Abstract
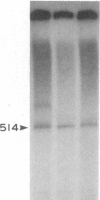
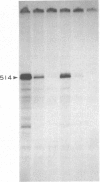
Six cell-free extracts have been used to characterize the nature of DNA signals and trans-acting factors responsible for the transcription enhancement of the Bombyx mori fibroin gene. The upstream element of the fibroin gene involved in the enhancement can be divided into two regions. The proximal region, -72 to -32, is recognized as a common enhancing signal by all B. mori extracts from the posterior silk gland, the middle silk gland, the ovarian tissue, and an embryonic cell line. It is weakly recognized by an Antheraea silkworm cell line extract but not by a HeLa cell extract. The distal region, -238 to -73, appears to be a tissue-specific enhancing signal that is recognized more effectively by the posterior silk gland extract than by the middle silk gland extract. These observations suggest that the use of these cell-free systems can offer a means for the biochemical characterization of the trans-acting factors involved in the tissue-specific regulation of the fibroin gene.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dudler R., Travers A. A. Upstream elements necessary for optimal function of the hsp 70 promoter in transformed flies. Cell. 1984 Sep;38(2):391–398. doi: 10.1016/0092-8674(84)90494-x. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Garabedian M. J., Shepherd B. M., Wensink P. C. A tissue-specific transcription enhancer from the Drosophila yolk protein 1 gene. Cell. 1986 Jun 20;45(6):859–867. doi: 10.1016/0092-8674(86)90560-x. [DOI] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Delimitation of far upstream sequences required for maximal in vitro transcription of an H2A histone gene. Proc Natl Acad Sci U S A. 1982 Jan;79(2):297–301. doi: 10.1073/pnas.79.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen U., Sharp P. A. Sequences controlling in vitro transcription of SV40 promoters. EMBO J. 1983;2(12):2293–2303. doi: 10.1002/j.1460-2075.1983.tb01737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein U., England B., Tjian R. Characterization of Drosophila transcription factors that activate the tandem promoters of the alcohol dehydrogenase gene. Cell. 1985 Jul;41(3):965–977. doi: 10.1016/s0092-8674(85)80077-5. [DOI] [PubMed] [Google Scholar]
- Hen R., Sassone-Corsi P., Corden J., Gaub M. P., Chambon P. Sequences upstream from the T-A-T-A box are required in vivo and in vitro for efficient transcription from the adenovirus serotype 2 major late promoter. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7132–7136. doi: 10.1073/pnas.79.23.7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose S., Tsuda M., Suzuki Y. Enhanced transcription of fibroin gene in vitro on covalently closed circular templates. J Biol Chem. 1985 Sep 5;260(19):10557–10562. [PubMed] [Google Scholar]
- Jove R., Manley J. L. In vitro transcription from the adenovirus 2 major late promoter utilizing templates truncated at promoter-proximal sites. J Biol Chem. 1984 Jul 10;259(13):8513–8521. [PubMed] [Google Scholar]
- Ladiges W. C., Raff R. F., Brown S., Deeg H. J., Storb R. The canine major histocompatibility complex. Supertypic specificities defined by the primed lymphocyte test (PLT). Immunogenetics. 1984;19(4):359–365. doi: 10.1007/BF00345410. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H., Ishikawa E., Suzuki Y. Structural analysis of sericin genes. Homologies with fibroin gene in the 5' flanking nucleotide sequences. J Biol Chem. 1982 Dec 25;257(24):15192–15199. [PubMed] [Google Scholar]
- Sassone-Corsi P., Dougherty J. P., Wasylyk B., Chambon P. Stimulation of in vitro transcription from heterologous promoters by the simian virus 40 enhancer. Proc Natl Acad Sci U S A. 1984 Jan;81(2):308–312. doi: 10.1073/pnas.81.2.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Sergeant A., Bohmann D., Zentgraf H., Weiher H., Keller W. A transcription enhancer acts in vitro over distances of hundreds of base-pairs on both circular and linear templates but not on chromatin-reconstituted DNA. J Mol Biol. 1984 Dec 15;180(3):577–600. doi: 10.1016/0022-2836(84)90028-7. [DOI] [PubMed] [Google Scholar]
- Topol J., Ruden D. M., Parker C. S. Sequences required for in vitro transcriptional activation of a Drosophila hsp 70 gene. Cell. 1985 Sep;42(2):527–537. doi: 10.1016/0092-8674(85)90110-2. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Suzuki Y. Faithful transcription initiation of fibroin gene in a homologous cell-free system reveals an enhancing effect of 5' flanking sequence far upstream. Cell. 1981 Nov;27(1 Pt 2):175–182. doi: 10.1016/0092-8674(81)90371-8. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Suzuki Y. Transcription modulation in vitro of the fibroin gene exerted by a 200-base-pair region upstream from the "TATA" box. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7442–7446. doi: 10.1073/pnas.80.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y., Hirose S., Tsuda M., Suzuki Y. Promoter sequence of fibroin gene assigned by in vitro transcription system. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4838–4842. doi: 10.1073/pnas.78.8.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y., Suzuki Y. Natural fibroin genes purified without using cloning procedures from fibroin-producing and -nonproducing tissues reveal indistinguishable structure and function. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1644–1648. doi: 10.1073/pnas.81.6.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M. D., Edlund T., Boulet A. M., Rutter W. J. Cell-specific expression controlled by the 5'-flanking region of insulin and chymotrypsin genes. Nature. 1983 Dec 8;306(5943):557–561. doi: 10.1038/306557a0. [DOI] [PubMed] [Google Scholar]
- Weil P. A., Luse D. S., Segall J., Roeder R. G. Selective and accurate initiation of transcription at the Ad2 major late promotor in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979 Oct;18(2):469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]
- Wildeman A. G., Sassone-Corsi P., Grundström T., Zenke M., Chambon P. Stimulation of in vitro transcription from the SV40 early promoter by the enhancer involves a specific trans-acting factor. EMBO J. 1984 Dec 20;3(13):3129–3133. doi: 10.1002/j.1460-2075.1984.tb02269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]