Abstract
The Sry-containing protein Sox2 initially was known to regulate the self-renewal of the mouse and human embryonic stem cells (ESCs). It is also important for the maintenance of stem cells in multiple adult tissues including the brain and trachea, and it is one of the key transcription factors for establishing induced pluripotent stem cells. Recently, overexpression and gene amplification of Sox2 has been associated with the development of squamous cell carcinoma in multiple tissues such as the lung and esophagus. These different roles for Sox2 involve a complicated regulatory networks consisting of microRNAs, kinases and signaling molecules. While the levels of Sox2 are modulated transcriptionally and transnationally, post-translational modification is also important for the various functions of Sox2. In clinics, high levels of Sox2 are correlated with poor prognosis and increased proliferation of cancer stem cells. Therefore targeting Sox2 can be potentially explored for new therapeutic avenue to treat cancers. This review will focus on the different roles for Sox2 in stem cell maintenance and its oncogenic roles in the context of signal transcription and microRNA regulation. We will also review the main upstream and downstream targets of Sox2, which can be potentially used as therapeutic measures to treat cancer with abnormal levels of Sox2
Keywords: Sox2, signal transduction, cancer therapy
Introduction
Sox2, a protein belongs to the family of high-mobility group transcription factors, is pivotal for early development and maintenance of undifferentiated ESCs (embryonic stem cells). It is also one of the key transcription factors initially used to derive induced pluripotent stem (iPS) cells from fibroblast cells (REF). Interestingly, recent studies suggest that SOX2 overexpression or gene amplification have been associated with cancer development in several tissues such as the lung, esophagus and breast.
As an extremely important transcription factor, Sox2 regulates an array of genes expression involved in normal development and malignant processes, thereby plays its complicated effect in physiological network. To better understand the oncogenic roles and the corresponding signal transduction pathways involved in Sox2 protein, here, in this study, we not only emphasize the role of Sox2 in cancer and its correlated upstream or downstream molecules in signal transduction, but also propose some strategies for cancer therapy based previous studies.
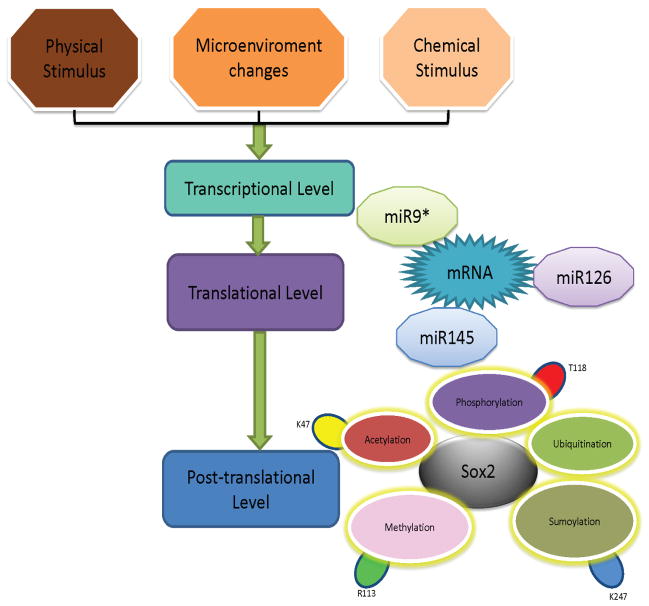
1 Variable activities of Sox2 protein are controlled at translational and post-translational level
Although there is only one transcript after Sox2 expression, activities of Sox2 are also variable due to translational and post-translational modifications. At translational level, several studies reported that microRNA could inhibit Sox2 and thus impact its role and downstream events. The low level of microRNA-145 expression in self-renewing human ESCs whereas improved expression during differentiation, and they demonstrated that microRNA-145 could inhibit human ESCs self-renewal and expression of pluripotency genes through direct repression of OCT4, SOX2, and KLF4, and microRNA-145 could promote lineage-restricted differentiation. Moreover, they also demonstrated that the promoter of microRNA-145 was repressed by OCT4 in human ESCs and thus proposed a model of the double-negative feedback loop by miR-145 and three factors[1]. Otsubo et al showed microRNA-126 could inhibit Sox2 expression by targeting two binding sites in the 3′-untranslated region of Sox2 mRNA in multiple cell lines through gain- and loss-function assays, furthermore, they revealed that PLAC1 was a downstream molecules of Sox2 and the inhibitive effect of microRNA-126 on Sox2 contributed to gastric carcinogenesis[2]. Another studies performed by Jeon et al indicated that miR-9* could target binding to the 3′-untranslated region of Sox2 mRNA and thereby influence the expression of ABCC3 and ABCC6 [3]. Liu et al found HuAECs (human amniotic epithelial cells) could provide a good source of feeder cells for mouse and human embryonic stem cells, or spermatogonial stem cells and the underlying mechanism is that Sox2 expression may be regulated by microRNA-145 in human iPS with HuAECs feeder cells[4].
Besides translational regulation, another regulatory layer is posttranslational modifications and thus impacts the activities of Sox2 protein. Sox2 possesses 317amino acids and many sites can be modified through phosphorylation, acetylation, ubiquitination, methylation and SUMOylation. Based on these modifications, Sox2 displays different activities. Tsuruzoe et al reported that mouse Sox2 could be modified at lysine 247 through SUMOylation and the SUMOylation of Sox2 at lysine247 or carboxyl terminus impaired its binding to the Fgf4 enhancer, therefore, transcriptional activities of Sox2 were negatively regulated through impaired DNA binding[5]. Baltus et al identified a lysine47 residue of Sox2 located in DNA binding domain could be acetylated by p300/CBP, and the status of acetylated lysine47 promoted the nuclear export of Sox2, meanwhile, increased level of acetylated Sox2 resulted in ubiquitination and proteasomal degradation of Sox2, additionally, they explored that blocking acetylation at this site sustained the expression of its target genes under hyperacetylation or differentiation conditions[6]. Jeong et al reported that Akt could directly interact with Sox2 and resulted in the phosphorylation of Thr118 at Sox2 protein, the phosphorylated Sox2 improved its stability and thus enhanced its transcriptional activities, eventually contributed to the reprogramming of somatic cells[7]. Zhao et al verified the existence of interactions between Sox2 and CARM1 (coactivator-associated arginine methyltransferase1), a protein possessing arginine methyltransferase activity and required for maintaining the identity of ESCs. In this study, they found CARM1 facilitated transactivation mediated by Sox2 through methylation on Arg113 of Sox2 and this event enhanced its self-association[8]. Zhang et al showed the negative effect of SUMOylation on Nanog expression and SUMOylation of Sox2 protein inhibited the Nanog expression[9]. All these modifications on Sox2 were summarized in Fig. 1, concluding from Fig. 1, we know that the activities of Sox2 are variable through translational and posttranslational modifications and thereby regulate the pluripotency and differentiation.
Fig. 1.
Multiple modifications for regulating Sox2 activities
2 Sox2 is an amplified gene in cancer
Besides the pivotal role in maintaining stemness of cells, emerging studies showed Sox2 was closely related to many types of cancer and amplified in these cancers. In SCCs (squamous cell carcinomas) of the lung and esophagus, Bass et al found a peak of genomic amplification on chromosome 3q26.33 containing Sox2 gene, which is requisite for proliferation and anchorage-independent growth of lung and esophageal cell lines verified by using RNA interference. In addition, Sox2 could cooperate with FOXE1 or FGFR2 to transform immortalized tracheobronchial epithelial cells [10]. Hussenet et al also demonstrated the occurrence of Sox2 amplification in human lung squamous cell carcinomas by using array comparative genomic hybridization, at the same time, they found that Sox2 overexpression not only deregulated genes in malignant processes but also cellular migration and anchorage-independent growth, additionally, it could also induce tumor-formation[11]. In preinvasive squamous lung cancer, McCaughan et al revealed that all high-grade lesions had 3q amplification and these lesions were prone to progress to cancer, eventually, they concluded that Sox2 and PIK3CA was amplified in high-grade dysplastic lesions through MCC (molecular copy-number counting) and increased level of Sox2 contributed to the malignant process [12]. Wilbertz et al discovered that Sox2 amplification and its corresponding up-regulation were frequent events in lung squamous cell carcinomas and were associated with indicators of favorable prognosis[13]. Updated studies indicated that Sox2 amplification in about 27% of human SCLC (small-cell lung cancer) sample, meanwhile, Sox2 knockdown blocked the proliferation of Sox2-amplified SCLC lines[14, 15].
3 Correlations between Sox2 and clinical phenomenon and its contribution to multiple processes of cancer cells
As mentioned above, amplified Sox2 in some types of cancer leads to high level of Sox2. Therefore, there might be correlations between Sox2, and clinical phenomenon and multiple processes of cancer cells were involved in the roles of Sox2. To better elucidate the relationships between them and contributions of Sox2, we explain them as following.
3.1 Clinical outcome is more deleterious along with Sox2 overexpression leading to lower survival rates of patient
At present, several studies have showed that Sox2 overexpression in cancer cells displayed more deleterious outcome, finally, giving rise to lower survival rates of patient. To investigate the roles of CD133, Oct 4, and Sox2 and the clinical outcome derived from their levels in rectal cancer patients with preoperative CRT (chemoradiotherapy), by using real time PCR, Saigusa et al discovered that patients who developed distant recurrence possessing higher levels of these three genes comparing to those without recurrence in residual cancer after CRT, and increasing levels of these genes were significantly associated with poor disease-free survival. In addition, up-regulation of these genes was also found in LoVo and SW480 cells after radiation [16]. Aim to study the relationships between Sox2 and SCLC, Maddison and his colleagues detected the Sox2 antibodies in a group of patients with SCLC, among 212 unselected SCLC patients, they found Sox2 showed with a sensitivity of 33% and specificity of 97% comparing to control. Hence, they concluded that Sox2 antibodies were specific markers in SCLC comparing to matched non-tumor controls, however, their presence did not have a significant increase in survival rate of SCLC compared with Sox2-negative patients[17]. Li et al observed Sox2 and Oct 4 exclusively expressed in the nuclei of human NSCLC (non-small-cell lung cancer) cells and the expression of Oct 4 was significantly associated with poor prognosis of lung cancer, then they concluded that Sox2 and Oct 4 could be served as promising markers in directing NSCLC diagnosis and therapy[18]. Lu et al showed the evidence that Sox2 overexpression was oncogenic in human squamous cell lung tumors and some adenocarcinomas, meanwhile, patients with high level of Sox2 were more difficult to survive[19]. However, opposite viewpoint from Wilbertz’s group supported that high-level of Sox2 was significantly associated with better survival [13].
3.2 Sox2 positively contributed to the stemness of cancer stem cells and multiple processes of cancer cells
As in embryonic stem cells, Sox2 plays the similar role in CSCs (cancer stem cells). Rodriguez-Pinilla et al found Sox2 expression in 16.7% of sporadic node-negative invasive breast carcinomas and was significantly more frequently expressed in basal-like breast carcinomas through tissue microarrays, therefore, Sox2 was preferentially expressed in tumors with basal-like phenotype and might contribute to define the characteristics of less differentiated/‘stem cell’ phenotype[20]. To distinguish cancer cells into different subsets. Hagerstrand and his colleagues identified two subsets of high-grade glioma cultures based on their expression patterns, they found type A subset possessing high level of Sox2 was more prone to induce xenograft tumors and neurospheres and displayed low or no sensitivity to monotreatment with PDGF- and IGF-1–receptor inhibitors, however, the growth of these tumors and neurospheres were efficiently inhibited by combinational treatment with low doses of these two inhibitors. Furthermore, Sox2 knockdown had significant impact on sphere formation, expression of defining gene and resistance to PDGF- and IGF-1–receptor inhibitors [21]. Wu et al identified two novel phenotypically distinct cell subsets in MCF7 and ZR751 breast cancer cell lines based on their differential Sox2 transcription activity, and subset possessing Sox2 transcription could significantly form more colonies in methylcellulose and more mammospheres in vitro compared to GFP negative cells, moreover, Sox2 knockdown in the subset expressing Sox2 abolished these abilities. Eventually, they concluded that Sox2 transcription activity was responsible for the tumorigenicity and cancer stem cell-like phenotypes in breast cancers [22].
Besides its application in identifying different subsets of cancer cells, Sox2 also plays its key role in maintaining the stemness of CSCs or TICs (tumor-initiating cells). GANGEMI et al demonstrated that Sox2 knockdown in TICs derived from adult human glioblastoma led to inhibition of proliferation and loss of tumorigenicity in immunodeficient mice, indicating that Sox2 was also fundamental for maintaining the self-renewal capacity of TICs and Sox2 or its downstream molecules might be served as an ideal target for glioblastoma therapy[23]. Basu-Roy et al also drew a conclusion that Sox2 maintained self-renewal of TICs in osteosarcomas because down-regulating Sox2 drastically reduced their transformed properties in vitro and their ability to form tumors, furthermore, osteosarcoma cells with depletion of Sox2 couldn’t form osteospheres and differentiate into mature osteoblasts any longer. Finally, they defined Sox2 as a survival factor and a novel biomarker of self-renewal in osteosarcomas[24]. The same role of Sox2 in maintaining stemness of GSC (Glioma stem cells) through miR-9*[3], CSCs in HNSCC (Head and Neck squamous cell carcinoma) through miR-302[25], CSCs in breast tumors[26] and SP (side population) cells in NSCLC were demonstrated [27]. All these studies indicated that Sox2 expression promoted and maintained the stemness of CSCs or TICs with the similar manner.
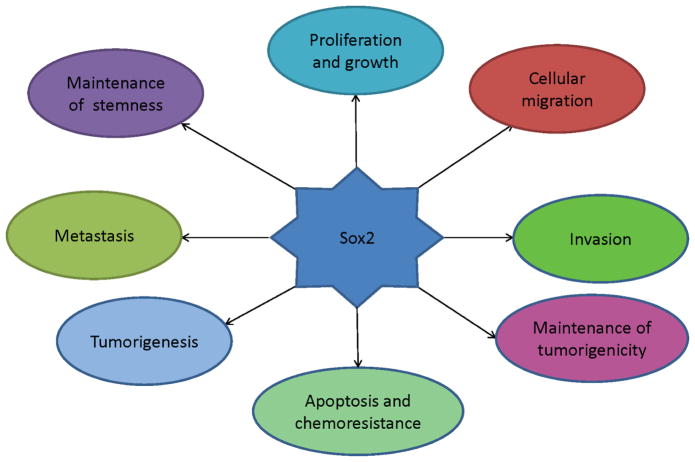
Other important roles of Sox2 in cancer progression focused on its positive contribution to many physiological processes of cancer cells. BASS et al reported that Sox2 expression was required for proliferation and anchorage-independent growth of SCCs of lung and esophageal cell lines through Sox2 knockdown experiments, moreover, Sox2 could transform immortal tracheobronchial epithelial cells with FOXE1 or FGFR2[10]. Other properties of cancer cells involved in Sox2 protein comprised proliferation and growth[23],[28],[29],[30]; cellular migration and invasion[11],[31],[32],[33]; maintenance of stemness and tumorigenicity[3],[23]; apoptosis and chemoresistance[3],[28],[25],[30]; metastasis and tumorigenesis[27],[34],[29],[28],[35]. All these roles of Sox2 in cancers were summarized in Fig. 2 and Table 1, from Table 1, we concluded that oncogenic roles were studied in many types of cancer.
Fig. 2.
Sox2 protein impacted many processes of cancer cells
Table 1.
Relationships between Sox2 and properties of cancer cells
| Cases | Multiple roles of Sox2 in cancer cells | Types of cancer | References |
|---|---|---|---|
| 1 | Promotes proliferation and growth | Lung and esophageal SCCs | [10] |
| 2 | Promotes Cellular migration | Lung SCCs | [11] |
| 3 | Promotes Cellular migration | Colorectal cancer | [31] |
| 4 | Cancer stemness and chemoresistant | Glioma | [3] |
| 5 | Promotes proliferation and tumorigenicity | Glioblastoma Tics | [23] |
| 6 | Increases anti-apoptotic property and promotes tumorigenesis | Prostate cancer | [28] |
| 7 | Invasion and migration | Glioma and BTSCs (Brain tumor stem cells) | [32] |
| 8 | Metastases | Colon cancer | [34] |
| 9 | Invasion | Gastric cancer | [33] |
| 10 | Growth and metastasis | Lung cancer | [29] |
| 11 | Chemoresistance | HNSCC | [25] |
| 12 | Proliferation and apoptosis | Prostate cancer | [30] |
| 13 | Tumorigenesis | Lung cancer | [35] |
| 14 | Proliferation | SCLC | [14] |
| 15 | Proliferation and colony formation | LN229 GBM cells | [36] |
| 16 | Proliferation and tumorigenesis | Breast cancer | [43] |
| 17 | Proliferation and colony formation | Breast cancer | [50] |
4 Associations between Sox2 and microRNAs
As a key regulator in development and carcinogenesis, Sox2 also displayed close associations with microRNAs. As mentioned above, at translational level, activities of Sox2 were controlled by several microRNAs, namely microRNA-145, microRNA-126 and microRNA-9*[1],[2], [3], [4].
Additionally, microRNAs could also be regulated by Sox2. Lin et al utilized integrated technologies including ChIP-seq, microarray profiling, and microRNA sequencing to analyze the downstream targets of Sox2. Altered expression level of 489 genes were found after Sox knockdown in GBM (glioblastoma multiforme) cells through microarray analysis, meanwhile, they also identified 105 precursor microRNAs, corresponding to 95 mature microRNAs, were regulated by Sox2, including down-regulation of miR-143, -145, -253-5p and miR-452. On the other hand, they found the occurrence of a double negative feedback loop in GBM cells between miR-145 and Sox2, potentially creating a bistable system in GBM cells[36]. Bourguignon et al found that a subset of CSCs in HSC-3 cell, derived from HNSCC, expressed high level of CD44v3 and ALDH1 (aldehyde dehydrogenase-1), Oct 4, Sox2 and Nanog, meanwhile, these cells also displayed hallmark CSCs properties of self-renewal/clonal formation and the ability to generate heterogeneous cell populations. In their study, they also observed HA (hyaluronan) promoted the interactions between CD44v3 and Oct4-Sox2-Nanog and eventually led to a complex formation and the nuclear translocation of Oct4-Sox2-Nanog. Underlying mechanism of self-renewal, clonal formation and cisplatin resistance in CSCs was microRNA-302 expression stimulated by HA-CD44 and corresponding down-regulation of several epigenetic regulators and up-regulation of several survival proteins[25].
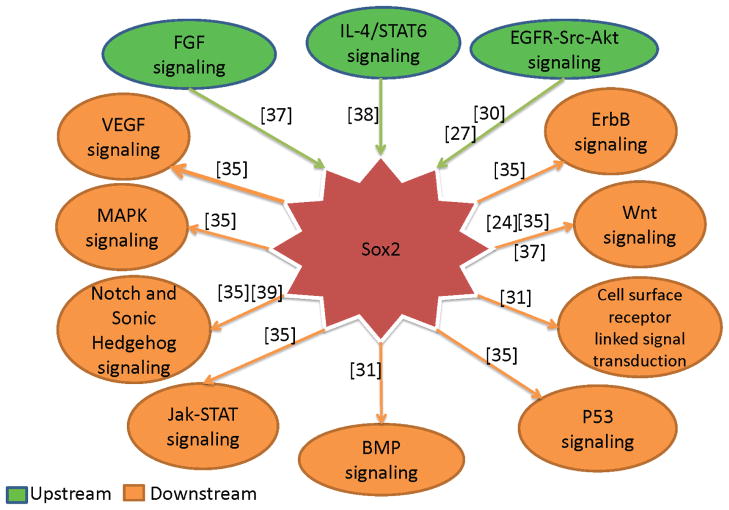
5 Sox2 is involved in complicated signal transduction pathways
As a key transcriptional regulator, Sox2 was involved in many physiological actions including normal development and pathological processes. Hence, Sox2 must be closely correlated with some signal transduction pathways. Previous studies demonstrated that several signal pathways regulated the Sox2 expression and thereby impacted cell behaviors. Moreover, Based on Sox2 overexpression and knockdown, other evidences showed that Sox2 was relevant to many signal pathways directly through its targets or indirectly through its interacting proteins. According to upstream molecules and downstream molecules of Sox2 and its interacting proteins, main signal pathways of Sox2 were summarized in Fig. 3, and also from Fig. 3, we got the information that Sox2 was involved in complicated signal pathways[24], [30],[31], [35], [37], [38],[39]. Meanwhile, in different types of cells, upstream molecules and downstream molecules of Sox2 or interacting proteins are variable.
Fig. 3.
Signal transduction pathways were directly or indirectly correlated with Sox2. FGF, IL-4/STAT6 and EGFR-Src-Akt signal pathways directly regulated Sox2 expression, additionally, other potential signal pathways were regulated by Sox2 through its targets or interacting partners.
5.1 Upstream molecules regulates Sox2 expression
Evidences that Sox2 expression was regulated by several molecules were demonstrated in previous studies. Mansukhani and colleagues found that Sox2 expression was induced by FGF in osteoblasts and thus down-regulated WNT signaling [37]. Asonuma et al found that Sox2 expressed in both human oxyntic and pyloric glands with H.pylori infection except of intestinal metaplasia. Meanwhile, they showed interleukin-4 dose dependently improved Sox2 expression among H.pylori infection-medicated cytokines in gastric epithelial cells and the induction of Sox2 was suppressed through decreasing the level of STAT6 or inhibiting STAT6 phosphorylation mediated by H.pylori and IFN-γ[38]. Riggi et al discovered that combined effect, derived from EWS-FLI-1 on its target gene expression and the inhibitive activity of microRNA-145 promoter, induced the ESFTs (Ewing sarcoma family tumors) cancer stem cell phenotype. In addition, they demonstrated that the existence of a mutually repressive feedback loop between EWS-FLI-1 and microRNA-145, and their common target gene Sox2 played key roles in determing the differentiation and tumorigenicity of EFST cells[40]. In lung cancer, Hajime et al showed another factor, hypoxia condition, induced the expression of CD133 through up-regulation of Oct3/4 and Sox2 and thereby enhanced their capability of DNA-binding to CD133 P1 promoter[41]. In SP of NSCLC, Singh et al demonstrated that the ability of self-renewal in these cells depended on EGFR-Src-Akt signaling. They also showed that the expression level of stem cell markers such as Oct 4, Sox2 and Nanog increased as well as verified EMT characteristics in SP cells. EGFR-Src-Akt signaling and their targets Sox2 were responsible for self-renewal and expansion of stem-like cells from NSCLC. Comparing to Oct 4 and Nanog, Sox2 played a more important role in modulating self-renewal[27]. Lin et al also demonstrated that EGFR/PI3K/AKT pathway regulated Sox2 expression, however, the upstream molecule of Sox2 was TGF-α [30].
5.2 Downstream molecules controlled by Sox2 protein
Previous studies have reported that an array of genes regulated by Sox2. Fang et al obtained an expression profile, including down-regulation of 1,715 probes (corresponding to 1,205 genes and 124 unannotated probes) and up-regulation of 2,109 probes (corresponding to 1,381 annotated genes, and 182 unannotated probes) in Sox2 knockdown SW620 cells compared with the mock control cells, through Sox2 knockdown and microarrays[31]. In LN229 GBM cells, by using ChIP-seq, microarray profiling, microRNA sequencing and Knockdown technologies, Fang et al discovered 4883 Sox2 binding regions in the GBM cancer genome and altered expression of 489 genes after Sox2 knockdown, Meanwhile, they identified 105 precursor microRNAs (corresponding to 95 mature microRNAs) regulated by SOX2, including down-regulation of miR-143, -145, -253-5p and miR-452 by next generation sequencing [36]. Chen et al found that silence of Sox2 with RNA interference could reduce the tumorigenic property of A549 cells with decreased expression of C-MYC, WNT1, WNT2, and NOTCH1 in xenografted NOD/SCID mice. Moreover, additional 246 target cancer genes of Sox2 were revealed through the RNA-Seq method[35]. In lung cancers, Hajime et al showed that HIF1alpha/HIF2alpha induced expression of OCT4 and SOX2 and thus promoted CD133 expression through direct interaction with P1 promoter of CD133[41]. Lin et al explored other downstream molecules of Sox2, such as Cyclin E, p27 and survivin, through RNA interference and overexpression in human prostate cancer [30]. Asonuma et al showed that the down-regulation of Sox2 led to intestinal phenotype in gastric epithelial cells AGS through its downstream targets Cdx2, MUC2 [38]. Engelen et al discovered that Jag1, Gli3 and Mycn are regulated by Sox2 and Chd7 complex through advanced technologies [39]. Mallanna et al revealed that the ectopic expression of Sox21 was induced after Sox2 overexpression in ESC and thereby promoted their differentiation into specific cell types and expression of the markers of neurectoderm and heart development [42]. Chen and colleagues demonstrated that Sox2 cooperated with β-catenin to up-regulate the expression of Cyclin D1 in breast cancer cells, and thus promoted cell proliferation and tumorigenesis. Moreover, they found 145 up-regulated genes and 41 down-regulated genes through Sox2 overexpression and microarray assays [43]. Other studies also revealed that Cyclin D1 and Nanog[22], Orai1[28], TGF-β and Oct 4[29] were also served as targets of Sox2 protein. Main genes mentioned above were summarized in Table 2. As shown in Table 2, we concluded that Sox2 is involved in many physiological processes based on the roles of partial genes regulated by Sox2.
Table 2.
Genes regulated by Sox2 and their corresponding roles
| Genes | Roles of genes regulated by Sox2 | References |
|---|---|---|
|
| ||
| C-Myc | Promotes cell proliferation, regulates cell growth, apoptosis, differentiation and self-renewal of stem cell | |
| WNT1 | Acts in oncogenesis and several developmental processes including regulation of cell fate and patterning during embryogenesis | |
| WNT2 | Plays an essential role in carcinogenesis of the esophagus, suppresses anoikis, enhances anchorage-independent sphere formation, and increases metastatic propensity in pancreatic cancer | |
| WNT7B | Plays important roles in the development and progression of gastric cancer, esophageal cancer, and pancreatic cancer | |
| NOTCH1 | A receptor of membrane bound ligands that promotes differentiation of progenitor cells into astroglia | |
| NOTCH3 | Associated with CADASIL(cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy) | |
| KLF4 | A good indicator of stem-like capacity and required for the accumulation of β-catenin at the Tert promoter | [35] |
| EGFR | Autophosphorylation elicits downstream activation and signaling that modulate DNA synthesis, cell proliferation, cell migration and adhesion | |
| BCL10 | Induces apoptosis and activates NF-kappaB | |
| JUN | Transcriptional factor associated with transformation | |
| JAK1 | Essential for signaling for certain type I and type II cytokines and important in initiating responses to multiple major cytokine receptor families | |
| YAP1 | Plays an important role in the clonogenicity and growth of pancreatic cancer cells and recruits c-Abl to protect AMOTL1 against Nedd 4.2-mediated degradation | |
|
| ||
| Sox1 | Involved in development of early central nervous system and maintenance of neural progenitor cell identity | |
| Sox18 | Plays roles in hair, blood vessel, and lymphatic vessel development and act as a transcriptional regulator | |
| GPR37 | Induces cellular autophagy | |
| PCDH10 | Involved in angiogenesis and methylation and acts as a tumor suppressor gene | [36] |
| PCDH9 | May involved in specific neuronal connections and signal transduction | |
| PCDHB11 | Unknown | |
| PCDHGA3 | Unknown | |
| PCDHGC3 | Unknown | |
|
| ||
| ST14 | Plays its roles in cancer invasion and metastasis | |
| MTSS1 | Promotes cell-cell junction assembly and stability, regulates EGF signaling in HNSSC and function as tumor suppressor | |
| NGFRAP1 | Unknown | |
| FGFR2 | Plays important roles in embryonic development and tissue repair, especially bone and blood vessels, mediates cell division, growth and differentiation | [31] |
| JUN | As described above | |
| CDKN2C | Prevents the activation of the CDK kinases and controls cell cycle G1 progression, regulates spermatogenesis and suppresses tumorigenesis | |
|
| ||
| Oct 4 | Regulates pluripotency and early cell differentiation | [29] |
| TGF-β | Controls tumor progression and metastasis | |
|
| ||
| Cyclin E | Plays critical roles in in the G1-S phase transition and tumorigenesis, phosphorylates retinoblastoma, p27, p21, Smad 3 and p220 | |
| p27 | Binds and inhibits activity of cyclin kinase, causes cells arrest in the G1 phase of the cell cycle | [30] |
| Survivin | Inhibits caspase activation and Bax and Fas-induced apoptosis | |
|
| ||
| Cyclin D1 | Regulates CDK kinases and thus alters cell cycle progression, may contribute to tumorigenesis | [22] |
| Nanog | Transcriptional activator for the Rex1 promoter and involved with self-renewal of undifferentiated embryonic stem cells | |
|
| ||
| Cdx2 | Associated with intestinal inflammation and tumorigenesis, required to form the placenta, directs early embryogenesis and induces acute myeloid leukemia | [38] |
| MUC2 | Component of a gel that protects the intestinal epithelium | |
|
| ||
| Jag1 | Plays a role in hematopoiesis and associated with Alagille syndrome | |
| Gli3 | A mediators of Sonic hedgehog (Shh) signaling and play a role during embryogenesis, associated with several diseases, including Greig cephalopolysyndactyly syndrome, Pallister-Hall syndrome | [39] |
| Mycn | Associated with Feingold syndromes and a variety of tumors including neuroblastomas | |
|
| ||
| CD133 | Unknown | [41] |
|
| ||
| Sox21 | Promotes neuronal cellular differentiation and prevents hair loss | [42] |
|
| ||
| Cyclin D1 | As described above | |
| FGFR2 | As described above | |
| SMARCA1 | Regulates transcription of certain genes by altering the chromatin structure | |
| CXCL10 | Attributes to chemoattraction for monocytes/macrophages, T cells, NK cells, and dendritic cells, and promotes T cell adhesion to endothelial cells, antitumor activity, and inhibits bone marrow colony formation and angiogenesis | [43] |
| CSTA | Functions as a cysteine protease inhibitor and plays roles in epidermal development and maintenance | |
| MME | Endopeptidase that cleaves peptides at the amino side of hydrophobic residues and inactivates several peptide hormones including glucagon, enkephalins, substance P, neurotensin, oxytocin, and bradykinin | |
| ZNF407 | Unknown | |
5.3 Interactions between Sox2 protein and other proteins
Besides the role of regulating gene expression through direct DNA binding, Sox2 can also impact the cell activities, including DNA processing, chromatin organization and assembly, and RNA processing, via complexes that resulted from protein interactions. Mallanna et al identified many proteins interacting with Sox2 through MudPIT (multidimensional protein identification technology) [42]. Subsequently, they demonstrated that, in Sox2 protein complex, Sox2 interacted with other proteins through multiple domains [44]. Ambrosetti and colleagues found that a protein complex, including Sox2 and Oct-3, bound to adjacent sites within the enhancer of FGF-4 and synergistically activated its expression[45],[46]. Engelen et al purified a Sox2 protein complex in mouse neural stem cells and verified that Chd7, a member of the chromodomain helicase DNA-binding domain family of ATP-dependent chromatin remodelers, could be served as a Sox2 binding partner through immunoprecipitation, mass spectrometry. Moreover, among these genes regulated by Sox2 and Chd7 complex, Jag1, Gli3 and Mycn are extremely important because their mutations always lead to Alagille, Pallister-Hall and Feingold syndromes respectively [39],[47]. Fang et al analyzed the Sox2 interactome in glioma cells by using immunoprecipitation and mass spectrometry, their results indicated that Sox2 interactome consisted of many proteins including heterogeneous nuclear ribonucleoprotein family proteins, other ribonucleoproteins, DNA repair proteins and helicases[48]. Bourguignon et al found HA promoted the interactions between CD44v3 and Oct4-Sox2-Nanog leading to a complex formation and the nuclear translocation of Oct4-Sox2-Nanog[25]. Although we cannot find a large number of common partners of Sox2 based on integrated studies mentioned above, however, there are still some overlap proteins among them. Concluded from [39] and [48], only Supt16h is the common proteins in these two studies. However, there are seven common proteins interacting with Sox2 in [42] and [48], including RBM14, XRCC5, XRCC6, RPS11, H2AFY, ILF3 and SSRP1 proteins. Till now, only several studies published the roles of Sox2 complexes, and the roles of most complexes need to be elucidated. Explicit roles of Sox2 complexes and its interacting proteins were summarized in Table 3. From Table 3, we concluded that these complexes play their roles in many aspects of cell and still mainly through regulating gene expression.
Table 3.
Proteins interacting with Sox2 and the roles of complex
| Interacted proteins | Roles of Complex | References |
|---|---|---|
| Oct-3 | Synergistically activates the enhancer of FGF4 | [45] |
| Oct-3 | Generates a functional complex and activates FGF4 enhancer | [46] |
| HDAC1 HDAC2 Sall4 Lin28 |
HDAC1-Sox2 complex contributes to transcriptional repression of a subset of genes such as Sox21 | [42] [44] |
| Chd7 | Regulates many genes expression including Jag1, Gli3 and Mycn which could lead to Alagille, Pallister-Hall and Feingold syndromes | [39] [47] |
| CD44v3, Oct-4, Nanog | Promotes microRNA-302 expression and self-renewal, clonal formation and cisplatin resistance in CSCs | [25] |
| β-catenin | Modulates the expression of Cyclin D1 | [43] |
6 Prospect and conclusion
Sox2 was amplified in many types of cancer and enhanced expression level was found in many cancers, moreover, Sox2 played its role through many modifications including transcriptional level mediated by signal transduction, translational level mediated by microRNA, posttranslational level such as phosphorylation, acetylation, methylation, SUMOylation and ubiquitination. On the other hand, DNA-binding activity of Sox2 was also influenced through interacting proteins. Although we have obtained evidences that Sox2 could bind and influence an array of genes expression, whereas there are inconsistent results about the downstream targets in previous studies and integrated studies of deciding the universal targets of Sox2 in different types of cancers need to be developed.
Currently, there are some confused controversies about the role of Sox2 in cancers. Firstly, some scientists thought Sox2 had a role of suppressing tumor due to its reduced expression in some types of cancer such as gastric cancers, meanwhile, Sox2 overexpression inhibited cell proliferation and resulted in cell-cycle arrest and apoptosis, moreover, the lower level of Sox2 resulted from methylation showed a lower survival rate than those without methylation[49]. However, much more studies revealed that Sox2 overexpression could positively promote the cancer progression. Consequently, we deem that Sox2 could be served as a potential target for cancer therapy. Secondly, adverse viewpoints about the relationships between Sox2 and survival rate of patient were stated by different groups. Although most of cases showed that the survival rate of patient possessing high level of Sox2 is lower than those with low level of Sox2, adverse viewpoint from Wilbertz alleged that high-level Sox2 expression was significant associated with better survival[13], it suggests that the explanation of this phenomenon need to be much more studied.
Although previous studies demonstrated that the overexpression of Sox2 derived from amplification played pivotal roles in the tumor formation and promoting cancer progression, whereas the underlying mechanism and stimulus leading to amplification remain elusive. Moreover, occurrence of modifications or mutations on Sox2 or not, which could influence tumor suppressor gene through variable activities of Sox2, during tumorigenesis also need to be demonstrated. Therefore, studying single nucleotide polymorphism or modifications on Sox2 may be meaningful for us to deduce the reason of tumorigenesis. Additionally, as described above, there are strong correlations between Sox2 and microRNAs, among these microRNAs, several microRNAs inhibited the activities of Sox2 and an array of microRNAs regulated by Sox2. An interesting discovery is that microRNA-145 was up- regulated after Sox2 knockdown[36], however, Xu et al showed that microRNA-145 inhibited the Sox2 activities in human embryonic stem cells[1]. Based on these discoveries, they concluded that microRNA-145 and Sox2 generated a double negative feedback loop in GBM cells[36]. Besides microRNA-145, another microRNA, namely microRNA-9*, was also regulated by Sox2[36], meanwhile, the inhibitive role of microRNA-9* on Sox2 activity was also verified[3]. Therefore, we speculate that a double negative feedback loop may also occur between Sox2 and microRNA-9*. In addition, Sox2 may participate in RNA (including microRNAs) processing through its interacting proteins.
Due to the promoting role of Sox2 on cancer progression, developing novel reagents or tools that target against Sox2 is an effective attempt for cancer therapy. Xiang et al isolated SP from murine D121 lung carcinoma cells and demonstrated that these cells containing CSCs because of the up-regulation of Sox2 and Oct 4 in SP-D121 cells, moreover, Sox2 knockdown in SP-D121 cells resulted in decreasing migration and increasing apoptosis, at the same time, Sox2 silence dramatically suppressed the potential of metastasis in syngeneic mice[29]. In breast cancer, another study performed by Stolzenburg and his colleague utilized a more advanced tool, ZF-ATFs (Zinc-finger (ZF)-based artificial transcription factors), to silence Sox2 expression, their results indicated that tumor cell proliferation and colony formation decreased, and in vivo, the inhibitive effect of engineered ZF-ATFs on growth of breast cancer cells was also verified[50]. However, shRNA and ZF-ATFs in most studies were delivered by virus, the blemish of applying virus is the non-specific infection as long as infected cells have corresponding receptors. To achieve the aim of target delivering Sox2 siRNA, developing a novel tool, such as aptamers-siRNA complex, into pathological sites or combining Sox2 siRNA with other measures such as chemotherapy for synergistic therapy will be our future research interest. On the other hand, another tool, peptide aptamers, can also be used for inhibiting the activities of Sox2 protein or disrupting the interactions between Sox2 and other proteins. At present, we have identified several candidates of peptide aptamers against Sox2 protein from a peptide library, which possessing a constrained peptide expression cassettes in the active site loop of thioredoxin and a partial fragment of Venus protein. Through BiFC (bimolecular fluorescence complementation) and immunoprecipitation, we demonstrated their interactions and these peptide aptamers could decrease the proliferation of human esophageal cancer cells (unpublished).
Collectively, based on the phenomenon of abundant expression of Sox2 in many types of cancer and its roles of promoting cancer progression, we deem that application of Sox2 will throw light on the signal pathways in tumorigenesis and cancer therapy.
Acknowledgments
This work was initiated in Dr. Jianwen Que’s laboratory at the Department of Biomedical Genetics, University of Rochester. This study was supported by the National Science Foundation of China (No. 81271928).
Abbreviations
- ESCs
embryonic stem cells
- iPS
induced pluripotent stem
- HuAECs
human amniotic epithelial cells
- CARM1
coactivator-associated arginine methyltransferase1
- SCCs
squamous cell carcinomas
- MCC
molecular copy-number counting
- SCLC
small-cell lung cancer
- CRT
chemoradiotherapy
- NSCLC
non-small-cell lung cancer
- CSCs
cancer stem cells
- TICs
tumor-initiating cells
- GSC
Glioma stem cells
- HNSCC
Head and Neck squamous cell carcinoma
- SP
side population
- GBM
glioblastoma multiforme
- ALDH1
aldehyde dehydrogenase-1
- HA
hyaluronan
- ESFTs
Ewing sarcoma family tumors
- MudPIT
multidimensional protein identification technology
- ZF-ATFs
Zinc-finger (ZF)-based artificial transcription factors
- BiFC
bimolecular fluorescence complementation
- BTSCs
Brain tumor stem cells
- CADASIL
cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy
References
- 1.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 2.Otsubo T, Akiyama Y, Hashimoto Y, Shimada S, Goto K, Yuasa Y. MicroRNA-126 inhibits SOX2 expression and contributes to gastric carcinogenesis. PLoS One. 2011;6:e16617. doi: 10.1371/journal.pone.0016617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeon HM, Sohn YW, Oh SY, Kim SH, Beck S, Kim S, et al. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2011;71:3410–21. doi: 10.1158/0008-5472.CAN-10-3340. [DOI] [PubMed] [Google Scholar]
- 4.Liu T, Cheng W, Huang Y, Huang Q, Jiang L, Guo L. Human amniotic epithelial cell feeder layers maintain human iPS cell pluripotency via inhibited endogenous microRNA-145 and increased Sox2 expression. Exp Cell Res. 2012;318:424–34. doi: 10.1016/j.yexcr.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Tsuruzoe S, Ishihara K, Uchimura Y, Watanabe S, Sekita Y, Aoto T, et al. Inhibition of DNA binding of Sox2 by the SUMO conjugation. Biochem Biophys Res Commun. 2006;351:920–6. doi: 10.1016/j.bbrc.2006.10.130. [DOI] [PubMed] [Google Scholar]
- 6.Baltus GA, Kowalski MP, Zhai H, Tutter AV, Quinn D, Wall D, et al. Acetylation of sox2 induces its nuclear export in embryonic stem cells. Stem Cells. 2009;27:2175–84. doi: 10.1002/stem.168. [DOI] [PubMed] [Google Scholar]
- 7.Jeong CH, Cho YY, Kim MO, Kim SH, Cho EJ, Lee SY, et al. Phosphorylation of Sox2 cooperates in reprogramming to pluripotent stem cells. Stem Cells. 2010;28:2141–50. doi: 10.1002/stem.540. [DOI] [PubMed] [Google Scholar]
- 8.Zhao HY, Zhang YJ, Dai H, Zhang Y, Shen YF. CARM1 mediates modulation of Sox2. PLoS One. 2011;6:e27026. doi: 10.1371/journal.pone.0027026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y, Guo Z, Wu H, Wang X, Yang L, Shi X, et al. SUMOylation represses Nanog expression via modulating transcription factors Oct 4 and Sox2. PLoS One. 2012;7:e39606. doi: 10.1371/journal.pone.0039606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–42. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussenet T, Dali S, Exinger J, Monga B, Jost B, Dembele D, et al. SOX2 is an oncogene activated by recurrent 3q26. 3 amplifications in human lung squamous cell carcinomas. PLoS One. 2010;5:e8960. doi: 10.1371/journal.pone.0008960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCaughan F, Pole JC, Bankier AT, Konfortov BA, Carroll B, Falzon M, et al. Progressive 3q amplification consistently targets SOX2 in preinvasive squamous lung cancer. Am J Respir Crit Care Med. 2010;182:83–91. doi: 10.1164/rccm.201001-0005OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilbertz T, Wagner P, Petersen K, Stiedl AC, Scheble VJ, Maier S, et al. SOX2 gene amplification and protein overexpression are associated with better outcome in squamous cell lung cancer. Mod Pathol. 2011;24:944–53. doi: 10.1038/modpathol.2011.49. [DOI] [PubMed] [Google Scholar]
- 14.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–6. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genetics: SOX2 is amplified in small-cell lung cancer. Nat Rev Clin Oncol. 2012 [Google Scholar]
- 16.Saigusa S, Tanaka K, Toiyama Y, Yokoe T, Okugawa Y, Ioue Y, et al. Correlation of CD133, OCT4, and SOX2 in rectal cancer and their association with distant recurrence after chemoradiotherapy. Ann Surg Oncol. 2009;16:3488–98. doi: 10.1245/s10434-009-0617-z. [DOI] [PubMed] [Google Scholar]
- 17.Maddison P, Thorpe A, Silcocks P, Robertson JF, Chapman CJ. Autoimmunity to SOX2, clinical phenotype and survival in patients with small-cell lung cancer. Lung Cancer. 2010;70:335–9. doi: 10.1016/j.lungcan.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Wang J, Xu Z, Ahmad A, Li E, Wang Y, et al. Expression of sox2 and oct 4 and their clinical significance in human non-small-cell lung cancer. Int J Mol Sci. 2012;13:7663–75. doi: 10.3390/ijms13067663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y, Futtner C, Rock JR, Xu X, Whitworth W, Hogan BL, et al. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS One. 2010;5:e11022. doi: 10.1371/journal.pone.0011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez-Pinilla SM, Sarrio D, Moreno-Bueno G, Rodriguez-Gil Y, Martinez MA, Hernandez L, et al. Sox2: a possible driver of the basal-like phenotype in sporadic breast cancer. Mod Pathol. 2007;20:474–81. doi: 10.1038/modpathol.3800760. [DOI] [PubMed] [Google Scholar]
- 21.Hagerstrand D, He X, Bradic Lindh M, Hoefs S, Hesselager G, Ostman A, et al. Identification of a SOX2-dependent subset of tumor- and sphere-forming glioblastoma cells with a distinct tyrosine kinase inhibitor sensitivity profile. Neuro Oncol. 2011;13:1178–91. doi: 10.1093/neuonc/nor113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu F, Zhang J, Wang P, Ye X, Jung K, Bone KM, et al. Identification of two novel phenotypically distinct breast cancer cell subsets based on Sox2 transcription activity. Cell Signal. 2012;24:1989–98. doi: 10.1016/j.cellsig.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, et al. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–8. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 24.Basu-Roy U, Seo E, Ramanathapuram L, Rapp TB, Perry JA, Orkin SH, et al. Sox2 maintains self renewal of tumor-initiating cells in osteosarcomas. Oncogene. 2011 doi: 10.1038/onc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bourguignon LY, Wong G, Earle C, Chen L. Hyaluronan-CD44v3 Interaction with Oct4-Sox2-Nanog Promotes miR-302 Expression Leading to Self-renewal, Clonal Formation, and Cisplatin Resistance in Cancer Stem Cells from Head and Neck Squamous Cell Carcinoma. J Biol Chem. 2012;287:32800–24. doi: 10.1074/jbc.M111.308528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31:1354–65. doi: 10.1038/onc.2011.338. [DOI] [PubMed] [Google Scholar]
- 27.Singh S, Trevino JG, Bora-Singhal N, Coppola D, Haura E, Altiok S, et al. EGFR/Src/Akt signaling modulates Sox2 expression and self-renewal of stem-like side-population cells in non-small cell lung cancer. Mol Cancer. 2012;11:73. doi: 10.1186/1476-4598-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia X, Li X, Xu Y, Zhang S, Mou W, Liu Y, et al. SOX2 promotes tumorigenesis and increases the anti-apoptotic property of human prostate cancer cell. J Mol Cell Biol. 2011;3:230–8. doi: 10.1093/jmcb/mjr002. [DOI] [PubMed] [Google Scholar]
- 29.Xiang R, Liao D, Cheng T, Zhou H, Shi Q, Chuang TS, et al. Downregulation of transcription factor SOX2 in cancer stem cells suppresses growth and metastasis of lung cancer. Br J Cancer. 2011;104:1410–7. doi: 10.1038/bjc.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin F, Lin P, Zhao D, Chen Y, Xiao L, Qin W, et al. Sox2 targets cyclin E, p27 and survivin to regulate androgen-independent human prostate cancer cell proliferation and apoptosis. Cell Prolif. 2012;45:207–16. doi: 10.1111/j.1365-2184.2012.00812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang X, Yu W, Li L, Shao J, Zhao N, Chen Q, et al. ChIP-seq and functional analysis of the SOX2 gene in colorectal cancers. OMICS. 2010;14:369–84. doi: 10.1089/omi.2010.0053. [DOI] [PubMed] [Google Scholar]
- 32.Alonso MM, Diez-Valle R, Manterola L, Rubio A, Liu D, Cortes-Santiago N, et al. Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PLoS One. 2011;6:e26740. doi: 10.1371/journal.pone.0026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uozaki H, Barua RR, Minhua S, Ushiku T, Hino R, Shinozaki A, et al. Transcriptional factor typing with SOX2, HNF4aP1, and CDX2 closely relates to tumor invasion and Epstein-Barr virus status in gastric cancer. Int J Clin Exp Pathol. 2011;4:230–40. [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann J, Bahr F, Horst D, Kriegl L, Engel J, Luque RM, et al. SOX2 expression correlates with lymph-node metastases and distant spread in right-sided colon cancer. BMC Cancer. 2011;11:518. doi: 10.1186/1471-2407-11-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S, Xu Y, Chen Y, Li X, Mou W, Wang L, et al. SOX2 gene regulates the transcriptional network of oncogenes and affects tumorigenesis of human lung cancer cells. PLoS One. 2012;7:e36326. doi: 10.1371/journal.pone.0036326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, et al. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics. 2011;12:11. doi: 10.1186/1471-2164-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mansukhani A, Ambrosetti D, Holmes G, Cornivelli L, Basilico C. Sox2 induction by FGF and FGFR2 activating mutations inhibits Wnt signaling and osteoblast differentiation. J Cell Biol. 2005;168:1065–76. doi: 10.1083/jcb.200409182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asonuma S, Imatani A, Asano N, Oikawa T, Konishi H, Iijima K, et al. Helicobacter pylori induces gastric mucosal intestinal metaplasia through the inhibition of interleukin-4-mediated HMG box protein Sox2 expression. Am J Physiol Gastrointest Liver Physiol. 2009;297:G312–22. doi: 10.1152/ajpgi.00518.2007. [DOI] [PubMed] [Google Scholar]
- 39.Engelen E, Akinci U, Bryne JC, Hou J, Gontan C, Moen M, et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43:607–11. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- 40.Riggi N, Suva ML, De Vito C, Provero P, Stehle JC, Baumer K, et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010;24:916–32. doi: 10.1101/gad.1899710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iida H, Suzuki M, Goitsuka R, Ueno H. Hypoxia induces CD133 expression in human lung cancer cells by up-regulation of OCT3/4 and SOX2. Int J Oncol. 2012;40:71–9. doi: 10.3892/ijo.2011.1207. [DOI] [PubMed] [Google Scholar]
- 42.Mallanna SK, Ormsbee BD, Iacovino M, Gilmore JM, Cox JL, Kyba M, et al. Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells. 2010;28:1715–27. doi: 10.1002/stem.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, et al. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem. 2008;283:17969–78. doi: 10.1074/jbc.M802917200. [DOI] [PubMed] [Google Scholar]
- 44.Cox JL, Mallanna SK, Luo X, Rizzino A. Sox2 uses multiple domains to associate with proteins present in Sox2-protein complexes. PLoS One. 2010;5:e15486. doi: 10.1371/journal.pone.0015486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol Cell Biol. 1997;17:6321–9. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ambrosetti DC, Scholer HR, Dailey L, Basilico C. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J Biol Chem. 2000;275:23387–97. doi: 10.1074/jbc.M000932200. [DOI] [PubMed] [Google Scholar]
- 47.Puc J, Rosenfeld MG. SOX2 and CHD7 cooperatively regulate human disease genes. Nat Genet. 2011;43:505–6. doi: 10.1038/ng.843. [DOI] [PubMed] [Google Scholar]
- 48.Fang X, Yoon JG, Li L, Tsai YS, Zheng S, Hood L, et al. Landscape of the SOX2 protein-protein interactome. Proteomics. 2011;11:921–34. doi: 10.1002/pmic.201000419. [DOI] [PubMed] [Google Scholar]
- 49.Otsubo T, Akiyama Y, Yanagihara K, Yuasa Y. SOX2 is frequently downregulated in gastric cancers and inhibits cell growth through cell-cycle arrest and apoptosis. Br J Cancer. 2008;98:824–31. doi: 10.1038/sj.bjc.6604193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stolzenburg S, Rots MG, Beltran AS, Rivenbark AG, Yuan X, Qian H, et al. Targeted silencing of the oncogenic transcription factor SOX2 in breast cancer. Nucleic Acids Res. 2012;40:6725–40. doi: 10.1093/nar/gks360. [DOI] [PMC free article] [PubMed] [Google Scholar]