The kinetochore scaffold KNL1 contains an extensive array of short linear sequence modules that each independently can localize BUB1 but that together provide enhanced efficiency of chromosome segregation.
Abstract
Fidelity of chromosome segregation relies on coordination of chromosome biorientation and the spindle checkpoint. Central to this is the kinetochore scaffold KNL1 that integrates the functions of various mitotic regulators including BUB1 and BUBR1. We show that KNL1 contains an extensive array of short linear sequence modules that encompass TxxΩ and MELT motifs and that can independently localize BUB1. Engineered KNL1 variants with few modules recruit low levels of BUB1 to kinetochores but support a robust checkpoint. Increasing numbers of modules concomitantly increase kinetochore BUB1 levels and progressively enhance efficiency of chromosome biorientation. Remarkably, normal KNL1 function is maintained by replacing all modules with a short array of naturally occurring or identical, artificially designed ones. A minimal array of generic BUB recruitment modules in KNL1 thus suffices for accurate chromosome segregation. Widespread divergence in the amount and sequence of these modules in KNL1 homologues may represent flexibility in adapting regulation of mitotic processes to altered requirements for chromosome segregation during evolution.
Introduction
Equal distribution of the replicated genome during mitosis is essential for accurate propagation of genetic information and the maintenance of healthy tissues. Large multiprotein complexes known as kinetochores perform several essential functions in this process (Cheeseman and Desai, 2008; Foley and Kapoor, 2013). These include generating and maintaining physical attachment between chromatids and microtubules of the mitotic spindle, and signaling to the spindle assembly checkpoint (SAC, also known as the mitotic checkpoint) when kinetochores are unbound by microtubules. Such checkpoint signaling involves production of a diffusible inhibitor of anaphase onset (Chao et al., 2012; Vleugel et al., 2012).
Chromosome biorientation as well as SAC activity critically rely on the kinetochore scaffold KNL1/CASC5/AF15q14/Blinkin (hereafter referred to as KNL1; Cheeseman et al., 2006, 2008; Kiyomitsu et al., 2007). This long, largely unstructured protein is a member of the KNL1/MIS12 complex/NDC80 complex (KMN) network that constitutes the microtubule-binding site of kinetochores (Cheeseman and Desai, 2008). KNL1 itself directly contributes to this through its N-terminal microtubule-binding region (Welburn et al., 2010; Espeut et al., 2012), but also by localizing the paralogues BUB1 and BUBR1 to kinetochores. The pseudokinase BUBR1 (Suijkerbuijk et al., 2012a) is a component of the mitotic checkpoint complex (Chao et al., 2012) and additionally binds the PP2A-B56 phosphatase that is required for stabilizing kinetochore–microtubule interactions (Foley et al., 2011; Suijkerbuijk et al., 2012b; Kruse et al., 2013; Xu et al., 2013). BUB1, in turn, promotes efficient chromosome biorientation by localizing the Aurora B kinase to inner centromere regions via phosphorylation of H2A-T120 (Kawashima et al., 2010; Yamagishi et al., 2010). Its contribution to checkpoint signaling, although important, is not entirely clear (Tang et al., 2004; Klebig et al., 2009).
Although recruitment of BUB1 and BUBR1 (the BUBs) to kinetochores is critical for error-free chromosome segregation, the mechanism by which KNL1 accomplishes this is unknown. Both BUBs directly interact via their conserved TPR domains with two so-called KI motifs in the N-terminal 250 amino acids of human KNL1 (Bolanos-Garcia and Blundell, 2011; Kiyomitsu et al., 2011; Krenn et al., 2012). These interactions may, however, not be required for BUB1/BUBR1 kinetochore localization (Krenn et al., 2012), and the KI motifs are not apparent in nonvertebrate eukaryotic KNL1 homologues (Vleugel et al., 2012). In contrast, kinetochore binding of at least BUB1 relies on MPS1-mediated phosphorylation of the threonine within MELT-like sequences of KNL1 in humans and yeasts (Shepperd et al., 2012; London et al., 2012; Yamagishi et al., 2012). Such MELT-like sequences can be identified in numerous KNL1 homologues (Vleugel et al., 2012).
In this study, we set out to investigate the mode of BUB recruitment to kinetochores, and show that KNL1 is an assembly of previously unrecognized repeating modules. These modules operate in a generic fashion to recruit sufficient BUB proteins to kinetochores to ensure high-fidelity chromosome segregation.
Results
The N-terminal MDLT-KI module in KNL1 independently recruits BUB proteins
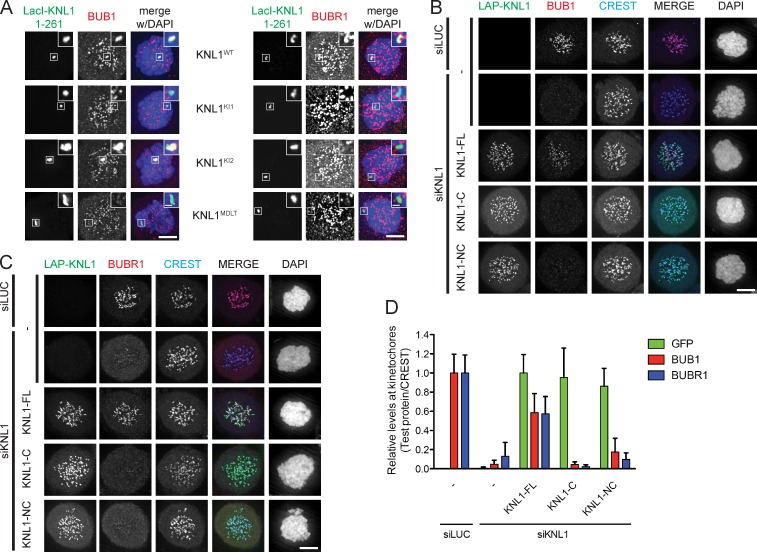
BUB1 and BUBR1 directly bind to KI motifs (KI1 and KI2) that are located near the N terminus of KNL1 (Bolanos-Garcia and Blundell, 2011; Kiyomitsu et al., 2011; Krenn et al., 2012). Their localization to kinetochores additionally requires MPS1-dependent phosphorylation of MELT-like sequences (London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012), although it is unknown which of these sequences are phosphorylated and which ones are important for BUB recruitment and KNL1 function. Because one such MELT-like sequence (MDLT) is located close to the two KI motifs, we examined whether the N-terminal region (1–261) of KNL1 encompassing MDLT-KI1-KI2 is sufficient to bind BUB1 and BUBR1. To this end, the KNL1 fragment was fused to LacI and tethered to an ectopic Lac operator (LacO) array that is stably integrated in the short arm of chromosome 1, distant to the centromere (1p36) in U2OS cells (Fig. S1 A; Janicki et al., 2004). LacI-LAP-KNL11–261 recruited endogenous BUB1 and BUBR1 to the LacO array in mitotic cells. (Fig. 1 A; Fig. S1 B). This required the MDLT and KI1 sequences because mutation of these motifs (MDLT to MDLA [KNL1MDLT] or KIDTTSF to KIDATSA [KNL1KI1]; Krenn et al., 2012) prevented both BUBs from localizing to the LacO array (Fig. 1 A; Fig. S1 B). In addition, BUBR1 but not BUB1 localization was also lost after mutating the KI2 motif (KIDFNDF to KIDANDA [KNL1KI2]; Bolanos-Garcia et al., 2011; Krenn et al., 2012). Thus, at least in the context of the ectopic KNL1 fragment, BUBR1 recruitment to KNL1 is dependent on all three motifs (Fig. 1 A; Fig. S1 B).
Figure 1.
The N-terminal MDLT-KI module in KNL1 independently recruits BUB proteins. (A) Immunolocalization of BUB1 (left panels, red) and BUBR1 (right panels, red) in LacI-LAP-KNL11–261–transfected, nocodazole-treated U2OS-LacO cells. LacI-LAP-KNL11–261 is shown in green and DNA (DAPI) is in blue. Insets show magnifications of the boxed regions. KNL1KI1 denotes LacI-LAP-KNL11–261 in which KIDTTSF is mutated to KIDATSA, KNL1KI2 is KIDFNDF mutated to KIDANDA, and KNL1MDLT mutated is MDLT to MDLA. Bars, 5 µm (insets, 0.5 µm). (B–D) Representative images (B and C) and quantification (D) of LAP-KNL1–expressing Flp-in HeLa cells transfected with siRNAs to luciferase (siLUC) or to KNL1 (siKNL1) and treated with nocodazole. LAP-KNL1 is shown in green, BUB proteins in red, centromeres (CREST) in blue, and DNA (DAPI) in white. Bars, 5 µm. Quantification in D shows total kinetochore signal intensity (+SD) of LAP-KNL1 and BUB proteins over CREST. Data are from >15 cells and representative of 3 experiments. Levels of kinetochore BUBs in control cells and of kinetochore LAP-KNL1 in KNL1-FL–expressing cells are set to 1.
The N-terminal MDLT-KI module in KNL1 is sufficient to support SAC activity but not chromosome biorientation
To next assess the contribution of the N-terminal module to KNL1’s function in the SAC and chromosome biorientation, we generated a LAP-tagged KNL1 variant in which this region was directly fused to the C-terminal kinetochore localization domain of KNL1 (aa 1834–2342: generating KNL1-NC; Fig. S1 C). This ensured maintenance of proper KMN network integrity, KNL1 position on the outer kinetochore, and Zwint-1 and HP1 kinetochore localization (Kiyomitsu et al., 2007; Petrovic et al., 2010). Full-length KNL1 (KNL1-FL) and the C-terminal domain of KNL1 (KNL1-C) were used as controls. To ensure comparable genetic background and expression levels, siRNA-resistant KNL1 variants were expressed from a doxycycline-inducible promoter at a single integration site in HeLa cells (Klebig et al., 2009). All KNL1 variants efficiently incorporated into the outer kinetochore to similar levels, as judged by immunofluorescence (Fig. 1, B–D). Functionality of these proteins was assayed by their ability to restore KNL1 function upon siRNA-mediated depletion of endogenous KNL1. Depletion of KNL1 removed BUB1 and BUBR1 from kinetochores (Fig. 1, B–D), and this was restored by expression of KNL1-FL and weakly by KNL1-NC, but not by KNL1-C (Fig. 1, B–D). In support of this, comparative proteomics analysis of LAP-KNL1 pull-downs showed strong reduction in BUB co-precipitation with KNL1-NC compared with KNL1-FL (Fig. S1 D). The observation that KMN network members were present in roughly equal amounts in both pull-downs, and that MPS1 kinetochore localization was similar in cells expressing the KNL1 variants, further verified that KMN network integrity was unaffected in the various cell lines (Fig. S1, D–F; Nijenhuis et al., 2013).
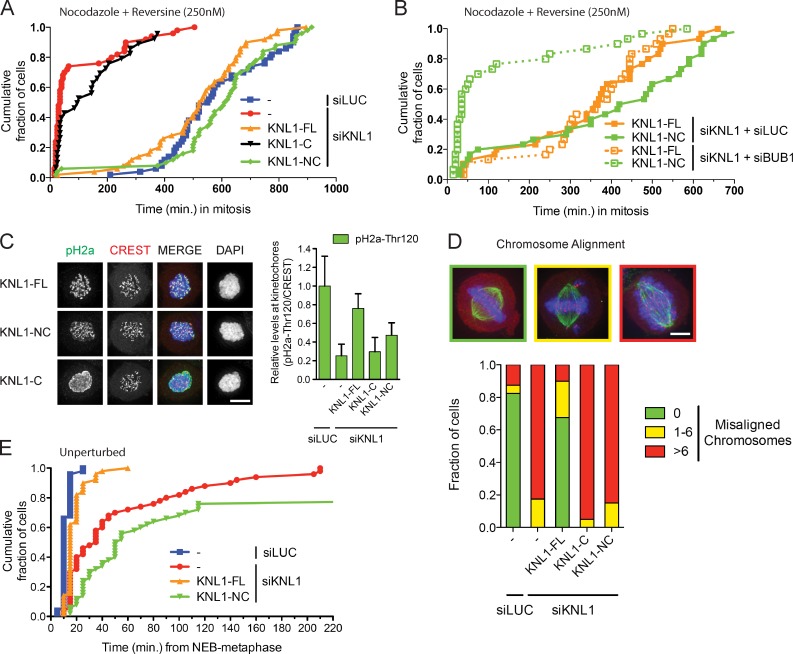
KNL1 depletion severely weakened the SAC: nocodazole-treated cells depleted of KNL1 rapidly exited mitosis when MPS1 kinase activity was slightly reduced with a low dose of reversine (250 nM; Santaguida et al., 2011; Saurin et al., 2011), whereas control cells maintained mitotic delays for many hours (Fig. 2 A; Fig. S2 A). Incomplete penetrance of RNAi or a nonessential role for KNL1 in the SAC can account for the residual weak SAC response in KNL1-depleted cells (Fig. S2 B), and we were unable to distinguish between these possibilities because no residual kinetochore KNL1 or BUB1/BUBR1 was detectable in siKNL cells. Regardless, the high sensitivity of nocodazole-treated, KNL1-depleted cells to low concentrations of reversine allowed us to examine functionality of KNL1 variants in the SAC. Somewhat unexpectedly, KNL1-NC was equally efficient as KNL1-FL in restoring SAC signaling to KNL-depleted cells (Fig. 2 A). In agreement with this, KNL1-NC was able to recruit significantly more MAD1 to kinetochores than KNL1-C (Fig. S1, G and H). Checkpoint activity of KNL1-NC depended on the MDLT and KI motifs (Fig. S2 C), indicating that KNL1-NC was able to recruit sufficient amounts of BUB proteins to perform SAC signaling. In support of this, SAC activity in KNL1-NC– but not KNL1-FL–expressing cells was highly sensitive to BUB1 levels (Fig. 2 B), and weak but detectable H2A-Thr120 phosphorylation (a mark that depends on BUB1 activity; Kawashima et al., 2010) was restored on centromeric chromatin by KNL1-NC (Fig. 2 B). We thus conclude that KNL1-NC can support robust SAC function by recruiting low levels of BUB proteins to kinetochores.
Figure 2.
The N-terminal MDLT-KI module in KNL1 is sufficient to support SAC activity but not chromosome biorientation. (A) Time-lapse analysis of Flp-in HeLa cells expressing LAP-KNL1 variants, transfected with siLUC or siKNL1, and treated with nocodazole and 250 nM reversine. Data (n = 40 representative of 3 independent experiments) indicate cumulative fraction of cells that exit from mitosis (as scored by cell morphology using DIC) at the indicated time after NEB. (B) As in A, but with transfection of the indicated siRNAs. (C) Immunostaining and quantification of centromeric H2A-Thr120 phosphorylation in Flp-in HeLa cells expressing LAP-KNL1 variants and transfected with siLUC or siKNL1. pH2A-Thr120 is shown in green, centromeres (CREST) in red, and DNA (DAPI) in blue. Bars, 5 µm. pH2A-Thr120 is quantified over CREST (n = 10 representative of 3 independent experiments). (D) Immunostaining and quantification of chromosome alignment in Flp-in HeLa cells expressing LAP-KNL1 variants, transfected with siLUC or siKNL1, and treated with MG132 for 45 min. Tubulin is shown in green, centromeres (CREST) in red, and DNA (DAPI) in blue. Bars, 5 µm. The data shown are from a single representative experiment out of three repeats. For the experiment shown, n = 50. (E) Time-lapse analysis of Flp-in HeLa cells expressing LAP-KNL1 variants and transfected with siLUC or siKNL1. Data (n = 40 representative of 3 independent experiments) indicate cumulative fraction of cells that exit from mitosis at the indicated time after NEB (as scored by GFP-H2B).
Two observations indicated that unlike the SAC, chromosome biorientation was not efficiently restored in cells expressing KNL1-NC. First, KNL1-NC was unable to support chromosome alignment in cells that were prevented from exiting mitosis by addition of the proteasome inhibitor MG132 (Fig. 2 D). Second, KNL1-NC expression caused long mitotic delays, likely due to absence of proper kinetochore–microtubule attachment that prevents SAC silencing (Fig. 2 E; Fig. S2, D and E).
Together, these data indicate that the N-terminal MDLT-KI1-KI2 motifs in KNL1 function as an independent module that is capable of activating the SAC by recruiting low BUB levels to kinetochores, but is insufficient for proper chromosome biorientation.
KNL1 contains multiple independent BUB recruitment modules
Our analyses of KNL-NC function showed that the N-terminal MDLT-KI1-KI2 fragment of KNL1 recruited low amounts of BUB1 to kinetochores but was sufficient to maintain a robust SAC. To examine if the N-terminal fragment was also required for full-length KNL1 to promote SAC activity, we analyzed function of KNL1 carrying mutations in this fragment. BUB1 localization as well as SAC activity were indistinguishable in KNL1-depleted cells expressing KNL1 with mutations in the MDLT or KI motifs (KNL1-FLMDLA or KNL1-FLKI1) or lacking the module altogether (KNL1Δ261; Fig. S2, F and G). In addition, cells expressing full-length KNL1 with mutations in either of the KI motifs (KNL1-FLKI1 or KNL1-FLKI2) restored chromosome alignment as efficiently as wild-type KNL1 and progressed through an unperturbed mitosis with similar kinetics, even in a sensitized situation (Fig. S2, H–J; Kiyomitsu et al., 2007, 2011). We thus conclude that, although able to bind BUBs (Fig. 1 A) and support SAC activity (Fig. 2 A), the N-terminal KI-containing module is dispensable for KNL1 function, at least in our assays. Most likely, therefore, KNL1 can recruit BUBs by alternative means.
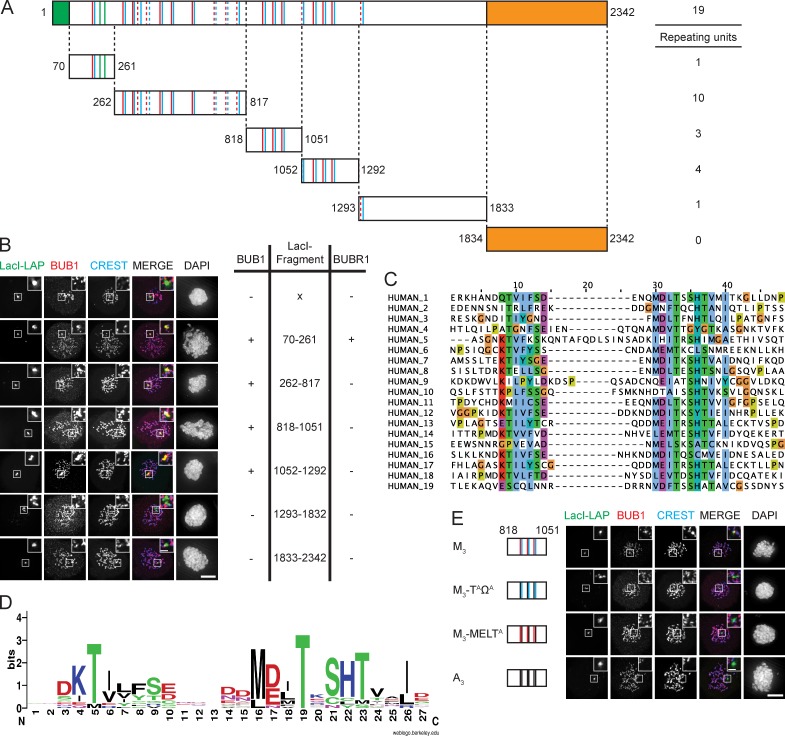
To test whether additional regions in KNL1 could also function as independent BUB recruitment modules, we analyzed the ability of various KNL1 fragments to recruit BUBs to LacO arrays. The LacI-LAP-KNL170–261, the LacI-LAP fusions of KNL1262–817, KNL1818–1051, and KNL11052–1292 were sufficient to recruit BUB1 to the LacO array, whereas LacI-LAP, LacI-LAP-KNL11293–1833, and LacI-LAP-KNL1-C were not (Fig. 3, A and B; Fig. S3 B). Interestingly, LacI-LAP-KNL170–261 was the only fragment that could recruit detectable amounts of BUBR1 to the LacO array (Fig. S3, A and B). Using the repeat-finding algorithm MEME (Bailey et al., 2009), we noticed that KNL1 consists of 19 repeating modules that include but are not limited to MELT-like sequences (Fig. 3, A and C). MELT-like sequences, when phosphorylated by MPS1, are thought to participate in BUB recruitment to unattached kinetochores (London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012). The repeating modules uncovered by MEME consist of a MELT-like sequence flanked on the C-terminal side by SHT and on the N-terminal side by the sequence TΦΦΩ[ST][DE] (where Φ denotes a hydrophobic residue and Ω denotes F or Y), which we will refer to as “TΩ” motifs (Fig. 3 D). Although overall quite different in sequence, the TΩ motifs have resemblance to KI1, in which the threonine and phenylalanine in the TxxF sequence (KIDTTSFLA) directly interact with BUB1 and are indispensable for KNL1–BUB1 interaction (Krenn et al., 2012). For convenience, we will refer to these repeating modules as “TΩ-MELT.” 10 modules adhere closely to the TΩ-MELT sequence (1, 4, 6, 8, 12–14, 16–18), whereas the remaining nine deviate to some degree in either the TΩ, the MELT, or both motifs (Fig. 3 C).
Figure 3.
KNL1 contains multiple independent BUB recruitment modules. (A) Schematic representation of KNL1 showing the microtubule- and PP1-binding domain in green and the kinetochore recruitment domain in orange. KI1 and KI2 motifs are shown as green bars, MELT-like sequences in red, and TΩ-like sequences in blue. Dashed lines indicate the generated LacI-LAP-KNL1 fragments used in B. (B) Immunolocalization of BUB1 (red) in nocodazole-treated U2OS-LacO cells transfected with LacI-LAP-KNL1 fragments. LacI-LAP-KNL1 fragments are shown in green, centromeres (CREST) in blue, and DNA (DAPI) in white. Insets show magnifications of the boxed regions. Bars, 5 µm (insets, 0.5 µm). Table indicates the ability (− or +) to recruit BUB1 and BUBR1 by the indicated KNL1 fragments (see also Fig. S3, A and B). (C) Alignment of identified TΩ-MELT modules showing conserved (green/purple/red/blue) and atypical (orange/yellow) amino acids. (D) Sequence logo of the 19 TΩ-MELT units. (E) As in B, but with LacI-LAP-KNL1818–1051 (M3) or mutant variants thereof. These variants are: M3-TAΩA (TxxΩ to AxxA), M3-MELTA (MELT to MELA), and A3 (TxxΩ-MELT to AxxA-AELA), as shown in Fig. S3 C.
To study functionality of the TΩ-MELT motifs, we analyzed their contribution to the ability of the KNL1818–1051 fragment to recruit BUB1. This fragment contains three TΩ-MELT repeat modules, and will be referred to as the M3 fragment. A mutated version of this fragment, in which four amino acids in each TΩ-MELT module were substituted for alanine (TΩ-MELT to AA-AELA), will be referred to as A3. BUB1 recruitment to LacI-KNL1818–1051 depended on TΩ-MELT motifs, as the A3 fragment was unable to localize BUB1 to the LacO arrays (Fig. 3 E; Fig. S3, C and D). Furthermore, the TΩ and the MELT motifs were each indispensable for the ability of LacI-KNL1818–1051 to recruit BUB1 because mutating either TΩ or MELT abolished BUB1 localization (Fig. 3 E; Fig. S3, C and D). The TΩ motif was also critical for the N-terminal module that uniquely contains KI1 and KI2 (Fig. S3, E and F).
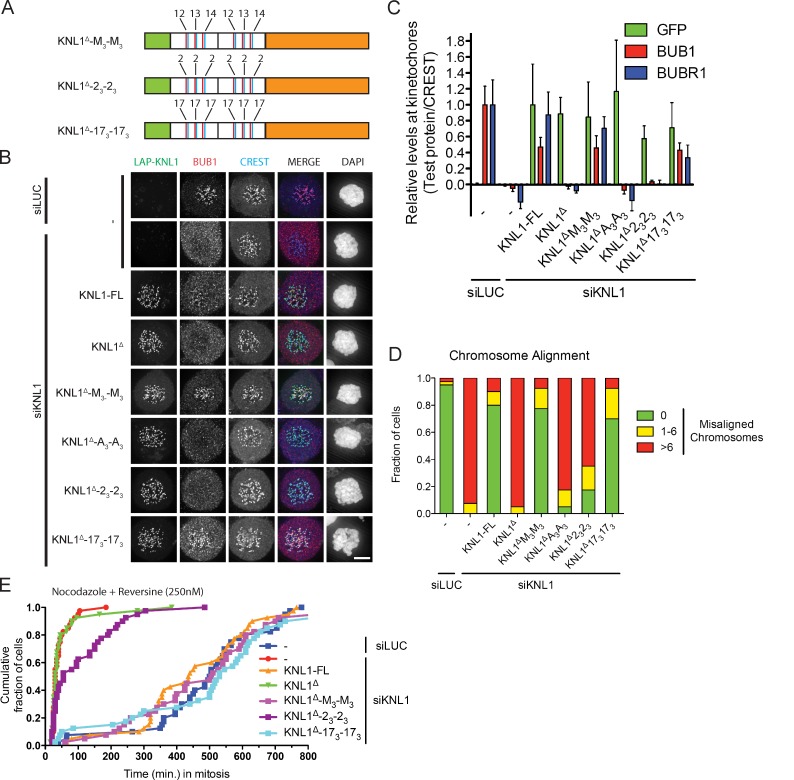
Engineered KNL1 proteins reveal differential requirements for TΩ-MELT modules in the SAC and chromosome biorientation
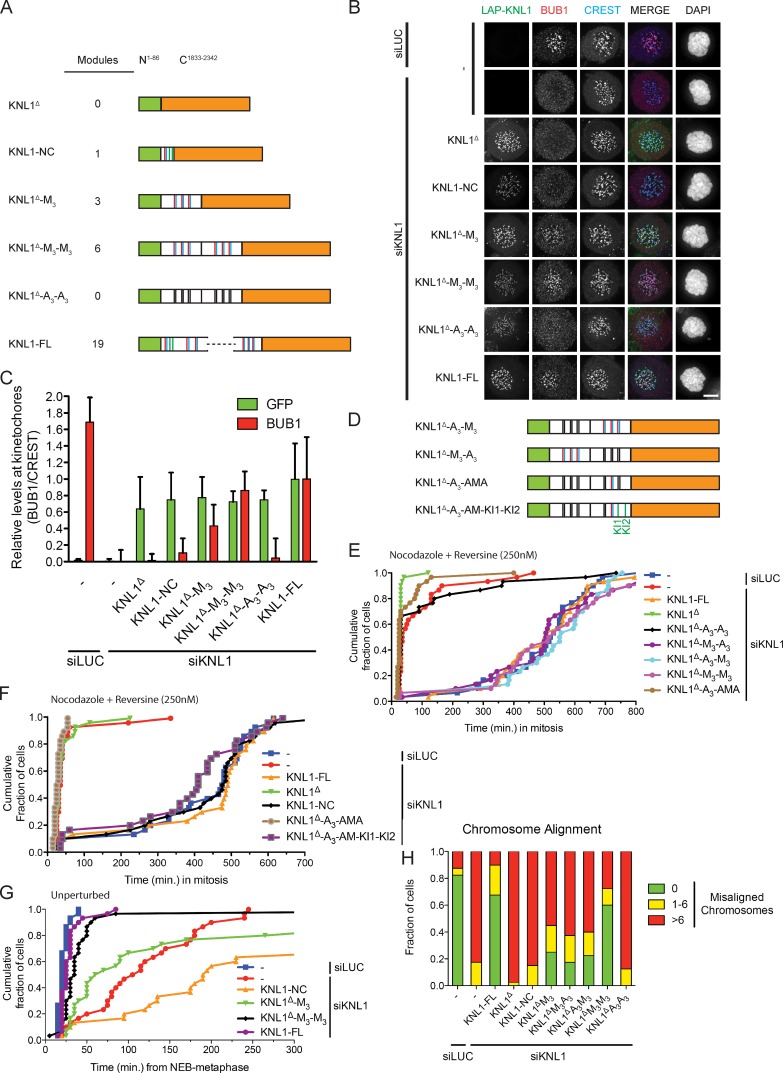
Our observations that the N-terminal module is sufficient but not required for the SAC, that this module is insufficient for proper chromosome biorientation, and that other modules in KNL1 highly resemble this N-terminal module raised the question of whether there is functional redundancy between modules or whether some modules have specialized. To examine this, we generated a KNL1 protein devoid of all TΩ-MELT–like modules but containing the N-terminal-most 86 amino acids (responsible for microtubule and PP1 binding) fused to KNL1-C (Kiyomitsu et al., 2007; Liu et al., 2010; Welburn et al., 2010). Into this protein, named KNL1Δ, we inserted one or two of the M3 fragments to create KNL1Δ-M3 and KNL1Δ-M3-M3, respectively (Fig. 4 A). A3 fragments were used as control, as well as combinations of M3 and A3 fragments, giving rise to KNL1Δ-A3, KNL1Δ-A3-A3, KNL1Δ-A3-M3, and KNL1Δ-M3-A3 (Fig. 3 E; Fig. 4, A and D). Isogenic cell lines with inducible expression of these engineered KNL proteins were generated and analyzed for functionality of various processes upon depletion of endogenous KNL1. Strikingly, the amount of BUB1 detectable at unattached kinetochores followed the amount of repeat modules present in KNL1: a single module (KNL1-NC) recruited low amounts of BUB1, one block of three modules (KNL1Δ-M3) recruited approximately threefold more BUB1, and two blocks totaling six modules (KNL1Δ-M3-M3) doubled that to close to the levels observed in KNL-FL reconstituted cells (Fig. 4, B and C). Absence of any module (KNL1Δ or KNL1Δ-A3-A3) eliminated BUB1 kinetochore binding. These data are indicative of a direct correlation between the number of functional TΩ-MELT modules and the amount of BUB1 protein at mitotic kinetochores. Interestingly, although BUBR1 did not interact with amino acids 818–1051 in KNL1 in the context of the LacO array (Fig. S3, A and B), KNL1Δ-M3-M3 was able to recruit BUBR1 to kinetochores (Fig. S4, A and B). This suggested that that BUBR1 recruitment to KNL1 requires the context of kinetochores.
Figure 4.
Engineered KNL1 proteins reveal differential requirements for TΩ-MELT modules in the SAC and chromosome biorientation. (A) Schematic representation of synthetic LAP-KNL1 constructs, showing the microtubule- and PP1-binding domain in green and the kinetochore recruitment domain in orange. KI1 and KI2 motifs are shown as green bars, MELT-like sequences in blue, and TxxΩ-like sequences in red. See main text for details about constructs. (B and C) Representative images (B) and quantification (C) of LAP-KNL1-expressing Flp-in HeLa cells transfected with siRNAs to luciferase (siLUC) or to KNL1 (siKNL1) and treated with nocodazole. LAP-KNL1 is shown in green, BUB1 in red, centromeres (CREST) in blue, and DNA (DAPI) in white. Bars, 5 µm. Quantification in C shows total kinetochore signal intensity (+SD) of LAP-KNL1 and BUB proteins over CREST. Data are from >15 cells and representative of 3 experiments. Levels of kinetochore BUBs in control cells and of kinetochore LAP-KNL1 in KNL1-FL–expressing cells are set to 1. (D) Schematic as in A. See main text for details about constructs. (E) Time-lapse analysis of Flp-in HeLa cells expressing LAP-KNL1 variants, transfected with siLUC or siKNL1, and treated with nocodazole and 250 nM reversine. Data (n = 40 representative of 3 independent experiments) indicate cumulative fraction of cells that exit from mitosis (as scored by cell morphology using DIC) at the indicated time after NEB. (F) As in E, with the indicated constructs. (G) Time-lapse analysis of Flp-in HeLa cells expressing LAP-KNL1 variants and transfected with siLUC or siKNL1. Data (n = 40 representative of 3 independent experiments) indicate cumulative fraction of cells that exit from mitosis at the indicated time after NEB (as scored by GFP-H2B). (H) Quantification of chromosome alignment in Flp-in HeLa cells expressing LAP-KNL1 variants, transfected with siLUC or siKNL1, and treated with MG132 for 45 min. The data shown are from a single representative experiment out of three repeats. For the experiment shown, n = 40.
All KNL1 variants that contained at least one M3 fragment (M3, M3-M3, A3-M3, and M3-A3) were proficient in recruiting MAD1 and supporting the SAC (Fig. S1, G and H; Fig. 4 E). Because SAC activity was also supported by a single module in the context of the N-terminal fragment (KNL1-NC; see Fig. 2 A), we next examined whether any single module could support the SAC. To this end, one TΩ-MELT module was restored in KNL1Δ-A3-A3, creating the KNL1Δ-A3-AMA protein (Fig. 4 D). Although able to recruit low levels of kinetochore BUB1 and promote partial H2A-T120 phosphorylation (Fig. S4, C–E), KNL1Δ-A3-AMA could not recover SAC activity (Fig. 4 E). We thus conclude that a single module recruits sufficient BUB1 for SAC activation only in the context of the N-terminal fragment, whereas more than one is needed in the context of other KNL1 fragments.
The N-terminal BUB recruitment module is unique in two ways: it is close to the PP1- and microtubule-binding site on KNL1, and it contains the KI motifs that in the context of KNL1-NC significantly contribute to BUB recruitment and SAC activity (Fig. 1 A; Fig. S2 C). To examine if either of these is the cause of the difference in ability of KNL1-NC and KNL1Δ-A3-AMA to support SAC activity, we placed KI1 and KI2 downstream of the TΩ-MELT module in KNL1Δ-A3-AMA (resulting in KNL1Δ-A3-AM-KI1-KI2; see Fig. 4 D). Strikingly, adding KI1 and KI2 to KNL1Δ-A3-AMA endowed the protein with SAC function (Fig. 4 F) and this correlated with a slight increase in kinetochore BUB1 to close to the levels attained by KNL1-NC (Fig. S4, F and G). These data therefore indicate that the KI motifs enhance BUB recruitment potential of individual TΩ-MELT modules, and as such allow the N-terminal module to be sufficient for SAC function.
Time-lapse imaging of mitotic progression in the different cell lines showed that increasing amounts of repeat modules gradually decreased the time from nuclear envelope breakdown (NEB) to metaphase (Fig. 4 G). This corresponded to increased efficiency of chromosome alignment, as assayed in fixed MG132-treated mitotic cells (Fig. 4 H). Directly in line with BUB1 levels, three modules were more efficient than one, whereas six modules were more efficient than three. In fact, cells expressing KNL1Δ-M3-M3 were almost as efficient in chromosome alignment as control cells or cells expressing KNL1-FL and displayed comparable mitotic timing (Fig. 4, G and H). High fidelity chromosome segregation in human cells therefore requires between four and six TΩ-MELT modules that combine to recruit sufficient BUBs.
Functional TΩ-MELT modules in KNL1 are redundant and exchangeable
Our observations with the engineered KNL1 proteins suggested that the modules within the M3 fragment may be redundant and that their functionality in SAC activity is independent of exact position in relation to the kinetochore or to the microtubule- and PP1-binding sites. This raised the possibility that redundancy is relatively widespread across the 19 identified repeat modules. To examine this, we designed artificial fragments, based on existing TΩ-MELT modules, and tested their functionality in the context of KNL1Δ. We swapped module 12, 13, and 14 within the M3 fragment for either module 2 or module 17 to create KNL1Δ-23-23 or KNL1Δ-173-173, respectively (Fig. 3 C; Fig. 5 A). We chose module 17 because its sequence adheres closely to the “consensus” TΩ-MELT module sequence (TILYSCGQDDMEITRSHTTAL), and module 2 was chosen because its sequence deviates from that consensus (TRLFREKDDGMNFTQCHTANI) but maintains the TxxΩ and MxxT characteristics (Fig. 3, C and D; Fig. 5 A). KNL1Δ-173-173 fully restored BUB localization, chromosome alignment, and SAC activity in KNL1-depleted cells (Fig. 5, B–E; Fig. S5 A). Interestingly, KNL1Δ-23-23 could not support chromosome alignment and SAC activity, which correlated with low levels of BUB1/BUBR1 recruitment to kinetochores and incomplete restoration of centromeric pH2A-T120 (Fig. 5, B–E; Fig. S5 B). We therefore conclude that neither chromosome biorientation nor the SAC relies on any specific TΩ-MELT module but that both processes require any combination of modules that can recruit sufficient BUB1. We thus propose that different TΩ-MELT modules have redundant functions and that any array of functional modules that can recruit sufficient BUB1 will promote high fidelity chromosome segregation.
Figure 5.
TΩ-MELT modules in KNL1 are redundant and exchangeable. (A) Schematic representation of synthetic LAP-KNL1 constructs. For color codes, see Fig. 4 A. See main text for details about constructs. (B and C) Representative images (B) and quantification (C) of LAP-KNL1–expressing Flp-in HeLa cells transfected with siRNAs to luciferase (siLUC) or to KNL1 (siKNL1) and treated with nocodazole. LAP-KNL1 is shown in green, BUB1 in red, centromeres (CREST) in blue, and DNA (DAPI) in white. Bars, 5 µm. Quantification in C shows total kinetochore signal intensity (+SD) of LAP-KNL1 and BUB proteins over CREST. Data are from >15 cells and representative of 3 experiments. Levels of kinetochore BUBs in control cells and of kinetochore LAP-KNL1 in KNL1-FL–expressing cells are set to 1. (D) Quantification of chromosome alignment in Flp-in HeLa cells expressing LAP-KNL1 variants, transfected with siLUC or siKNL1, and treated with MG132 for 45 min. The data shown are from a single representative experiment out of three repeats. For the experiment shown, n = 40. (E) Time-lapse analysis of Flp-in HeLa cells expressing LAP-KNL1 variants, transfected with siLUC or siKNL1, and treated with nocodazole and 250 nM reversine. Data (n = 40 representative of 3 independent experiments) indicate cumulative fraction of cells that exit from mitosis (as scored by cell morphology using DIC) at the indicated time after NEB.
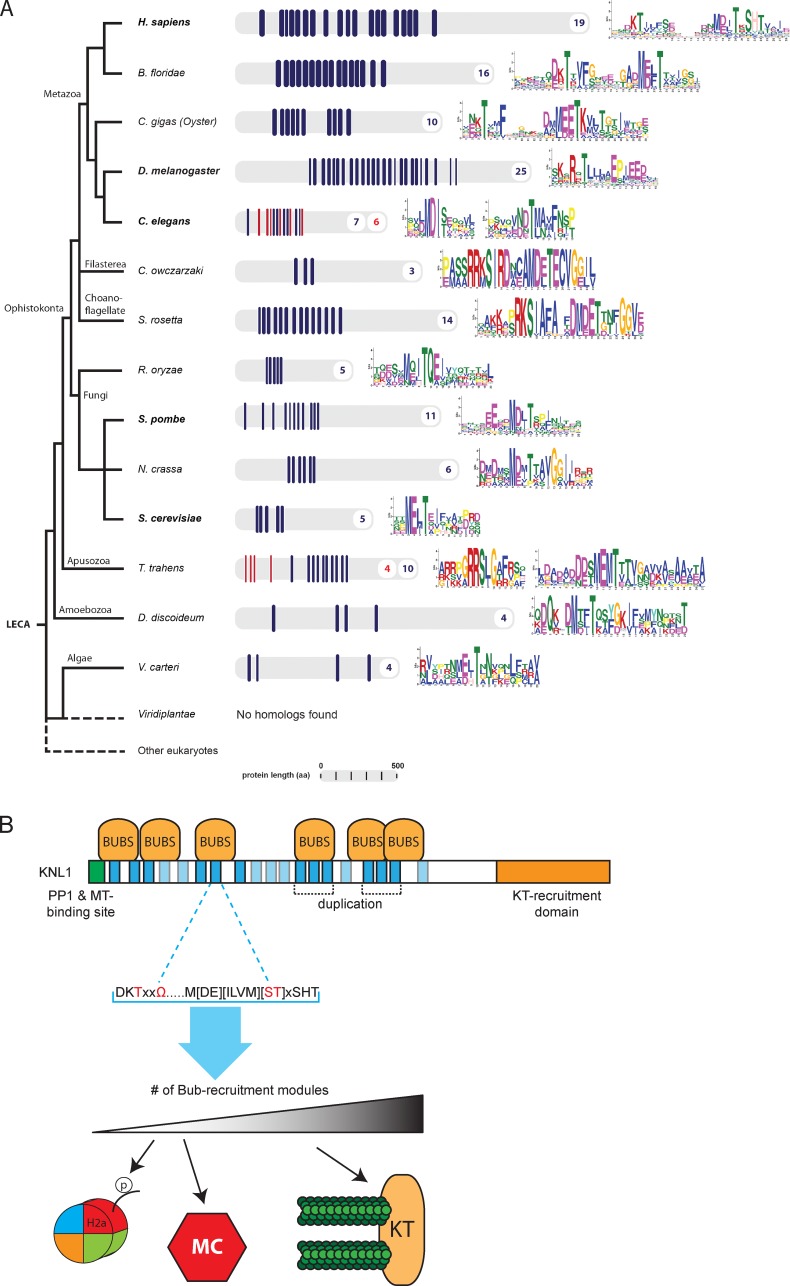
Extensive divergence in sequence and amount of repeat modules in eukaryotic KNL1 homologues
Our findings suggest that human KNL1 has evolved by extensive duplications of the TΩ-MELT modules, possibly followed by degeneration of a number of these sequences. Furthermore, our recent analysis of selected eukaryotic KNL1 homologues (Vleugel et al., 2012) showed that the amount of MELT-like sequences varies quite extensively from species to species. To examine if repeating modules exist in these and other KNL1-like sequences, we applied MEME on predicted KNL1 homologues from 15 species across three supergroups of eukaryotic evolution (Fig. 6 A). Predicted homologues were identified by similarity in the C-terminal coiled-coil region and homology was further strengthened by the presence of an N-terminal PP1-binding RVSF motif. Interestingly, all homologues contained repeating modules, but they diverged extensively in sequence and number. The methionine of the MELT motif is conserved in most species, but the “LT” sequence is often replaced by additional negative charges. A striking example of this are the drosophilids in which the repeating module is based around a MEED-like sequence (Fig. 6 A) (Schittenhelm et al., 2009). TΩ-like sequences were apparent in KNL1 homologues of Branchiostoma floridae and Crassostrea gigas, but MELT-like sequences of most other organisms were complemented with different motifs. In some species (Nematostella vectensis, Thecamonas trahens), KNL1 homologues contained two different types of repeating modules. We conclude that KNL1 is a rapidly evolving protein, with extensive variations in the number and sequence of repeating modules across different eukaryotic KNL1 homologues.
Figure 6.
TΩ-MELT module evolution and model. (A) Schematic representation of eukaryotic tree of life showing KNL1 homologues from indicated species. Repeating units are shown in blue and red with the number of repeats in corresponding colors. Repeat sequences are shown as sequence logos. (B) Model for TΩ-MELT function in human KNL1. Conserved (dark blue) and degenerated (light blue) TΩ-MELT modules (essential amino acids in red) in KNL1 can independently recruit BUB protein complexes (BUBs) to promote H2A-Thr120 phosphorylation and SAC activity (few modules, low BUB levels) and chromosome biorientation (increasing fidelity with increasing modules and BUB levels).
Discussion
An extensive array of generic BUB recruitment modules in KNL1
Our data demonstrate that KNL1 is a scaffold that contains multiple independent and redundant repeating modules, which together ensure recruitment of sufficient amounts of BUB proteins to kinetochores (Fig. 6 B). The ability of KNL1 to recruit BUB proteins and ensure efficient chromosome alignment is independent of protein length, of localization of the recruitment modules within KNL1, and of any particular module, per se: KNL1 function is maintained when only two copies of modules 12-13-14 or six copies of module 17 are present. Moreover, compared with the SAC, more modules seem required for chromosome alignment, and the efficiency of chromosome alignment directly follows the amount of functional modules present in KNL1, suggesting that the modules act in an additive fashion (Fig. 6 B).
Mutational analysis of the N-terminal module shows that the TxxΩ motif that we identified as part of the repeating module is critical for BUB recruitment. Previous structural work has shown direct interactions between the TPR domain of BUB1 and the TxxF sequence in the KI1 motif of KNL1 (Krenn et al., 2012). Considering that there is only one potential TxxF interaction groove within the TPR domain of BUB1, we hypothesize that one functional TΩ-MELT module is capable of recruiting one BUB1 molecule. The contribution of the MELT-like sequences was recently described, and involves the BUB1-interacting protein BUB3 (Primorac et al., 2013).
Unlike BUB1, all LacO-targeted KNL1 fragments except for KNL170–261 failed to recruit BUBR1. This may be related to a difference by which the BUBs interact with KNL1. Whereas the TxxF motif in KI1 is critical for interaction with BUB1, a similarly positioned FxxF motif in KI2 is critical for interaction with BUBR1, and neither motif can substitute for loss of the other. The repeating modules present in the KNL1 fragments all contain TxxΩ or variants thereof, but never an aromatic residue in the T position. Interestingly, however, a KNL1 fragment that was unable to recruit BUBR1 to LacO arrays was able to recruit BUBR1 to kinetochores. In fact, KNL1Δ-M3-M3 restored BUBR1 kinetochore levels to the same extent as KNL1-FL. BUBR1 binding to KNL1 under these conditions is therefore likely indirect and requires one or more kinetochore-localized proteins or activities. Because BUB1 is normally indispensable for BUBR1 localization (Johnson et al., 2004; Klebig et al., 2009) and KI2 is not (this study), and because mutations in the TΩ-MELT motifs abolished BUBR1 localization, we hypothesize that the predominant mode of BUBR1 kinetochore binding is indirect via TΩ-MELT–mediated KNL1–BUB1 interaction, aided by an unidentified kinetochore-localized activity.
In contrast to the TΩ-MELT modules, the role of the N-terminal KI1 and KI2 modules in KNL1 function is unclear. Our recent bioinformatics analysis has indicated that the KI motifs are a recent invention of the vertebrate lineage (Vleugel et al., 2012). Furthermore, the interaction of KI1 with the TPR domain of BUB1 is dispensable for BUB1 recruitment to kinetochores (Krenn et al., 2012), and we show here that in the context of the full-length protein and with our assays, KI1 and KI2 are not required for SAC activity, chromosome alignment, and mitotic progression. These observations raise the question of what the functionality of the KI motifs is. Both within the N-terminal module and the synthetic KNL1Δ-A3-AM-KI1-KI2 construct, the KI motifs enhance BUB recruitment potential of the TΩ-MELT motifs to levels that support SAC activity. It is therefore likely that the KI motifs contribute to some extent to the BUB recruitment ability of full-length KNL1. This may become beneficial under conditions that require maximal BUB levels at kinetochores.
TΩ-MELT module function
KNL1Δ-173-173 and KNL1Δ-M3-M3 contain six recruitment modules, yet were able to recruit roughly the same amount of BUB1 to kinetochores as KNL1-FL with its 19 modules. One possible explanation for why KNL1-FL does not recruit more BUB1 is that not all modules are functional in KNL1-FL. Consistent with this, our analysis of KNL1Δ-23-23 showed that module 2 is less capable of binding BUB1 than modules 12, 13, 14, and 17. Module 2 contains the motif TF-MNFT with relatively significant substitutions within the MELT-like motif. Besides module 2, modules 3, 5, 7, 9, 10, 11, 15, and 19, and to a lesser extent module 8, have alterations in either the TΩ and/or the MELT-like motifs, possibly rendering them less or not functional. In addition to sequence, phosphorylation of the motifs also likely contributes to BUB-binding affinity. Some TΩ- (16/18) and MELT-like (12/15/16/17/18) sequences can be phosphorylated by MPS1 in vitro (Yamagishi et al., 2012), and one was found phosphorylated in mitotic cells (7: MDIpTKSHpT [bold/underlined letters represent phosphorylated residues]; Hegemann et al., 2011). A KNL1-8A mutant in which all the in vitro phosphorylation sites were mutated to alanine reduced BUB1 kinetochore localization by ∼50% (Yamagishi et al., 2012), showing that TΩ-MELT phosphorylation enhances BUB recruitment. Non-phosphorylatable TΩ- (11/15/19) and MELT-like (9/10) sequences are therefore likely to be less functional than phosphorylatable ones. This additionally raises the question of how many functional modules are phosphorylated at any given moment on one KNL1 molecule on an unattached kinetochore. It is conceivable that expanding the amount of functional modules simply increases the chance that a certain, small number of modules is phosphorylated at steady state, and that the actual amount of KNL1-bound BUBs required for K-MT attachment and the SAC is lower than the amount of modules that we have engineered into KNL1. A systematic biochemical survey of TΩ-MELT functionality and phosphorylation, combined with cell biological analyses will be required to elucidate which TΩ-MELT modules are functional and how they contribute to BUB recruitment.
TΩ-MELT module evolution
Our present and past (Vleugel et al., 2012) surveys of eukaryotic homologues of KNL1 have revealed striking differences between species. Most homologues contain an array of repeating modules that is unique to KNL1, but the number and sequence of those modules varies quite extensively. It will be interesting to examine whether BUB–KNL1 interactions in different species require the species-specific repeat module characteristics, or whether these additional motifs contribute to other, unknown module functionality. More in-depth analysis has provided evidence of rapid evolution of the modules in eukaryotes (unpublished data). This, combined with the conserved roles for BUBs in chromosome segregation and our demonstration that the modules in human KNL1 are generic in nature may thus indicate that the extensive species-specific differences in module sequence may not affect BUB binding per se, but may reflect other evolutionary important roles for the modules. Assaying function in human cells of KNL1 containing modules of other species might start to provide some answers to these questions.
Besides sequence, the number of modules per KNL1 homologue also differs strongly. Green algae like Volvox carteri have KNL1 homologues with only a few modules, whereas those of species like Drosophila melanogaster and Xenopus tropicalis have more than 20 (Fig. 6 A; Vleugel et al., 2012). Possibly, the amount of modules correlates with the amount of BUB signaling required for high fidelity chromosome segregation. Phosphorylation of H2A-T120 is significantly restored with a single module in human cells, and this role of BUB1 in chromosome segregation is conserved also in more primitive species (Kawashima et al., 2010). Perhaps, therefore, H2A phosphorylation and SAC activity require only one or a few modules and this allows more primitive species to survive with few modules in KNL1. More challenging requirements in mitosis for the more complex organisms (for instance, expanding complexity of kinetochores and increasing numbers of microtubules bound per kinetochore) may thus have spurred multiplication of modules to enable recruitment of more BUBs to kinetochores. An exciting possibility therefore is that altering BUB signaling by module expansion and degeneration during evolution is a relatively facile mechanism for adapting the chromosome segregation machinery to changing requirements during mitosis.
Materials and methods
Plasmids
pCDNA5-LAP-KNL1FL encodes full-length, N-terminally LAP-tagged, and siRNA-resistant wild-type KNL1 (modified codons 258 and 259) and was created by digestion of pEYFP-LAP-KNL1FL (a gift from I. Cheeseman, Whitehead Institute, Cambridge, MA) with XhoI and HpaI to isolate the full-length KNL1 cassette, which was ligated into the XhoI and PmeI sites of pCDNA5/FRT/TO (Invitrogen). An N-terminal LAP-tag was introduced by subcloning the LAP-tag cassette from pCDNA3-LAP-MPS1Δ200 (Nijenhuis et al., 2013) into the KpnI and XhoI sites of the resulting plasmid. KNL1-NC was generated by PCR and subcloning of KNL1-C (aa 1833–2342, using Xho1–BamH1) and KNL1-N (aa 1–261, using Xho1–Xho1) and subsequent ligation into pCDNA5/FRT/TO-LAP. LacI-KNL1 fragments were generated by PCR and cloned into pLacI-LAP. MELT-block aa 818–1051 and aa 1052–1228 and corresponding variants were synthesized by GenScript and cloned into the Xho1 site of KNL1Δ (GenScript) using Sal1 and Xho1. Additional blocks were inserted in the Xho1 site of KNL1-NM/A3C.
Cell culture and transfection
U2OS LacO cells (a gift from S. Janicki, The Wistar Institute, Philadelphia, PA) were grown in DMEM supplemented with 8% FBS (Takara Bio Inc.), 200 µg/ml hygromycin, 50 µg/ml penicillin/streptomycin, and 2 mM l-glutamine. HeLa Flp-in cells were grown in 8% Tet-approved FBS (Takara Bio Inc.) supplemented with 200 µg/ml hygromycin and 4 µg/ml blasticidin. Plasmids were transfected using FuGENE HD (Roche) according to the manufacturer’s instructions. To generate stably integrated HeLa Flp-In cells, pCDNA5 constructs were cotransfected with Ogg44 recombinase in a 10:1 ratio (Klebig et al., 2009). Constructs were expressed by addition of 1 µg/ml doxycycline for 24 h. siKNL1 (CASC5#5, J-015673-05, Thermo Fisher Scientific; 5′-GCAUGUAUCUCUUAAGGAA-3′) and siBUB1 (5′-GAAUGUAAGCGUUCACGAA-3′) were transfected using HiPerFect (QIAGEN) at 20 nM for 2 d according to the manufacturer’s instructions.
Live-cell imaging
For live-cell imaging experiments, cells were transfected with 20 nM siRNA for 24 h, after which cells were arrested in early S-phase for 24 h by addition of 2 mM thymidine, and expression was induced by addition of 1 µg/ml doxycycline. Subsequently, cells were released from thymidine for 8–10 h and arrested in prometaphase by the addition of 830 nM nocodazole with or without 250 nM reversine. Unperturbed mitotic progression was assayed after a 24-h infection with BacMam-H2B-GFP virus (BioTek) followed by a release from thymidine into normal media. Cells were imaged in a heated chamber (37°C and 5% CO2) using a 20×/0.5 NA UPLFLN objective on a microscope (model IX-81; Olympus) controlled by Cell-M software (Olympus). Images were acquired using a CCD camera (ORCA-ER; Hamamatsu Photonics) and processed using Cell-M software.
Immunofluorescence and antibodies
Asynchronously growing cells were arrested in prometaphase by the addition of 830 nM nocodazole for 2–3 h. Cells plated on 12-mm coverslips were fixed (with 3.7% paraformaldehyde, 0.1% Triton X-100, 100 mM Pipes, pH 6.8, 1 mM MgCl2, and 5 mM EGTA) for 5–10 min. Coverslips were washed with PBS and blocked with 3% BSA in PBS for 1 h, incubated with primary antibodies for 16 h at 4°C, washed with PBS containing 0.1% Triton X-100, and incubated with secondary antibodies for an additional hour at room temperature. Coverslips were then washed, incubated with DAPI for 2 min, and mounted using ProLong Antifade (Molecular Probes). All images were acquired on a deconvolution system (DeltaVision RT; Applied Precision) with a 100×/1.40 NA U Plan S Apochromat objective (Olympus) using softWoRx software (Applied Precision). Images are maximum intensity projections of deconvolved stacks. For quantification of immunostainings, all images of similarly stained experiments were acquired with identical illumination settings; cells expressing comparable levels of exogenous protein were selected for analysis and analyzed using ImageJ (National Institutes of Health). An ImageJ macro was used to threshold and select all centromeres and all chromosome areas (excluding centromeres) using the DAPI and anticentromere antibody channels as described previously (Saurin et al., 2011). This was used to calculate the relative mean kinetochore intensity of various proteins ([centromeres–chromosome arm intensity (test protein)]/[centromeres–chromosome arm intensity (CREST)]).
Cells were stained using GFP-booster (ChromoTek), BUB1 (Bethyl Laboratories, Inc.), BUBR1 (Bethyl Laboratories, Inc.), H2A-pT120 (ActiveMotif), MPS1 (EMD Millipore), Mad1 (a gift from A. Musacchio, MPI, Dortmund, Germany), CREST/anti-centromere antibodies (Cortex Biochem, Inc.), and/or tubulin (Sigma-Aldrich). Secondary antibodies were goat anti–human Alexa Fluor 647 and goat anti–rabbit and anti–mouse Alexa Fluor 568 (Molecular Probes) for immunofluorescence experiments.
SILAC mass spectrometry
For SILAC mass spectrometry, LAP-KNL1-FL and -NC cells were adapted to light (C12N14 lysine/arginine) and heavy (C13N15 lysine/arginine) medium, respectively. Cells were synchronized in mitosis by a 24-h thymidine block, followed by overnight treatment with nocodazole. KNL1 expression was induced for 24 h using doxycycline and cells were harvested followed by immunoprecipitation and mass spectrometry. Cells were lysed at 4°C in hypertonic lysis buffer (500 mM NaCl, 50 mM Tris-HCl, pH 7.6, 0.1% sodium deoxycholate, and 1 mM DTT) including phosphatase inhibitors (1 mM sodium orthovanadate, 5 mM sodium fluoride, and 1 mM β-glycerophosphate), sonicated, and LAP-KNL1 proteins were coupled to GFP-trap (ChromoTek) for 1 h at 4°C. Purifications were washed three times with high-salt (2 M NaCl, 50 mM Tris-HCl, pH 7.6, 0.1% sodium deoxycholate, and 1 mM DTT) and low-salt wash buffers (50 mM NaCl, 50 mM Tris-HCl, pH 7.6, and 1 mM DTT) and subsequently eluted in 2 M urea, 50 mM Tris-HCL, pH 7.6, and 5 mM IAA. Samples were loaded on a C18 column and run on a nano-LC system coupled to a mass spectrometer (LTQ-Orbitrap Velos; Thermo Fisher Scientific) via a nanoscale LC interface (Thermo Fisher Scientific), as described in Suijkerbuijk et al. (2012a).
Repeat identification
For all KNL1 orthologues used in this study, separate MEME (Bailey et al., 2009) analyses (option “any number of repeats”) were performed to detect repeating motifs for which HMMR3 (Eddy, 2011) profiles were created. Significant motifs were added to the profiles and searches were repeated until no novel repeats were found. Searches were manually inspected for consistency and significance; clear false-positives were discarded. The resulting repeats were aligned by hand and the alignment was used to construct sequence logos using WebLogo, which is embedded in the MEME package. Due to the degenerate nature for some the repeats, we introduced multiple gaps in the alignments. Therefore the sequence logos do not fully reflect the true spacing for the conserved amino acid positions within the repeats.
Online supplemental material
Figs. S1–S5 are related to Figs. 1–5. Fig. S1 shows a quantification of the LacO-targeted KNL1 fragments, a schematic of KNL1-NC, comparative mass spectrometry data of KNL1-FL vs. KNL1-NC, and MPS1 and MAD1 kinetochore localization (and quantification thereof) in cells expressing the different KNL1 constructs. Fig. S2 shows representative still images of checkpoint assays, quantification of checkpoint assays without sensitization and in cells expressing KNL-NC mutants, unperturbed metaphase–anaphase timing, representative still images of unperturbed mitosis for the different KNL1 constructs, and immunofluorescence of BUB1 localization (sensitized), mitotic progression, alignment, and checkpoint assays of KI mutant KNL1 constructs. Fig. S3 shows BUBR1 recruitment to indicated LacO-targeted KNL1 fragments and quantification of BUB1 and BUBR1 recruitment to these loci, an overview of the described M3 mutations and a quantification of BUB1 recruitment to LacO-targeted M3 blocks, and immunofluorescence and quantification of LacI-KNL1-N–containing TΩ mutations. Fig. S4 shows immunofluorescence and quantification of BUBR1 recruitment to KNL1Δ-M3-M3 and immunofluorescence and quantification of pH2A-Thr120 and BUB1 kinetochore localization in KNL1Δ-M3-AMA and BUB1 in KNL1Δ-M3-AM-KI1-KI2 cells. Fig. S5 shows immunofluorescence of BUBR1 and pH2A-Thr120 kinetochore localization in KNL1Δ-23-23 and KNL1Δ-173-173 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201307016/DC1.
Supplementary Material
Acknowledgments
We thank I. Cheeseman, S. Taylor, and S. Janicki for providing reagents; A. Musacchio and J. Nilsson for sharing unpublished observations; A. Schram for critical reading of the manuscript; and the Kops and Lens laboratories for discussions.
This work was supported by grants from the Netherlands Organization for Scientific Research (NWO-Vici 016.130.661), from the European Research Council (ERC-StG KINSIGN), and from KWF Kankerbestrijding (UU-2012-5427) to G.J.P.L. Kops.
Footnotes
Abbreviations used in this paper:
- KMN
- KNL1/MIS12 complex/NDC80 complex
- LacO
- Lac operator
- NEB
- nuclear envelope breakdown
- SAC
- spindle assembly checkpoint
References
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37(Web Server issue):W202–W208 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Garcia V.M., Blundell T.L. 2011. BUB1 and BUBR1: multifaceted kinases of the cell cycle. Trends Biochem. Sci. 36:141–150 10.1016/j.tibs.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Garcia V.M., Lischetti T., Matak-Vinković D., Cota E., Simpson P.J., Chirgadze D.Y., Spring D.R., Robinson C.V., Nilsson J., Blundell T.L. 2011. Structure of a Blinkin-BUBR1 complex reveals an interaction crucial for kinetochore-mitotic checkpoint regulation via an unanticipated binding Site. Structure. 19:1691–1700 10.1016/j.str.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W.C., Kulkarni K., Zhang Z., Kong E.H., Barford D. 2012. Structure of the mitotic checkpoint complex. Nature. 484:208–213 10.1038/nature10896 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. 2011. Accelerated profile HMM searches. PLOS Comput. Biol. 7:e1002195 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J., Cheerambathur D.K., Krenning L., Oegema K., Desai A. 2012. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J. Cell Biol. 196:469–482 10.1083/jcb.201111107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E.A., Kapoor T.M. 2013. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 14:25–37 10.1038/nrm3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E.A., Maldonado M., Kapoor T.M. 2011. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 13:1265–1271 10.1038/ncb2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann B., Hutchins J.R., Hudecz O., Novatchkova M., Rameseder J., Sykora M.M., Liu S., Mazanek M., Lénárt P., Hériché J.K., et al. 2011. Systematic phosphorylation analysis of human mitotic protein complexes. Sci. Signal. 4:rs12 10.1126/scisignal.2001993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki S.M., Tsukamoto T., Salghetti S.E., Tansey W.P., Sachidanandam R., Prasanth K.V., Ried T., Shav-Tal Y., Bertrand E., Singer R.H., Spector D.L. 2004. From silencing to gene expression: real-time analysis in single cells. Cell. 116:683–698 10.1016/S0092-8674(04)00171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson V.L., Scott M.I., Holt S.V., Hussein D., Taylor S.S. 2004. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 117:1577–1589 10.1242/jcs.01006 [DOI] [PubMed] [Google Scholar]
- Kawashima S.A., Yamagishi Y., Honda T., Ishiguro K.I., Watanabe Y. 2010. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 327:172–177 10.1126/science.1180189 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 13:663–676 10.1016/j.devcel.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Murakami H., Yanagida M. 2011. Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol. Cell. Biol. 31:998–1011 10.1128/MCB.00815-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebig C., Korinth D., Meraldi P. 2009. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 185:841–858 10.1083/jcb.200902128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn V., Wehenkel A., Li X., Santaguida S., Musacchio A. 2012. Structural analysis reveals features of the spindle checkpoint kinase Bub1-kinetochore subunit Knl1 interaction. J. Cell Biol. 196:451–467 10.1083/jcb.201110013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T., Zhang G., Larsen M.S., Lischetti T., Streicher W., Kragh Nielsen T., Bjørn S.P., Nilsson J. 2013. Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J. Cell Sci. 126:1086–1092 10.1242/jcs.122481 [DOI] [PubMed] [Google Scholar]
- Liu D., Vleugel M., Backer C.B., Hori T., Fukagawa T., Cheeseman I.M., Lampson M.A. 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188:809–820 10.1083/jcb.201001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., Ceto S., Ranish J.A., Biggins S. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22:900–906 10.1016/j.cub.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijenhuis W., von Castelmur E., Littler D., De Marco V., Tromer E., Vleugel M., van Osch M.H., Snel B., Perrakis A., Kops G.J. 2013. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J. Cell Biol. 201:217–231 10.1083/jcb.201210033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A., Pasqualato S., Dube P., Krenn V., Santaguida S., Cittaro D., Monzani S., Massimiliano L., Keller J., Tarricone A., et al. 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190:835–852 10.1083/jcb.201002070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primorac I., Weir J.R., Chiroli E., Gross F., Hoffmann I., van Gerwen S., Ciliberto A., Musacchio A. 2013. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. Elife. 2:e01030 10.7554/eLife.01030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Vernieri C., Villa F., Ciliberto A., Musacchio A. 2011. Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J. 30:1508–1519 10.1038/emboj.2011.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.T., van der Waal M.S., Medema R.H., Lens S.M., Kops G.J. 2011. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat Commun. 2:316 10.1038/ncomms1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm R.B., Chaleckis R., Lehner C.F. 2009. Intrakinetochore localization and essential functional domains of Drosophila Spc105. EMBO J. 28:2374–2386 10.1038/emboj.2009.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepperd L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., Millar J.B. 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22:891–899 10.1016/j.cub.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk S.J., van Dam T.J., Karagöz G.E., von Castelmur E., Hubner N.C., Duarte A.M., Vleugel M., Perrakis A., Rüdiger S.G., Snel B., Kops G.J. 2012a. The vertebrate mitotic checkpoint protein BUBR1 is an unusual pseudokinase. Dev. Cell. 22:1321–1329 10.1016/j.devcel.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Suijkerbuijk S.J., Vleugel M., Teixeira A., Kops G.J. 2012b. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev. Cell. 23:745–755 10.1016/j.devcel.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Tang Z., Shu H., Oncel D., Chen S., Yu H. 2004. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell. 16:387–397 10.1016/j.molcel.2004.09.031 [DOI] [PubMed] [Google Scholar]
- Vleugel M., Hoogendoorn E., Snel B., Kops G.J. 2012. Evolution and function of the mitotic checkpoint. Dev. Cell. 23:239–250 10.1016/j.devcel.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Welburn J.P., Vleugel M., Liu D., Yates J.R., III, Lampson M.A., Fukagawa T., Cheeseman I.M. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Raetz E.A., Kitagawa M., Virshup D.M., Lee S.H. 2013. BUBR1 recruits PP2A via the B56 family of targeting subunits to promote chromosome congression. Biol. Open. 2:479–486 10.1242/bio.20134051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., Watanabe Y. 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 330:239–243 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Yang C.H., Tanno Y., Watanabe Y. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14:746–752 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.