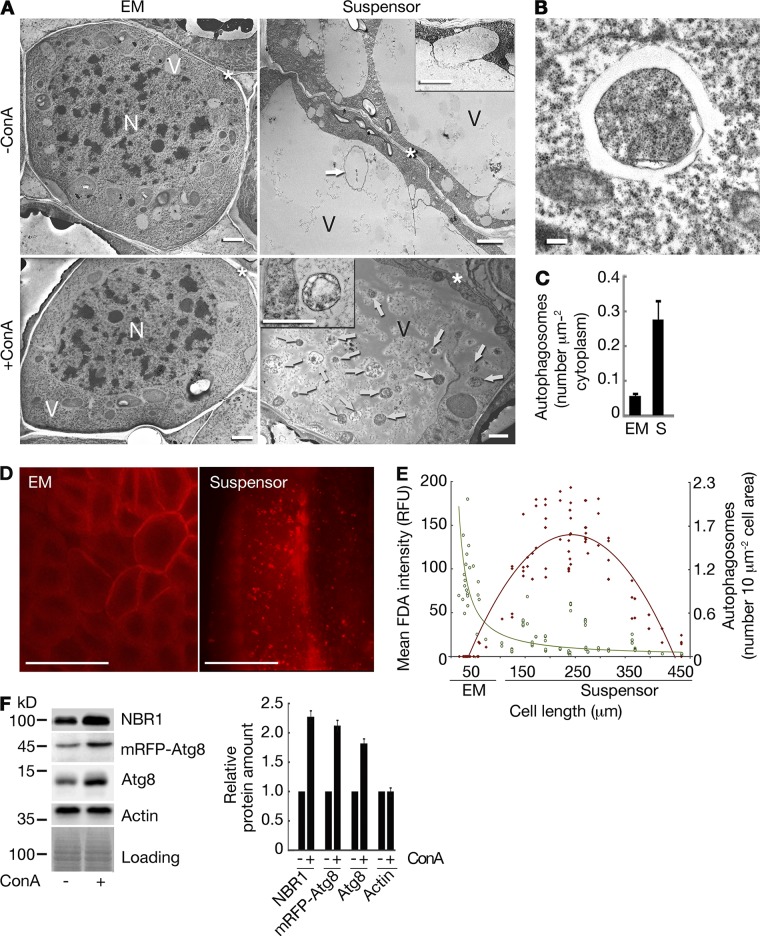
We obtained three lines of evidence that vacuolar PCD in the P. abies suspensor is associated with increased autophagic activity. First, transmission electron microscopy (TEM) revealed accumulation of autophagic bodies in the vacuoles of suspensor cells upon inhibition of vacuolar acidification using concanamycin A (ConA; Fig. 2 A) as well as increased amounts of double membrane–bound autophagosomes in the cytoplasm of suspensor cells as compared with EM cells (Fig. 2, A–C; Filonova et al., 2000). Second, transgenic mRFP-Atg8 lines showed cytoplasmic localization of mRFP-Atg8 in the EM cells and punctate localization in the suspensor cells (Fig. 2 D; Klionsky et al., 2012). Simultaneous measurement of fluorescein diacetate (FDA) staining intensity, cell length, and amount of mRFP-Atg8 puncta per cell area in the EM and suspensor cells confirmed that progression of vacuolar PCD in the suspensors correlates with cell elongation and enhanced autophagy (Fig. 2 E). Finally, abrogation of autophagic flux by ConA led to dramatic increase in the levels of autophagic target proteins Atg8 and NBR1 (Fig. 2 F; Svenning et al., 2011; Klionsky et al., 2012; Minina et al., 2013b).
Figure 2.
Enhanced autophagy in the P. abies embryo suspensor. (A) Assessment of autophagic flux in the EM and suspensor cells using ConA treatment. Arrows denote autophagic bodies. Insets depict autophagosome docking to the vacuole (−ConA) and autophagic body (+ConA). N, nucleus; V, vacuole; asterisks, cell wall. Bars, 2 µm. (B) Typical autophagosome in the suspensor cell. Bar, 0.2 µm. (C) Number of autophagosomes in the cytoplasm estimated from the micrographs of EM and suspensor (S) cells. Data represent the means ± SEM from three independent experiments, each including at least four cells per cell type. (D) mRFP-Atg8 accumulates in puncta in the suspensor cells but remains cytoplasmic in the EM cells upon ConA treatment. Bars, 50 µm. (E) Correlation between anisotropic cell expansion (cell length), progression of cell death (mean FDA intensity; green symbols and green power trendline, R2 = 0.5402), and accumulation of autophagosomes (number of mRFP-Atg8–positive puncta; red symbols and red polynomial trendline, R2 = 0.8202) in the embryos. RFU, relative fluorescence units. (F) Accumulation of NBR1, mRFP-Atg8, and Atg8 upon ConA treatment detected by Western blot analysis of total protein extracts from the embryos. Actin was used as a reference control. Graph represents relative protein amounts shown as means ± SEM from three independent measurements. For each protein, the integrated band intensities were first normalized to corresponding intensities of Coomassie staining (loading) and then to the sample without ConA. The amounts of all three proteins were significantly increased upon ConA treatment (P < 0.001; Student’s t test).
Silencing of ATG5 or ATG6 abrogates embryogenesis and phenocopies mcII-Pa depletion
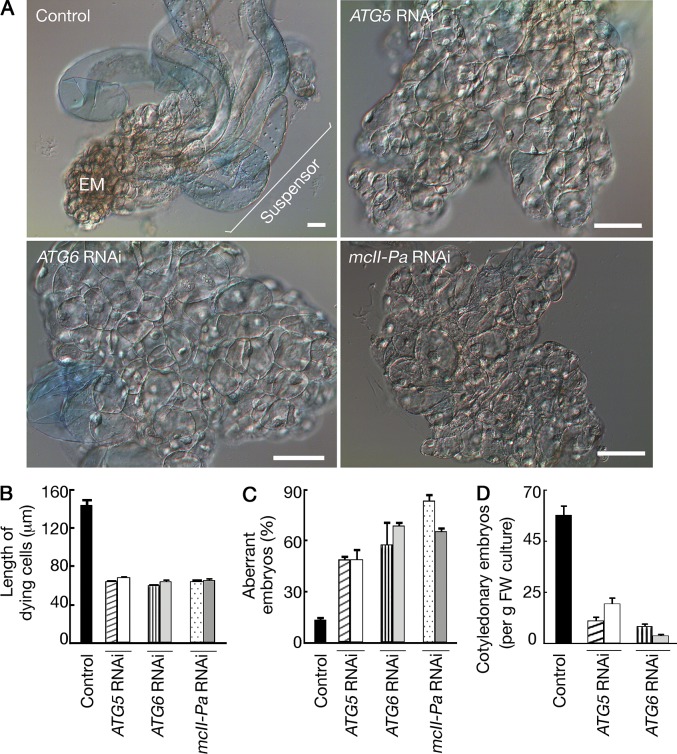
The vast body of genetic data demonstrates conservation of the major functional groups of Atg (autophagy related) proteins in plants (Doelling et al., 2002; Hanaoka et al., 2002; Liu and Bassham, 2012). We addressed the requirement of autophagy for vacuolar PCD in the suspensor by silencing two single-copy ATG genes, ATG5 and ATG6, by RNAi (Figs. S1 and S2). Silencing either gene abrogated suspensor formation and apical–basal patterning (Fig. 3 A), the phenotype being similar to lines deficient for mcII-Pa, the only type II metacaspase expressed in the P. abies embryos (Figs. 3 A, S1, and S2; Suarez et al., 2004). The failure of normal suspensor formation in Atg5-, Atg6- and mcII-Pa–deficient lines was accompanied by reduced anisotropic expansion of dying cells as estimated by measuring the length of Evans blue–positive cells (Fig. 3 B). Genetic inhibition of autophagy or metacaspase increased the frequency of aberrant early embryos lacking suspensors in several independent lines by four- to sixfold (Fig. 3 C). Accordingly, the efficiency of subsequent embryo development to the cotyledonary stage was dramatically reduced (Fig. 3 D; Suarez et al., 2004).
Figure 3.
Autophagy and metacaspase are essential for embryo patterning. (A–D) Knockdown of ATG5, ATG6, or mcII-Pa results in suspensor-deficient phenotype (A) and inhibits anisotropic growth of dying cells (B), early (C), and late (D) embryogenesis. (A) The cultures were grown without GF for 7 d and stained with Evans blue to detect dead cells. Bars, 20 µm. (B) Length of Evans blue–positive cells measured in the same samples. Data represent the means ± SEM from three independent experiments, each including at least 200 Evans blue–positive cells from 10 embryos per line. (C) Frequency of aberrant early embryos lacking suspensors in the same samples as in A. Data represent the means ± SEM from three independent experiments, each including ≥100 embryos per line. (D) The yield of cotyledonary embryos after ABA treatment. Data represent the means ± SEM from four independent experiments, each including eight samples per line. FW, fresh weight. The mean values for RNAi lines in B–D significantly differ from control (P < 0.001; Dunnett’s test).
Deficiency of either autophagy or metacaspase switches the mode of cell death from vacuolar to necrotic
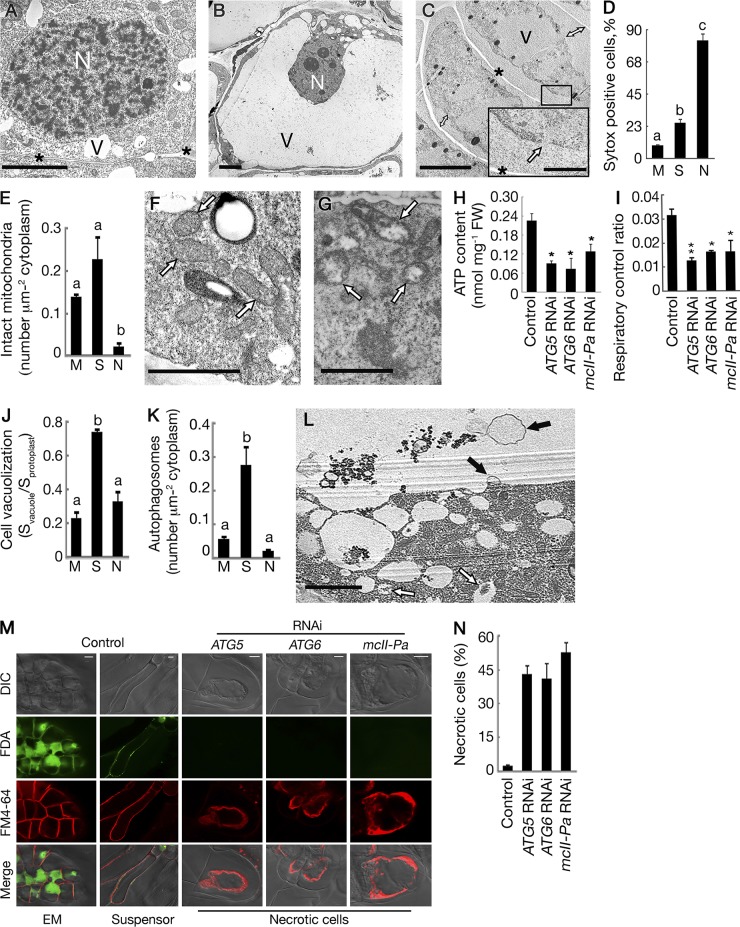
Because autophagy or metacaspase deficiency did not prevent cell death upon withdrawal of GF (Fig. 3 A), we compared the morphology of cell death in RNAi and control lines using TEM. In control lines, dying suspensor cells maintained intact plasma membranes and turgid protoplasts until almost all of the cytoplasm was digested by lytic vacuoles, whereas dying cells in RNAi lines contained shrunken and largely undigested protoplasts with ruptured plasma membranes (Fig. 4, A–D). This morphology was reminiscent of necrosis (Majno and Joris, 1995; van Doorn et al., 2011). The reduced number of morphologically intact mitochondria in the cytoplasm of dying cells from the RNAi lines as compared with both meristematic and suspensor cells (Fig. 4 E), correlated with the accumulation of swollen mitochondria (Fig. 4, F and G). Consistent with the altered morphology of mitochondria, the RNAi lines displayed a drop in intracellular ATP content (Fig. 4 H) and respiratory decline (Fig. 4 I; Eguchi et al., 1997; Bal-Price and Brown, 2000). These metabolic changes indicate mitochondrial dysfunction and, in combination with the loss of plasma membrane integrity, demonstrate that the cell death observed in autophagy- or metacaspase-deficient lines is necrosis.
Figure 4.
Deficiency of either autophagy or metacaspase switches the mode of cell death from vacuolar to necrotic. (A–C) Cell morphology in control and RNAi lines (exemplified by an ATG6 RNAi line). N, nucleus; V, vacuole; asterisks, cell wall. (A) A meristematic cell from the EM of the control line. (B) Vacuolated suspensor cell with intact plasma membrane and turgid protoplast. (C) Dead cells from an RNAi line with shrunken protoplasts (double arrows denote detachment of plasma membranes from the cell walls) and ruptured plasma membranes (inset, arrow). (D) Frequency of cells with compromised plasma membranes assessed by SYTOX orange staining of meristematic (M; control line), suspensor (S; control line), and necrotic (N; RNAi line) cells. Data represent the means ± SEM from assessing all cells in ≥10 embryogenic structures per line. (E–G) Mitochondria in control and RNAi lines (exemplified by an ATG6 RNAi line). (E) Number of intact mitochondria per micrometer squared of the cytoplasm counted on the micrographs of meristematic, suspensor, and necrotic cells. Data represent the means ± SEM from three independent experiments, each including at least four cells per cell type. (F) Intact mitochondria (arrows) in a suspensor cell from the control line. (G) Swollen and degraded mitochondria (arrows) in a cell from an RNAi line. (H and I) Intracellular ATP content (H) and oxygen consumption (I). Data represent the means ± SEM from three independent experiments. *, P < 0.05; **, P < 0.01 (vs. control, Dunnett’s test). FW, fresh weight. (J and K) Cell vacuolization (J) and number of autophagosomes in the cytoplasm (K) estimated from the micrographs of meristematic (control line), suspensor (control line), and necrotic (ATG6 RNAi line) cells. Data represent the means ± SEM from three independent experiments, each including at least four cells per cell type. (L) Autophagosomes (open arrows) in the cytoplasm and autophagic bodies (closed arrows) in the vacuole of the embryo suspensor cell from a control line. (M and N) Quantification of necrotic death in control and RNAi lines after staining with FDA and FM4-64. (M) Staining patterns of EM and suspensor cells (control line) and necrotic cells (RNAi lines). DIC, differential interference contrast microscopy. (N) Frequency of necrotic cells. Data represent the means ± SEM from two independent experiments, each including two lines per genotype and ≥50 cells per line (P < 0.001; vs. control, Dunnett’s test). Different letters in D, E, J, and K indicate statistically different means (P < 0.001; Student’s t test). Bars: (A–C, main image) 5 µm; (C, inset) 0.5 µm; (F, G, and L) 2 µm; (M) 10 µm.
The main reason for the incomplete clearance of cellular contents in necrotic cells (Fig. 4 C) was compromised growth of lytic vacuoles, as evidenced from the measurements of the ratios of vacuolar to the whole protoplast areas on the electron micrographs (Fig. 4 J). The failure of progressive vacuolization of necrotic cells correlated with decreased autophagic activity in the cytoplasm (Fig. 4 K) compared with the suspensor cells from control lines in which enhanced autophagy sustained vacuole-mediated cell clearance (Fig. 4, K and L).
To quantify necrotic death in different lines, embryogenic cultures were stained with FM4-64 to distinguish between intact and shrunken protoplasts and FDA to assess cell viability. Combination of both dyes provides robust diagnostic tool for identification of necrotic cells (Fig. 4 M; Minina et al., 2013a). Although the frequency of necrotic cells (as distinguished by collapsed protoplast and no FDA staining) was negligible in the control lines, it increased to 60% in the individual RNAi lines (Fig. 4 N). No statistical differences were found between lines silenced for ATG5, ATG6, or mcII-Pa. Collectively, our data demonstrate that autophagy and metacaspase mcII-Pa are required for both progression of vacuolar cell death and suppression of necrosis.
Metacaspase lies upstream of autophagy in the vacuolar cell death pathway
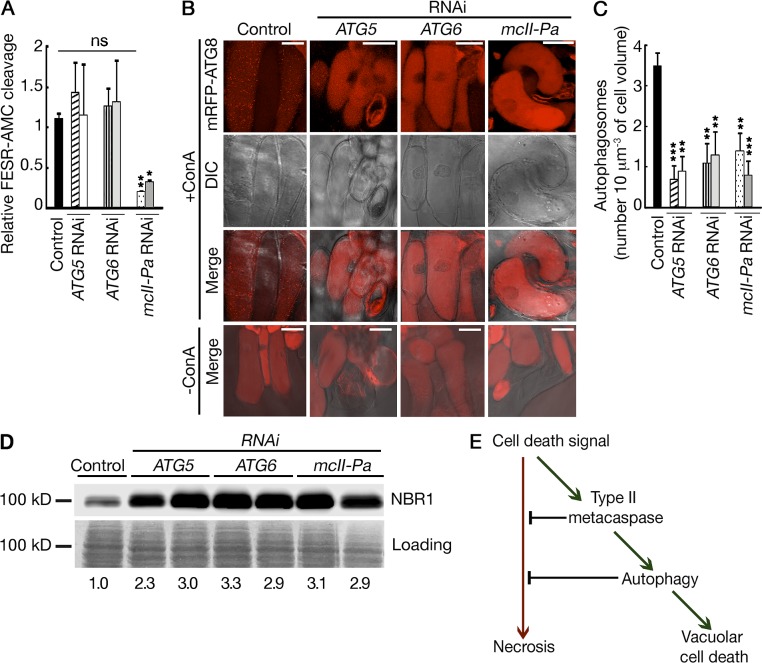
We have previously shown that the physiological effect of mcII-Pa in the control of suspensor PCD requires its proteolytic activity (Bozhkov et al., 2005b). To address whether metacaspase lies up- or downstream of autophagy in the vacuolar cell death pathway, we first compared proteolytic activity of mcII-Pa in the protein extracts from control, ATG5 RNAi, and ATG6 RNAi lines using three independent approaches: cleavage of peptidic substrate FESR–7-amino-4-methylcoumarin (AMC; Fig. 5 A), mcII-Pa autoprocessing, and cleavage of Tudor staphylococcal nuclease (TSN), a target of mcII-Pa (Fig. S3; Bozhkov et al., 2005b; Sundström et al., 2009). Because there was no difference in the levels of metacaspase activity observed between autophagy-deficient and control lines using all three approaches, we rejected the possibility that autophagy lies upstream of mcII-Pa in the pathway.
Figure 5.
Metacaspase lies upstream of autophagy in the vacuolar cell death pathway. (A) Proteolytic activity of mcII-Pa in control and RNAi lines measured using the FESR-AMC peptide. Data represent the means ± SEM from three independent experiments. The mean values were normalized to the control line. *, P < 0.05; **, P < 0.01 (vs. control, Dunnett’s test). (B) Localization of mRFP-Atg8 in dying cells from control and RNAi lines with and without ConA treatment. Bars, 50 µm. DIC, differential interference contrast microscopy. (C) Quantification of autophagosomes. Data represent the means ± SEM number of mRFP-Atg8–positive puncta counted for ≥10 cell volumes, 10 µm3 each, per line. **, P < 0.01; ***, P < 0.001 (vs. control, Dunnett’s test). (D) Western blot analysis of NBR1 degradation. Numbers shown below the blots correspond to the relative amount of NBR1. The integrated band intensities were first normalized to corresponding intensities of Coomassie staining (loading) and then to the control. (E) Schematic pathway showing the relationship between autophagy and metacaspase mcII-Pa in the regulation of vacuolar and necrotic modes of cell death.
To test an alternative scenario in which mcII-Pa is upstream of autophagy, we expressed mRFP-Atg8 in the wild-type (control) and RNAi lines and quantified the amount of autophagosomes per volume of dying cells by confocal microscopy after ConA treatment (Fig. 5, B and C). As shown in Fig. 5 C, silencing of mcII-Pa led to a dramatic decrease in the number of autophagosomes in the dying cells, to levels found in Atg5- and Atg6-deficient lines. Suppression of autophagy under mcII-Pa deficiency was further confirmed by a significant decrease in the efficiency of the degradation of NBR1 (Fig. 5 D). Together, these data establish metacaspase as an upstream mediator of autophagy in the vacuolar cell death pathway (Fig. 5 E).
Conclusions
Our combined data demonstrate a cell-autonomous requirement of autophagy and mcII-Pa for the execution of vacuolar cell death in the P. abies embryo suspensor, which in turn is essential for embryogenesis. Critical dependence of vacuolar death on autophagy and type II metacaspases seems to be independent of the experimental system because progression of xylem PCD (classic example of vacuolar cell death; van Doorn et al., 2011) in A. thaliana was shown to require autophagy (Kwon et al., 2010) and type II metacaspase AtMC9 (Bollhöner et al., 2013), although these two components have not been linked together in a pathway. The contribution of vacuolar cell death to developmental processes in different systems can vary. For example, the knockout mutants of ATG genes in A. thaliana are fertile and do not reveal massive embryo abortion caused by autophagy deficiency in the suspensor. The difference between A. thaliana and P. abies can be explained by the minute contribution of the seven-cell suspensor to the A. thaliana embryo in comparison to the giant suspensors of P. abies that exceed the EMs (Fig. 1 B). Furthermore, although the suspensor cells in P. abies do not divide and are subjected to PCD as soon as they are formed (Smertenko et al., 2003; Bozhkov et al., 2005a), the suspensor cells in A. thaliana undergo several rounds of divisions before PCD is initiated during transition from the late globular to the heart stages (Mansfield and Briarty, 1991; Kawashima and Goldberg, 2010; Wendrich and Weijers, 2013). This implies that in addition to size differences, the suspensor in P. abies embryos is composed entirely of dying and dead cells regardless of the developmental stage, whereas the fate of suspensor cells in A. thaliana is determined by the stage of embryo development.
Our work strengthens the idea about the evolutionarily conserved and context-independent role of autophagy in the suppression of necrosis and establishes a new paradigm for unraveling molecular mechanisms underlying relationships between two major classes of cell death in plants. In vacuolar cell death, autophagy is both an executioner of slow yet complete cell clearance and a protector against rapid cell death, necrosis, which leaves behind a largely unprocessed cell corpse. Autophagy was also shown to counteract the spread of necrotic lesions in plant leaves infected with microbial or fungal pathogens (Liu et al., 2005; Hofius et al., 2009; Yoshimoto et al., 2009; Lenz et al., 2011). Finally, in mammalian systems and Dictyostelium discoideum, autophagy differentially controls distinct modes of cell death and usually suppresses necrosis (Kosta et al., 2004; Degenhardt et al., 2006; Chen et al., 2008; Kroemer and Levine, 2008; Yuan and Kroemer, 2010; Shen et al., 2012). Interestingly, although autophagy-dependent vacuolar cell death in plants determines the cell fate (e.g., differentiation of the suspensor cells) and cannot be substituted by necrosis, the switch from autophagy-dependent to -independent cell death in mammals results in no major developmental defects (Golstein and Kroemer, 2005). This indicates that the role of autophagic cell death in development is not conserved throughout the evolution and can be redundant.
Our study shows that autophagy is an integral part of the suspensor cell death in P. abies; however, instead of being the effector mechanism of cell death, it performs cytoprotective role and orchestrates slow kinetics and morphological features of the vacuolar cell death. This demonstrates that the suspensor cells die with autophagy but not by autophagy (Kroemer and Levine, 2008). The primary and so far unknown cell death induction signal, as well as point of no return in the cell death pathway, lies apically to both autophagy and its upstream regulator metacaspase mcII-Pa (Fig. 5 E). A similar hierarchy was observed in PCD during Drosophila melanogaster oogenesis, in which caspases act upstream to autophagy (Hou et al., 2008). Further understanding of proteolytic processes mediated by evolutionarily related caspases and metacaspases is required to identify the molecular mechanisms activating autophagy during execution of PCD in animals and plants.
Materials and methods
Phylogenetic analysis of Atg5, Atg6, and metacaspase homologues
A search for proteins containing PF00656 (C14 peptidase), PF04106 (Apg5 domain), or PF04111 (Apg6 domain) was performed in Pfam database for species enlisted in the Table S1. An alternative search for proteins containing cl00042 (CASc superfamily) or cl04402 (APG5 superfamily) was performed using CDART (Conserved Domain Architecture Retrieval Tool) tool (Geer et al., 2002). Obtained hits were used for multiple alignments in ClustalW to confirm the presence of p20 and p10 domains in metacaspase homologues, Beclin-1 domain in Atg6 homologues, and Apg5 domain in Atg5 homologues. Estimated conserved sequence elements from the alignments were used as a query for additional search of homologues from gymnosperm species using TBLASTN tool in NCBI and ConGenIE databases. To confirm the absence of additional homologues, verified sequences for each individual species were cross-checked in proteome, transcriptome, and genome databases available at Phytozome, Ensembl, and ConGenIE. Multiple alignments of verified sequences were performed in ClustalW. Unrooted trees were constructed using the neighbor-joining method (Saitou and Nei, 1987) using the yeast homologue as an out group. Topology of each tree was confirmed by bootstrap analysis (2,000 repeats).
Cloning of hairpin and overexpression constructs
Full-length cDNA sequences of P. abies ATG5 (available from GenBank under accession no. HE793992.1) and ATG6 (GenBank accession no. HG326275) were obtained by rapid amplification of cDNA ends PCR using specific primers designed based on comparison of full-length or partial sequences of homologues from Oryza sativa, A. thaliana, Picea glauca, and Picea sitchensis. To produce the hairpin of ATG5, the corresponding part of ATG5 cDNA was amplified using primers hpATG5-P-sense/ATG5-reverse and hpATG5-M-antisense/ATG5-reverse (the sequences of all primers are listed in Table S2) and cut with BglII–AscI and PacI–BamHI (Thermo Fisher Scientific), respectively, producing 138–635-nucleoside triphosphate (ntp) and 138–847-ntp fragments of ATG5 coding DNA sequence (CDS). The fragments were ligated with T4 DNA ligase (Thermo Fisher Scientific) into a modified pAHC25 vector cut with PacI–AscI. The full-length hairpin was then amplified with primers attB1-pAHC25-forward/attB2-pAHC25-reverse and recombined into pDONR/Zeomycin (Zeo) vector (Invitrogen) followed by LR recombination into the pMDC32 vector (Curtis and Grossniklaus, 2003). To produce the hairpin of ATG6, the corresponding part of ATG6 cDNA was amplified using primers Beclin-AscI-sense/attB2-hpATG6 and attB1-hpATG6/Beclin-Ascl-Ant. Both PCR products were then cut with the AscI enzyme (Thermo Fisher Scientific) producing 689–1,298-ntp and 826–1,298-ntp fragments of ATG6 CDS, which were then ligated into pAHC25 and cloned into the pDONR/Zeo vector followed by recombination into pMDC32 vector as described for ATG5. To produce the hairpin of mcII-Pa, fragments of mcII-Pa cDNA (GenBank accession no. AJ534970.2) were amplified using primers attB1-hpmcII-Pa/as-hpmcII-Pa-EcoRI and s-hpmcII-Pa-EcoRI/attB2-hpmcII-Pa and cut with EcoRI (Thermo Fisher Scientific) producing 312–1,253-ntp and 537–1,253-ntp fragments of mcII-Pa CDS, respectively. The PCR products were then ligated, and the resulting construct was recombined into pDONR/Zeo vector followed by LR recombination into pMDC32 vector. Full-length CDS of ATG8 (IcIcomp43277_c0_seq1; Nystedt et al., 2013) was amplified using primers attB1-ATG8-Pa/attB2-ATG8-Pa, cloned into pDONR/Zeo vector and then into a modified pMDC32 vector containing mRFP. The GUS gene was amplified from the pAHC25 plasmid (Christensen and Quail, 1996) using primers attB1-GUS-forward/attB2-GUS-forward and cloned into the pDONR/Zeo vector and then into the pMDC32 vector.
P. abies embryogenesis system
P. abies embryogenic cell lines were subcultured every 7 d by transferring ∼3 ml of the cell pellet into 100 ml of fresh half-strength LP medium and incubated at 22°C in the darkness on a shaker at 120 rpm as described previously (Filonova et al., 2000; Bozhkov et al., 2002; Minina et al., 2013a). Early embryogenesis was induced by omitting GF auxin and cytokinin from the liquid medium (−GF medium) for 7 d. To assess the efficiency of early embryogenesis, 5 ml of 7-d-old −GF culture was stained with Evans blue, and the percentage of polarized and aberrant embryos was counted. To assess the efficiency of late embryogenesis, 7-d-old −GF suspension cultures were plated on filter papers placed on the top of solidified maturation medium containing ABA (Filonova et al., 2000) and incubated in the darkness at 22°C. Fully developed cotyledonary embryos were scored in week 4 and 6 of ABA treatment.
Transgenic lines
Cell lines expressing mRFP-Atg8 in ATG5 RNAi, ATG6 RNAi, mcII-Pa RNAi, or control (expressing GUS) background were established by agrotransformation of P. abies wild-type embryogenic cell line 06.28.05. The constructs were transformed into the Agrobacterium tumefaciens c58c1 strain carrying additional virulence plasmid pTOK47. Transformed bacteria were then grown overnight in YEP (yeast extract peptone) medium supplemented with 100 mg/liter rifampicin, 5 mg/liter tetracycline, 100 mg/liter carbenicillin, and 50 mg/liter kanamycin. Cells were centrifuged at 4,500 g for 5 min and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, pH 5.5, and 150 µM acetosyringone). The OD600 of the cultures was adjusted to 10 with the infiltration buffer. A. tumefaciens transformed with mRFP-Atg8 was added in a 1:1 ratio to the agrobacterial cultures transformed with GUS or RNAi constructs and incubated at room temperature for 1 h. 1 ml of the A. tumefaciens mixture was added to 5 ml of a cell pellet of 7-d-old suspension culture grown in the presence of GF. The volume of the mixture was adjusted to 10 ml with GF-containing medium (+GF medium), and acetosyringone was added to a final concentration at 150 µM. After 4 h, the cells were plated on a filter paper placed on the top of solidified +GF medium and incubated at room temperature in the darkness for 48 h. Then, filters were transferred on +GF plates containing 400 mg/ml timentin and 250 mg/ml cefotaxime and incubated under the same conditions for 5 d. Subsequently, filter papers were transferred onto fresh +GF plates containing 20 µg/ml hygromycin, 400 µg/ml timentin, and 250 µg/ml cefotaxime and subcultured onto the same medium every week. The transgenic calli were picked from the plates after 3 wk. The presence of hairpins was verified by PCR with genomic DNA. The genomic DNA was extracted using a modified hexadecyltrimethylammonium bromide method as previously described (Ciavatta et al., 2002) and used for PCR with primers attB1/PaATG5-forward, attB1/Beclin-AscI-antisense, and attB1/as-hpmcII-Pa-EcoRI for ATG5 RNAi, ATG6 RNAi, and mcII-Pa RNAi lines. Absence of A. tumefaciens in the cultures was confirmed by performing PCR with picA-forward/picA-reverse and VirG-forward/VirG-reverse primers. GUS expression was confirmed by a histochemical assay as previously described by Jefferson et al. (1987). mRFP-Atg8 expression was confirmed by detecting fluorescence using a confocal microscope (LSM 780; Carl Zeiss), with excitation of 561 nm and emission of 563–686 nm. The wild-type line and lines expressing both GUS and mRFP-Atg8 were used as controls.
Culture staining and confocal microscopy
All imaging was performed at room temperature. Samples were mounted in aqueous solutions. 5 ml of 7-d-old −GF suspension cultures were incubated with 0.0025% (wt/vol) Evans blue solution for 10 min on the bench, transferred onto a 50-µm nylon mesh, and washed twice with −GF medium. For quantification of normal and aberrant cell structures, the cultures were embedded in 6% (wt/vol) low melting temperature agarose. Phenotypes were assessed using an inverted microscope (5× Achrostigmat and 10× Plan-Neofluar objectives with numerical apertures 0.12 and 0.3, respectively; Axiovert 10; Carl Zeiss), and pictures were taken using a camera (DFC295; Leica) and LAS AF v3.2 software (Leica). For measuring the length of dying cells, an aliquot of stained embryos was transferred onto a glass slide, and 10 embryogenic structures per line were imaged using z-stack and tile-scan functions of a confocal microscope (LSM 780) and a 20× Plan-Apochromat objective with numerical aperture of 0.8. The length of the cells was measured using ZEN 2011 blue software (Carl Zeiss), and ≥50 cells per structure were analyzed.
FM4-64, FDA, and DAPI triple staining was performed on 7-d-old suspension cultures grown without GF. DAPI and FDA were added to final concentration 4 and 2 µg/ml, respectively. After 15 min, the samples were washed twice with −GF medium on a 50-µm nylon mesh. Then, 1 µM FM4-64 was added, and samples were scanned within 10 min using the sequential scanning mode of a confocal (LSM 780) microscope, with excitation of 405/488/561 nm, emission of 410–492/490–587/582–754 nm, and a 20× objective with numerical aperture of 0.8.
Before detection of mRFP-Atg8, samples were treated with 0.5 µM ConA (Fluka) for 14 h. If required, samples were additionally incubated in 2 µg/ml FDA for 15 min, washed twice with fresh −GF medium, and scanned within 15 min after staining. Fluorescence was detected using the sequential scanning mode of a confocal microscope (LSM 780), with excitation of 561 nm and emission of 563–686 nm for RFP and excitation of 488 nm and emission of 490–587 nm for FDA and an objective of 40× water Plan-Apochromat with numerical aperture of 1.2. A 10-µm-thick z stack was used to produce a maximal projection image, and mRFP puncta were quantified on a randomly selected 10-µm2 area in ZEN 2011 (black edition). The mean intensity of FDA was assessed by selecting at least three 10-µm2 areas per cell using ZEN 2012 blue edition, and autophagosomes were counted in the selected areas.
SYTOX orange staining was performed on 7-d-old suspension cultures grown without GF. SYTOX orange and FDA were added to final concentration 1 µM and 2 µg/ml, respectively. After 15 min, the samples were washed twice with −GF medium on a 50-µm nylon mesh, and samples were scanned within 10 min using the sequential scanning mode of a confocal microscope (LSM 780), with excitation of 488/561 nm, emission of 490–587/582–754 nm, and objectives of 10 and 20× Plan-Apochromat with numerical apertures of 0.45 and 0.8, respectively.
TEM
Samples for TEM were fixed in 2% of paraformaldehyde, 3% glutaraldehyde, and 100 mM sodium cacodylate, pH 7.2, for 3 h at room temperature followed by overnight fixation at 4°C. Fixed samples were washed three times in 100 mM sodium cacodylate, pH 7.2, and postfixed in 1% OsO4 aqueous solution for 1 h at room temperature. Postfixed samples were gradually dehydrated in a series of ethanol solutions from 25 to 100% and infiltrated with LR white resin as previously described in Minina et al. (2013a), the resin was cured at 65°C overnight and cut to obtain 99-nm-thick sections. Sections were examined using a transmission electron microscope (CM12; Philips), and images were recorded on film (4489; Kodak). Films were then scanned with a resolution of 3,200 pixels/inch and processed using Photoshop CS5 extended (Adobe). Raw data were rescaled according to magnification used for each image.
ATP content and oxygen consumption measurement
7-d-old suspension cultures grown in −GF medium were pelleted on a sterile 50-µm mesh and washed three times with fresh growth medium to remove extracellular ATP. Cells were ground in liquid nitrogen, and ATP was extracted by boiling 100 mg of the powdered material in 150 µl of boiling buffer (100 mM Tris, 4 mM EDTA, and phosphatase inhibitors, pH 7.5). Debris from the samples was removed by centrifugation in a table centrifuge for 20 min at maximum speed at 4°C. Supernatants were transferred into a 96-well plate, and luminescence intensity was measured upon adding the reaction buffer (50 mM Tris HCl, 1 mM EDTA, 2 mM MgSO4, 7.5 mM DTT, 100 µM d-luciferin, and 5 µg/ml luciferase from Photinus pyralis) in the plate reader (Omega FLUOstar; BMG Labtech). The raw data were then normalized using a standard curve as previously described in Minina et al. (2013a).
7-d-old suspension cultures incubated in −GF medium were gently pelleted by centrifugation and resuspended in 1.6 vol of fresh medium. 300 µl of the cell suspension was used to measure oxygen consumption rate (OCR) in a liquid-phase oxygraph (Oxygraph Plus System; Hansatech Instruments) in the absence (OCRnormal) or presence of 10 µM carbonyl cyanide m-chlorophenyl hydrazone (OCRdecoupled) or 1 mM KCN (OCRrespiration block). Respiratory control ratio was calculated as previously described in Minina et al. (2013a).
Metacaspase activity assay
Cells from 7-d-old −GF suspension cultures were filtered and ground in liquid nitrogen. The total protein was extracted using HNCD buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 0.1% CHAPS, 5 mM DTT, 100 µM PMSF, and 10 µM pepstatin). Fluorogenic substrate FESR-AMC (final concentration of 50 µM; Sundström et al., 2009) was added to 7.5 µg of total protein in the presence or absence of 50 mM CaCl2. Accumulation of fluorescence was detected at excitation/emission of 355/460 nm and converted into picomoles of AMC released per minute per milligram of total protein. Values obtained for reactions without calcium were subtracted from values obtained in the presence of 50 mM CaCl2. Results were then normalized relative to control.
Polyclonal antiserum and immunoblotting
Antibody production in rabbits was performed according to Canton et al. (1996). A total of 100 µg of recombinant mcII-Pa protein mixed 1:1 with Freund’s complete adjuvant was used for the first immunization. The second boost was performed after 40 d, in which incomplete adjuvant was used instead of the complete version. The antibody was affinity purified using a polyvinylidene difluoride membrane containing recombinant metacaspase.
7-d-old −GF suspension cultures were ground in liquid nitrogen, and total protein was extracted in HNCD buffer, mixed 1:1 with 2× Laemmli buffer (Laemmli, 1970), and boiled for 3 min. For Atg8 detection, extraction was performed in 50 mM Tris, pH 8.0, 150 mM NaCl, 1 mM PMSF, and protease cocktail (cOmplete; Roche), and debris from the samples was removed by spinning down at 7,000 g for 5 min at 4°C followed by filtration through four layers of Miracloth. Cleared supernatants were ultracentrifuged for 1 h at 100,000 g at 4°C as previously described in Chung et al. (2010). Pellets from ultracentrifugation were resuspended in extraction buffer containing 0.5% Triton X-100. Proteins were separated on a 12% polyacrylamide gel (15% polyacrylamide gel with 6 M urea for Atg8) and transferred on a polyvinylidene difluoride membrane. The membrane was probed with antibodies against NBR1 (kindly provided by T. Johansen, University of Tromsø, Tromsø, Norway; Svenning et al., 2011), Atg8 (Abcam), mRFP (Chromatec), actin (Santa Cruz Biotechnology, Inc.), mcII-Pa, or TSN (Sundström et al., 2009) diluted 1:1,000 followed by anti–rabbit, anti–rat, or anti–mouse HRP conjugates (GE Healthcare) diluted 1:3,000. The membranes were incubated with ECL Prime kit (GE Healthcare), and the signal was imaged using the luminescent image analyzer (LAS-3000; Fujifilm). To check protein loading, membranes were stained with Coomassie brilliant blue G-250.
Real-time quantitative PCR (qPCR)
Total RNA was extracted from 7-d-old suspension cultures grown without GF. 1 µg RNA was used per real-time reaction with the Maxima kit (Fermentas), and 1/80th of each real-time reaction was used for qPCR analysis. qPCR reactions were performed using PCR thermal cycler (iQ5; Bio-Rad Laboratories) and qPCR kit (DyNAmo Flash SYBR Green; Finnzymes). Primers used for qPCR are listed in Table S2. ΔΔCT method was used to quantify fold expression of genes of interest in the samples, with two genes, cell division control 2 and phosphoglucomutase, previously reported to show stable expression during early embryo development of P. abies (Vestman et al., 2011), being used for normalization.
Statistical analysis
All statistical analysis was performed in JMP version 10.1 (SAS Institute) using either a pairwise Student’s t test or comparison with control by Dunnett’s test.
Online supplemental material
Fig. S1 shows phylogenetic analysis of Atg5, Atg6, and metacaspase homologues from protozoa, fungi, plants, and animals, highlighting the position of P. abies homologues. Fig. S2 shows decreased expression of ATG5, ATG6, and mcII-Pa in RNAi lines determined by Western blotting (for mcII-Pa) and real-time qPCR (for all three genes). Fig. S3 supports the data presented in Fig. 5 A and shows no effect of the suppression of autophagy on mcII-Pa activity using mcII-Pa and TSN (proteolytic target of mcII-Pa) Western blot analyses. Table S1 presents a list of proteins used for phylogenetic analysis shown in Fig. S1. Table S2 shows primers used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201307082/DC1.
Supplementary Material
Acknowledgments
We thank A. Rakhmaninova, V. Kaminskyy, and J.S. Kim for help with phylogenetic analysis, confocal microscopy, and TEM, respectively, and T. Johansen for the kind gift of anti-NBR1. We also wish to thank E.-L. Eskelinen for fruitful discussions of TEM results.
This work was supported by grants from the Swedish Research Council (to P.V. Bozhkov), Pehrssons Fund (to P.V. Bozhkov), Knut and Alice Wallenberg Foundation (to P.V. Bozhkov), the Swedish Foundation for Strategic Research (to P.V. Bozhkov), August T. Larsson Foundation (to A. Smertenko and P.V. Bozhkov), Olle Engkvist Foundation (to P.V. Bozhkov), and the Spanish Ministry of Science and Innovation (AGL2010-15684; to M.F. Suarez). V. Sanchez-Vera was a recipient of a FPI fellowship from the Spanish Ministry of Science and Innovation (BES-2008-003592).
Footnotes
Abbreviations used in this paper:
- ABA
- abscisic acid
- AMC
- 7-amino-4-methylcoumarin
- CDS
- coding DNA sequence
- ConA
- concanamycin A
- EM
- embryonal mass
- FDA
- fluorescein diacetate
- GF
- growth factor
- ntp
- nucleoside triphosphate
- PCD
- programmed cell death
- qPCR
- quantitative PCR
- TEM
- transmission electron microscopy
- TSN
- Tudor staphylococcal nuclease
References
- Bal-Price A., Brown G.C. 2000. Nitric-oxide-induced necrosis and apoptosis in PC12 cells mediated by mitochondria. J. Neurochem. 75:1455–1464 10.1046/j.1471-4159.2000.0751455.x [DOI] [PubMed] [Google Scholar]
- Beers E.P. 1997. Programmed cell death during plant growth and development. Cell Death Differ. 4:649–661 10.1038/sj.cdd.4400297 [DOI] [PubMed] [Google Scholar]
- Beers E.P., McDowell J.M. 2001. Regulation and execution of programmed cell death in response to pathogens, stress and developmental cues. Curr. Opin. Plant Biol. 4:561–567 10.1016/S1369-5266(00)00216-8 [DOI] [PubMed] [Google Scholar]
- Bollhöner B., Prestele J., Tuominen H. 2012. Xylem cell death: emerging understanding of regulation and function. J. Exp. Bot. 63:1081–1094 10.1093/jxb/err438 [DOI] [PubMed] [Google Scholar]
- Bollhöner B., Zhang B., Stael S., Denancé N., Overmyer K., Goffner D., Van Breusegem F., Tuominen H. 2013. Post mortem function of AtMC9 in xylem vessel elements. New Phytol. 200:498–510 10.1111/nph.12387 [DOI] [PubMed] [Google Scholar]
- Bozhkov P.V., Filonova L.H., von Arnold S. 2002. A key developmental switch during Norway spruce somatic embryogenesis is induced by withdrawal of growth regulators and is associated with cell death and extracellular acidification. Biotechnol. Bioeng. 77:658–667 10.1002/bit.10228 [DOI] [PubMed] [Google Scholar]
- Bozhkov P.V., Filonova L.H., Suarez M.F. 2005a. Programmed cell death in plant embryogenesis. Curr. Top. Dev. Biol. 67:135–179 10.1016/S0070-2153(05)67004-4 [DOI] [PubMed] [Google Scholar]
- Bozhkov P.V., Suarez M.F., Filonova L.H., Daniel G., Zamyatnin A.A., Jr, Rodriguez-Nieto S., Zhivotovsky B., Smertenko A. 2005b. Cysteine protease mcII-Pa executes programmed cell death during plant embryogenesis. Proc. Natl. Acad. Sci. USA. 102:14463–14468 10.1073/pnas.0506948102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton F.R., Garcia-Gutierrez A., Crespillo R., Cánovas F.M. 1996. High-level expression of Pinus sylvestris glutamine synthetase in Escherichia coli. Production of polyclonal antibodies against the recombinant protein and expression studies in pine seedlings. FEBS Lett. 393:205–210 10.1016/0014-5793(96)00886-1 [DOI] [PubMed] [Google Scholar]
- Chen Y., McMillan-Ward E., Kong J., Israels S.J., Gibson S.B. 2008. Oxidative stress induces autophagic cell death independent of apoptosis in transformed and cancer cells. Cell Death Differ. 15:171–182 10.1038/sj.cdd.4402233 [DOI] [PubMed] [Google Scholar]
- Christensen A.H., Quail P.H. 1996. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5:213–218 10.1007/BF01969712 [DOI] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D. 2010. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 62:483–493 10.1111/j.1365-313X.2010.04166.x [DOI] [PubMed] [Google Scholar]
- Ciavatta V.T., Egertsdotter U., Clapham D., von Arnold S., Cairney J. 2002. A promoter from the loblolly pine PtNIP1;1 gene directs expression in an early-embryogenesis and suspensor-specific fashion. Planta. 215:694–698 10.1007/s00425-002-0822-5 [DOI] [PubMed] [Google Scholar]
- Coll N.S., Epple P., Dangl J.L. 2011. Programmed cell death in the plant immune system. Cell Death Differ. 18:1247–1256 10.1038/cdd.2011.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133:462–469 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gélinas C., Fan Y., et al. 2006. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 10:51–64 10.1016/j.ccr.2006.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. 2002. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277:33105–33114 10.1074/jbc.M204630200 [DOI] [PubMed] [Google Scholar]
- Eguchi Y., Shimizu S., Tsujimoto Y. 1997. Intracellular ATP levels determine cell death fate by apoptosis or necrosis. Cancer Res. 57:1835–1840 [PubMed] [Google Scholar]
- Filonova L.H., Bozhkov P.V., Brukhin V.B., Daniel G., Zhivotovsky B., von Arnold S. 2000. Two waves of programmed cell death occur during formation and development of somatic embryos in the gymnosperm, Norway spruce. J. Cell Sci. 113:4399–4411 [DOI] [PubMed] [Google Scholar]
- Galluzzi L., Vitale I., Abrams J.M., Alnemri E.S., Baehrecke E.H., Blagosklonny M.V., Dawson T.M., Dawson V.L., El-Deiry W.S., Fulda S., et al. 2012. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 19:107–120 10.1038/cdd.2011.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer L.Y., Domrachev M., Lipman D.J., Bryant S.H. 2002. CDART: protein homology by domain architecture. Genome Res. 12:1619–1623 10.1101/gr.278202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golstein P., Kroemer G. 2005. Redundant cell death mechanisms as relics and backups. Cell Death Differ. 12(Suppl. 2):1490–1496 10.1038/sj.cdd.4401607 [DOI] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. 2002. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129:1181–1193 10.1104/pp.011024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath M.C. 2000. Hypersensitive response-related death. Plant Mol. Biol. 44:321–334 10.1023/A:1026592509060 [DOI] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H., Mattsson O., Jørgensen L.B., Jones J.D., Mundy J., Petersen M. 2009. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 137:773–783 10.1016/j.cell.2009.02.036 [DOI] [PubMed] [Google Scholar]
- Hou Y.C., Chittaranjan S., Barbosa S.G., McCall K., Gorski S.M. 2008. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J. Cell Biol. 182:1127–1139 10.1083/jcb.200712091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6:3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M. 2001. Programmed cell death in development and defense. Plant Physiol. 125:94–97 10.1104/pp.125.1.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T., Goldberg R.B. 2010. The suspensor: not just suspending the embryo. Trends Plant Sci. 15:23–30 10.1016/j.tplants.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A., et al. 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 8:445–544 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Aravind L. 2002. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 9:394–404 10.1038/sj.cdd.4400991 [DOI] [PubMed] [Google Scholar]
- Kosta A., Roisin-Bouffay C., Luciani M.F., Otto G.P., Kessin R.H., Golstein P. 2004. Autophagy gene disruption reveals a non-vacuolar cell death pathway in Dictyostelium. J. Biol. Chem. 279:48404–48409 10.1074/jbc.M408924200 [DOI] [PubMed] [Google Scholar]
- Kroemer G., Levine B. 2008. Autophagic cell death: the story of a misnomer. Nat. Rev. Mol. Cell Biol. 9:1004–1010 10.1038/nrm2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S.I., Cho H.J., Jung J.H., Yoshimoto K., Shirasu K., Park O.K. 2010. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J. 64:151–164 [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Lam E. 2004. Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Biol. 5:305–315 10.1038/nrm1358 [DOI] [PubMed] [Google Scholar]
- Lenz H.D., Haller E., Melzer E., Kober K., Wurster K., Stahl M., Bassham D.C., Vierstra R.D., Parker J.E., Bautor J., et al. 2011. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66:818–830 10.1111/j.1365-313X.2011.04546.x [DOI] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. 2012. Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63:215–237 10.1146/annurev-arplant-042811-105441 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Tallóczy Z., Levine B., Dinesh-Kumar S.P. 2005. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 121:567–577 10.1016/j.cell.2005.03.007 [DOI] [PubMed] [Google Scholar]
- Majno G., Joris I. 1995. Apoptosis, oncosis, and necrosis. An overview of cell death. Am. J. Pathol. 146:3–15 [PMC free article] [PubMed] [Google Scholar]
- Mansfield S.G., Briarty L.G. 1991. Early embryogenesis in Arabidopsis thaliana. 2. The developing embryo. Can. J. Bot. 69:461–476 10.1139/b91-063 [DOI] [Google Scholar]
- Minina E.A., Filonova L.H., Daniel G., Bozhkov P.V. 2013a. Detection and measurement of necrosis in plants. Methods Mol. Biol. 1004:229–248 10.1007/978-1-62703-383-1_17 [DOI] [PubMed] [Google Scholar]
- Minina E.A., Sanchez-Vera V., Moschou P.N., Suarez M.F., Sundberg E., Weih M., Bozhkov P.V. 2013b. Autophagy mediates caloric restriction-induced lifespan extension in Arabidopsis. Aging Cell. 12:327–329 10.1111/acel.12048 [DOI] [PubMed] [Google Scholar]
- Nystedt B., Street N.R., Wetterbom A., Zuccolo A., Lin Y.C., Scofield D.G., Vezzi F., Delhomme N., Giacomello S., Alexeyenko A., et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature. 497:579–584 10.1038/nature12211 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- Shen S., Kepp O., Kroemer G. 2012. The end of autophagic cell death? Autophagy. 8:1–3 10.4161/auto.8.1.16618 [DOI] [PubMed] [Google Scholar]
- Singh H. 1978. Embryology of gymnosperms. Handbuch der Pflanzenanatomie. Vol. 10 Zimmermann W., Carlquist Z., Ozenda P., Wulff H.D., Gebrüder Borntraeger, Berlin: 187–241 [Google Scholar]
- Smertenko A.P., Bozhkov P.V., Filonova L.H., von Arnold S., Hussey P.J. 2003. Re-organisation of the cytoskeleton during developmental programmed cell death in Picea abies embryos. Plant J. 33:813–824 10.1046/j.1365-313X.2003.01670.x [DOI] [PubMed] [Google Scholar]
- Suarez M.F., Filonova L.H., Smertenko A., Savenkov E.I., Clapham D.H., von Arnold S., Zhivotovsky B., Bozhkov P.V. 2004. Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Curr. Biol. 14:R339–R340 10.1016/j.cub.2004.04.019 [DOI] [PubMed] [Google Scholar]
- Sundström J.F., Vaculova A., Smertenko A.P., Savenkov E.I., Golovko A., Minina E., Tiwari B.S., Rodriguez-Nieto S., Zamyatnin A.A., Jr, Välineva T., et al. 2009. Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat. Cell Biol. 11:1347–1354 10.1038/ncb1979 [DOI] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., Johansen T. 2011. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 7:993–1010 10.4161/auto.7.9.16389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatsiani L., Van Breusegem F., Gallois P., Zavialov A., Lam E., Bozhkov P.V. 2011. Metacaspases. Cell Death Differ. 18:1279–1288 10.1038/cdd.2011.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn W.G., Woltering E.J. 2005. Many ways to exit? Cell death categories in plants. Trends Plant Sci. 10:117–122 10.1016/j.tplants.2005.01.006 [DOI] [PubMed] [Google Scholar]
- van Doorn W.G., Beers E.P., Dangl J.L., Franklin-Tong V.E., Gallois P., Hara-Nishimura I., Jones A.M., Kawai-Yamada M., Lam E., Mundy J., et al. 2011. Morphological classification of plant cell deaths. Cell Death Differ. 18:1241–1246 10.1038/cdd.2011.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestman D., Larsson E., Uddenberg D., Cairney J., Clapham D., Sundberg E., von Arnold S. 2011. Important processes during differentiation and early development of somatic embryos of Norway spruce as revealed by changes in global gene expression. Tree Genet. Genomes. 7:347–362 10.1007/s11295-010-0336-4 [DOI] [Google Scholar]
- Wendrich J.R., Weijers D. 2013. The Arabidopsis embryo as a miniature morphogenesis model. New Phytol. 199:14–25 10.1111/nph.12267 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. 2009. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 21:2914–2927 10.1105/tpc.109.068635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Kroemer G. 2010. Alternative cell death mechanisms in development and beyond. Genes Dev. 24:2592–2602 10.1101/gad.1984410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.