Abstract
Renal tubulointerstitial fibrosis is the common end point of progressive renal disease. MicroRNA (miR)-214 and miR-21 are upregulated in models of renal injury, but the function of miR-214 in this setting and the effect of its manipulation remain unknown. We assessed the effect of inhibiting miR-214 in an animal model of renal fibrosis. In mice, genetic deletion of miR-214 significantly attenuated interstitial fibrosis induced by unilateral ureteral obstruction (UUO). Treatment of wild-type mice with an anti-miR directed against miR-214 (anti-miR-214) before UUO resulted in similar antifibrotic effects, and in vivo biodistribution studies demonstrated that anti–miR-214 accumulated at the highest levels in the kidney. Notably, in vivo inhibition of canonical TGF-β signaling did not alter the regulation of endogenous miR-214 or miR-21. Whereas miR-21 antagonism blocked Smad 2/3 activation, miR-214 antagonism did not, suggesting that miR-214 induces antifibrotic effects independent of Smad 2/3. Furthermore, TGF-β blockade combined with miR-214 deletion afforded additional renal protection. These phenotypic effects of miR-214 depletion were mediated through broad regulation of the transcriptional response to injury, as evidenced by microarray analysis. In human kidney tissue, miR-214 was detected in cells of the glomerulus and tubules as well as in infiltrating immune cells in diseased tissue. These studies demonstrate that miR-214 functions to promote fibrosis in renal injury independent of TGF-β signaling in vivo and that antagonism of miR-214 may represent a novel antifibrotic treatment in the kidney.
CKD is associated with substantial medical and socioeconomic burdens. Renal tubulointerstitial fibrosis is the common end point of chronic renal diseases and remains the best predictor of disease progression.1 Fibrosis results from accumulation of extracellular matrix proteins with replacement of normal tissue with scar tissue.2 Fibrosis is broadly considered irreversible in renal disease. In the clinic, progressive CKD leads to ESRD, with dialysis or transplantation being the only treatment options. At the molecular level, TGF-β is a fundamental mediator of renal fibrosis.3 Treatment with anti–TGF-β antibodies,4 small molecule inhibitors of TGF-β signaling,5 or mothers against decapentaplegic homolog 7 (Smad7)6 are protective against the development of experimental renal tubulointerstitial fibrosis.
Identification of novel antifibrotic therapies would be aided by improved understanding of pathways that promote fibrosis. Recent attention has focused on the role of TGF-β–dependent regulation of microRNA (miRNA). miRNAs are noncoding RNAs that have substantial post-transcriptional regulatory functions.7–10 Previous reports demonstrated that miR-214 and miR-21 are upregulated in models of renal disease11–14 and implicated miR-21 in human renal pathology.12 At least in vitro, expression of both miRNAs is TGF-β dependent.11,12,15 Whereas miR-21 downregulation is protective in renal fibrosis,12,13,16 little is known about miR-214. MiR-214 is increased in monocytes of patients with CKD.17 In cardiac tissue, miR-214 is a marker of stress18 and miR-214 deletion leads to increased fibrosis after ischemic injury,19 a result of target derepression of the sodium/calcium exchanger (NCX-1), leading to Ca2+ overloading of cardiomyocytes.19
This study aimed to investigate the molecular regulation and function of miR-214 in an experimental model of renal tubulointerstitial fibrosis.
Results
After our demonstration that miR-21 and miR-214 are both induced by TGF-β stimulation in vitro, an effect blocked by the ALK5 inhibitor SB52533411 (6-[2-tert-butyl-5-(6-methyl-pyridin-2-yl)-1H-imidazol-4-yl]-quinoxaline) (orally active5), we hypothesized that both miR-21 and miR-214 expression would be reduced by SB525334 in the unilateral ureteral obstruction (UUO) murine model. Administration of vehicle had no effect on sham or UUO-injured mice (Supplemental Figure 1). SB525334 administration resulted in a 62% reduction (P<0.001) in fibrosis, as measured by both picrosirius red and Masson’s trichrome staining (Figure 1A). Analysis of profibrotic gene expression markers demonstrated a significant decrease in Col1a (79%) and Col3a (74%) in drug-treated mice (Figure 1B). A significant decrease in Mmp2 and Mmp9 was also observed (Figure 1B). α-smooth muscle actin (αSMA) is indicative of activated myofibroblasts that proliferate in the interstitial space and are profibrotic.20 SB525334 reduced the induction of αSMA mRNA (Figure 1B), which was also reflected in a decrease in αSMA staining (Supplemental Figure 1B). SB525334 prevents phosphorylation of Smad 2/3 leading to a block in canonical TGF-β/Smad 2/3–mediated signaling.5 We next determined the phosphorylation status of Smad 2/3. UUO resulted in a significant increase in p-Smad 2 and p-Smad 3 (Figure 1C), an effect attenuated by SB525334 (Figure 1C). We next quantified p-Smad 2/3 nuclear translocation. UUO resulted in approximately 45% of nuclei staining positive for p-Smad 2/3, reduced to 10% by SB525334 treatment (Figure 1D). Therefore, SB525334 prevents TGF-β–mediated canonical smad signaling after UUO in vivo. Downstream mediators of this signaling cascade, including plasminogen activator inhibitor-1 and connective tissue growth factor (Ctgf), were blocked by SB525334 (Supplemental Figure 2).
Figure 1.
Effect of SB525334 treatment on tubulointerstitial fibrosis in mice. C57bl/6 mice are treated daily with SB525334 from day −1 to 7 days at a dose of 10 mg/kg by oral gavage. UUO or sham surgery is performed on day 0 and animals euthanized at +7 days. (A) FFPE kidney tissue in 3-µM sections. Picrosirius red and Masson’s trichrome staining is performed to assess the level of collagen deposition, and the extent of fibrosis (by Picrosirius red staining) is quantified by Image Pro Plus (n=6 per group, one-way ANOVA with Tukey’s post hoc test is used to analyze data). ###P<0.001 versus UUO + vehicle. (B) Total kidney RNA is extracted (n=6 per group) using a miRNeasy kit (Qiagen). Gene expression is assessed using specific probes (Life Technologies) and normalized to mouse Gapdh. ***P<0.001 versus sham; #P<0.05 versus UUO + vehicle. (C) Protein lysates from snap-frozen kidney tissue are prepared and quantified, and 50 μg is fractionated. Proteins are transferred onto nitrocellulose membranes and probed for p-Smad 2 (1:500), p-Smad 3 (1:500), and GAPDH (1:1000). Densitometry is performed using Quantity One software (n=3 per group). One-way ANOVA with Tukey’s post hoc is used to analyze data. ***P<0.001; *P<0.05 versus sham; #P<0.01 versus UUO + vehicle. (D) Nuclear translocation of p-Smad 2/3 is assessed on 3-µM FFPE sections (n=4 per group). Rabbit polyclonal antibody specific to p-Smad 2/3 is used, with a goat-anti-rabbit Alexa488 as a secondary detection antibody (n=6 fields of view are blind counted per section). Quantification is performed by counting the number of positive nuclei as a percentage of the total nuclei present. One-way ANOVA with Tukey’s post hoc is used to analyze data. ***P<0.001; *P<0.05 versus sham; #P<0.001 versus UUO + vehicle. Scale bar, 100 µM in A; 20 µM in D. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; RQ, relative quantification; MMP, matrix metalloproteinase; DAPI, 4',6-diamidino-2-phenylindole.
Because in vitro TGF-β regulates miR-21 and miR-214 expression,11 we assessed whether miR-21 and miR-214 expression levels were altered by SB525334 in vivo. Surprisingly, kidney levels of both miR-214 and miR-21 were unaffected by drug treatment (Figure 2, A and B). In contrast, depression of miR-29 family expression (a known TGF-β/Smad 3–sensitive miRNA family21) induced by TGF-β was completely reversed by SB525334 treatment (Figure 2C). In situ analysis of miRNA in the mouse kidneys revealed that miR-21 and miR-214 were prominent in dilated tubules, and that this localization was unaffected by SB525334 (Figure 2D). High levels of nuclear miR-214 staining were observed, particularly in sham-operated kidneys (Figure 2D). Because the probes detect all forms of the miRNA, this may indicate a high level of primary (pri-) and precursor (pre-) miR-214 being present. This result suggests that under these experimental conditions, both miR-214 and miR-21 are induced after UUO in a manner independent of ALK5 signaling.
Figure 2.
Effect of SB525334 treatment on miRNA expression in mice with experimentally induced tubulointerstitial fibrosis. (A) Total kidney RNA is extracted (n=6 per group) and miRNA expression is assessed using specific probes (Life Technologies) for miR-21 and miR-214 and is normalized to U6. One-way ANOVA with Tukey’s post hoc test is used to analyze data. ***P<0.001 versus sham animals. (B) Northern blots are performed on total kidney RNA extracted using the miRNeasy kit (Qiagen) from the kidney tissue of UUO and sham-operated animals at 7 days postsurgery and probed using 5′- digoxigenin-labeled LNA mercury probes (Exiqon) for miR-21 and miR-214. U6 is used as a loading control (Exiqon). (C) Total kidney RNA is extracted (n=6 per group) and miRNA expression is assessed using specific probes for the miR-29 family (Life Technologies) and normalized to U6. One-way ANOVA with Tukey’s post hoc test is used to analyze data. **P<0.01; ***P<0.001 versus sham animals; #P<0.05; ###P<0.001 versus UUO + vehicle. (D) In situ analysis of the location and expression of miR-21 and miR-214. 5′,3′-digoxigenin-labeled LNA mercury probes (Exiqon) are used to detect miR expression. Scale bar, 100 µM. Arrows indicate dilating tubules.
We next evaluated the role of miR-214 in renal fibrosis, comparing the severity of renal fibrosis in miR-214 and miR-21 knockout mice with that in wild-type mice after UUO. Global knockout was confirmed by quantitative RT-PCR (qRT-PCR) for the mature miRNA and by Northern blot analyses using specific locked nucleic acid (LNA) probes for the mature miRNA and its precursors (Supplemental Figure 3). In contrast to the profibrotic phenotype of miR-214 knockout in ischemic cardiac injury,19 miR-214−/− animals demonstrated a 93% reduction in renal fibrosis compared with controls (Figure 3A). A 75% reduction was observed in miR-21−/− animals (Figure 3A), concurring with published data.12 Profibrotic gene expression profiles were decreased (Col1a, Col3a, and αSMA) in miR-214−/− mice (Figure 3B). We investigated whether the observed responses were due to a difference in p-Smad 2/3 nuclear translocation. In contrast to the substantial reduction in p-Smad 2/3 nuclear localization observed in miR-21−/− mice, deletion of miR-214 had no effect on p-Smad 2/3 nuclear translocation (Figure 3C). Consistent with this, Ctgf expression levels were lower in miR-21−/− animals compared with controls but were not altered in miR-214−/− mice (Supplemental Figure 4). This suggests that the TGF-β/miR-21/Smad 3 axis is intrinsically linked, because genetic ablation of key components of this pathway all affect fibrotic outcome. However, the antifibrotic effect of miR-214 deletion is mediated through a Smad 2/3-independent pathway.
Figure 3.
Effect of genetic deletion of miR-214 on tubulointerstitial fibrosis induced by UUO. miR-21−/−, miR-214−/−, and wild-type animals are subjected to sham (n=5 per group) or UUO (n=6 per group) surgery. Seven days after surgery, animals are euthanized and kidney tissue is formalin fixed; FFPE kidneys are then sectioned into 3-µM sections. (A) Picrosirius red staining is performed to assess the level of fibrosis. Fibrosis is quantified by Image Pro Plus in a blind manner. One-way ANOVA with Tukey’s post hoc test is used to analyze data. ***P<0.001; *P<0.05 versus sham; ##P<0.01 versus WT UUO. (B) Total kidney RNA is extracted using the miRNeasy kit (Qiagen) (n=5–6 per group) and gene expression is assessed using specific probes (Life Technologies) and normalized to mouse GAPDH. One-way ANOVA with Tukey’s post hoc test is used to analyze data. #P<0.05; ##P<0.01 versus WT UUO. (C) Nuclear translocation of p-Smad 2/3 is assessed on 3-µM FFPE sections (n=4 per group). Rabbit polyclonal antibody specific to p-smad 2/3 is used with a goat-anti-rabbit Alexa488 as a secondary. Quantification is performed by counting the number of positive nuclei as a percentage of all nuclei present. One-way ANOVA with Tukey’s post hoc test is used to analyze data. ###P<0.001 versus WT UUO. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; NS, not significant; RQ, relative quantification; DAPI, 4',6-diamidino-2-phenylindole. Scale bar, 100 µM in A; 20 µM in C.
Logically, we evaluated whether a combination of miR-214 loss with concomitant SB525334 administration would show additional benefits, because we had shown a decrease in fibrosis in wild-type animals treated with this agent (Figure 1). Treatment of miR-214−/− animals with SB525334 resulted in a further reduction in fibrosis, as assessed by picrosirius red staining (Figure 4A), and a decrease in collagen expression (Figure 4B). However, it is not possible to ascertain whether this effect was greater than that which would be observed with SB525334 treatment alone. The decrease in fibrosis was paralleled by a blockade in nuclear translocation of p-Smad 2/3 (Figure 4C).
Figure 4.
Effect of combination of SB525334 treatment and genetic deletion of miR-214 on tubulointerstitial fibrosis induced by UUO. (A) miR-214−/− mice are treated daily with SB525334 from day −1 to day 7 at a dose of 10 mg/kg by oral gavage (n=6 per group). UUO or sham surgery is performed at day 0 and animals are euthanized at +7 days. Kidney tissue is formalin fixed and FFPE kidneys are then sectioned in 3-µM sections. Picrosirius red staining is performed to assess the level of fibrosis by collagen deposition. Fibrosis is quantified by Image Pro Plus in a blind manner. The unpaired t test is used to analyze the data. **P<0.01 versus miR-214−/− UUO + vehicle. (B) Total kidney RNA is extracted (n=5 sham/n=6 UUO) and gene expression is assessed using specific probes (Life Technologies) and normalized to mouse Gapdh. One-way ANOVA with Tukey’s post hoc test is used to analyze data. *P<0.05 versus miR-214−/− + vehicle-treated animals. (C) Nuclear translocation of p-smad 2/3 is assessed on 3-µM FFPE sections (n=4 per group). Rabbit polyclonal antibody specific to p-smad 2/3 is used with a goat-anti-rabbit Alexa488 as a secondary (n=6 fields of view are blind counted per section). Quantification is performed by counting the number of positive nuclei as a percentage of all nuclei present. The unpaired t test is used to analyze the data. ***P<0.001 versus miR-214−/− + vehicle UUO. DAPI, 4',6-diamidino-2-phenylindole. Scale bar, 20 µM in C.
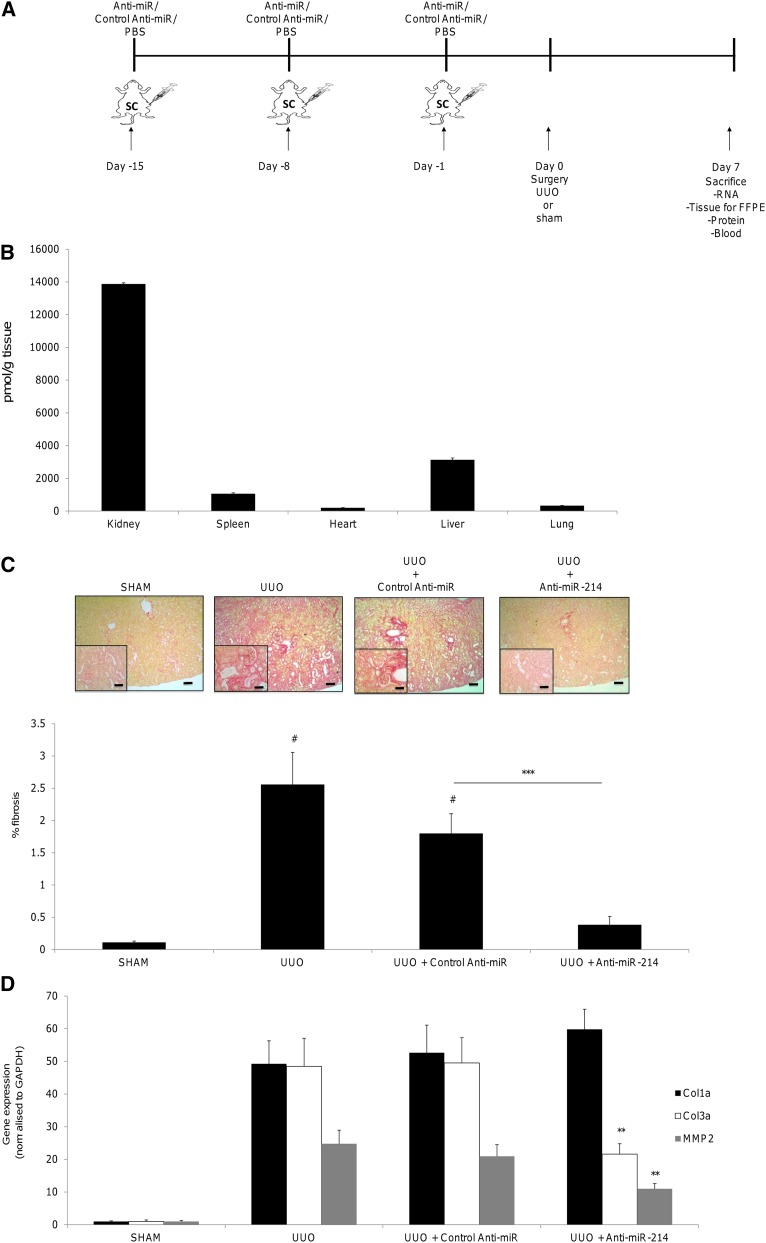
We next sought to establish whether pharmacological inhibition of miR-214 using anti-miRs would phenocopy these effects (Figure 5A). Subcutaneous delivery of anti-miR-214 was preferentially taken up by the kidney compared with other organs (Figure 5B) and qRT-PCR confirmed reduction of mature miR-214 (Supplemental Figure 5). Anti-miR-214–treated animals phenocopied miR-214−/− mice with a dramatic reduction in renal fibrosis (86%; Figure 5C). Expression of Col3a and Mmp2, but not Col1a, was reduced after anti-miR-214 treatment (Figure 5D). These data confirm that pharmacologic miR-214 antagonism results in effective inhibition of renal fibrosis.
Figure 5.
The affect of pharmacolgic deletion of miR-214 on tubulointerstital fibrosis. (A) C57bl/6 mice are subcutaneously injected with PBS, control anti-miR, anti–miR-21, or anti–miR-214 (n=10 per group) as outlined in Concise Methods, and are subjected to UUO or sham surgery. (B) Biodistribution of anti-miR-214 after subcutaneous administration is determined by a sandwich hybridization assay (see Concise Methods for details). (C) Seven days postsurgery, animals are euthanized and kidney tissue is formalin fixed. FFPE kidneys are sectioned in 3-µM sections. Picrosirius red staining is performed and the level of fibrosis is quantified by Image Pro Plus in a blind manner (n=8–10 animals). One-way ANOVA with Tukey’s post hoc test is used to analyze data. ***P<0.001 versus UUO + control anti-miR; #P<0.001 versus sham. (D) Total kidney RNA is extracted using the miRNeasy kit (Qiagen) (n=10 per group) and gene expression is assessed using specific probes (Life Technologies) and normalized to mouse Gapdh. One-way ANOVA with Tukey’s post hoc test is used to analyze data. **P<0.01 versus UUO + control anti-miR. Scale bar, 100 µM in C.
To understand the mechanism of the beneficial effects observed with anti–miR-214, we first examined expression of NCX-1 because it is a direct target of miR-214 implicated in cardiac fibrosis.19 NCX-1 is expressed on renal tubular epithelial cells,22 and mice heterozygous for NCX-1 (NCX-1 knockout is embryonic lethal23,24) are protected against renal ischemia reperfusion injury.22 However, neither protein nor mRNA expression levels of NCX-1 were derepressed in miR-214−/− animals (Figure 6A and Supplemental Figure 6). Similarly, NCX-1 mRNA was not increased in those animals treated with anti-miR-214 (Figure 6B). This suggests that miR-214 may have a tissue-specific phenotype, especially under conditions of different stress stimuli.
Figure 6.
Mechanism of beneficial effects of miR-214 deletion. (A) Protein lysates from snap-frozen kidney tissue are prepared and quantified, and 15 μg is fractionated. Proteins are transferred onto nitrocellulose membranes and probed for NCX-1 (1:500; Swant) (nonspecific band at 70 kD and specific NCX-1 band at 116 kD) and GAPDH (1:1000). Densitometry is performed using Quantity One software (n=3 per group). One-way ANOVA with Tukey’s post hoc test is used to analyze data. *P<0.05 versus sham and wild-type UUO. (B) Total kidney RNA is extracted using a miRNeasy kit (Qiagen) (n=10 per group) and gene expression is assessed using specific probes for NCX-1 (Life Technologies) and normalized to mouse Gapdh. One-way ANOVA with Tukey’s post hoc test is used to analyze data. ***P<0.01 versus all other experimental groups. (C) C57bl/6 mice are subcutaneously injected with PBS, control anti-miR, anti–miR-21, or anti–miR-214 (n=10 per group), as outlined in the Concise Methods, and are subjected to UUO or sham surgery. Total RNA is extracted using the miRNeasy kit (Qiagen) from kidneys of mice treated with control anti-miR (n=4) or anti–miR-214 (n=4). In vitro transcription is performed using the Ambion Illumina TotalPrep RNA Amplification Kit (Life Technologies). The microarray is performed using the Illumina MouseWG-6 v2.0 Expression BeadChip Kit and data are generated using GenomeStudio (Illumina). To assess the statistical significance of pairwise intergroup differences, RP is initially used; significance is assessed using the FDR multiple testing correction method with a FDR cut-off of 5%. A secondary analysis is conducted using linear models of microarray (FDR <0.05). (D) Ingenuity pathway analysis of predicted miR-214 target genes that are significantly altered in the microarray. The green color represents significant downregulation in anti-miR-214–treated animals compared with control anti-miR–treated animals. The red color represents significant upregulation in anti-miR-214–treated animals. Gray represents no change. (E) C57bl/6 mice are subcutaneously injected with PBS, control anti-miR, anti-miR-21, or anti–miR-214 (n=10 per group) as outlined in Concise Methods, and are subjected to UUO or sham surgery. Total RNA is extracted using miRNeasy kit (Qiagen) and gene expression is assessed using specific probes for Creb1 (Life Technologies) and normalized to mouse Gapdh. One-way ANOVA with Tukey’s post hoc test is used to analyze data. #P<0.05 versus UUO + control anti-miR. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; LIMMA, linear models of microarray; MMP, matrix metalloproteinase; TIMP, tissue inhibitor of metalloproteinase; UTR, untranslated region.
To evaluate the effect of miR-214 modulation on the transcriptome and reveal target genes involved in the antifibrotic effect of anti–miR-214, we performed a microarray in UUO kidneys treated with anti–miR-214 compared with control anti-miR–treated kidneys (Figure 6C). Using rank products (RP)–based analysis, we identified in 1911 significantly altered genes; of these, 751 genes were upregulated and 10.5% were predicted miR-214 targets (Figure 6C). A secondary analysis using linear models for microarray data–based analysis identified >5000 genes with significantly different expression, which demonstrates that a broad pleiotropic effect occurs after miR-214 loss on renal response to injury (Figure 6C). Similar to NCX-1, activating transcription factor 4 (ATF4) has been demonstrated to be a miR-214 target.25 However, ATF4 in this context was significantly downregulated, whereas predicted miR-214 targets cAMP responsive element binding protein 1 (CREB1) and Sloan-Kettering Institute proto-oncogen (SKI) were significantly upregulated (Figure 6D) and validated by qRT-PCR (Figure 6E). Creb1 is a nuclear transcription factor that is ubiquitously expressed and was previously shown specifically in tubular cells to promote ski-related novel gene N (SnoN), which can block the ability of Smads in the nucleus to transactivate gene expression and thus TGF-β signaling.26 SKI, along with SnoN, is a smad transcriptional corepressor but it blocks Smad 2 phosphorylation rather than Smad 3.27 This may explain why the levels of p-Smad 2/3 nuclear translocation are unaltered by the loss of miR-214 and suggest that it is mainly p-Smad 3 translocating. This would also imply that the further improvement observed in SB525334-treated miR-214−/− animals undergoing UUO is due to blockade of p-Smad 3.
Because tubular cell apoptosis is a feature of renal damage induced by UUO20 and is involved in human CKD,28,29 we hypothesized that reduced apoptosis might be an important mechanistic effect. We performed terminal deoxynucleotidyl transferase–mediated digoxigenin-deoxyuridine nick-end labeling (TUNEL) staining and observed that apoptosis levels (predominantly in tubular cells) were significantly reduced in miR-214−/− mice (Figure 7A), and were also phenocopied in anti-miR-214–treated mice (88%, P<0.01; Figure 7B). Caspase 3/7 activity assays confirmed this effect (Figure 7C). Analysis of potential miR-214 target genes that could be mediating this effect revealed proteasome (prosome, macropain) 26S subunit non-ATPase, 10 (PSMD10) which encodes gankyrin as a possible antiapoptotic mediator and recent verified target of miR-214.30 Increased expression of gankyrin inhibits apoptosis.31 In anti-miR-214–treated animals, we observed increased Psmd10 expression in the kidney (Figure 7D). These data suggest that multiple pathways have been altered in addition to apoptosis regulation and that the combined effect is protective in renal injury. This is expected because miRNAs exert their effects via multiple pathways.32
Figure 7.
Antiapoptotic effect of miR-214 antagonism in the kidney. (A) Wild-type and miR-214−/− animals are subjected to sham (n=5 per group) or UUO (n=6 per group) surgery. Animals are euthanized and kidney tissue is formalin fixed 7 days after surgery. Apoptosis is assessed by TUNEL staining on 3-µM FFPE sections (n=3 per group is assessed in a blind fashion with six fields of view counted per section). Quantification is performed by counting number of positive nuclei as a percentage of all nuclei present. One-way ANOVA with Tukey’s post hoc test is used to analyze data. ***P<0.001 versus wild-type UUO. (B) C57bl/6 mice are subcutaneously injected with PBS, control anti-miR, or anti–miR-214 (n=10 per group) as outlined in Concise Methods and had either UUO or sham surgery performed. Animals are euthanized and kidney tissue formalin fixed 7 days after surgery. Apoptosis is assessed by TUNEL staining on 3-µM FFPE sections. Quantification is performed by counting number of positive nuclei as a percentage of all nuclei present. One-way ANOVA with Tukey’s post hoc test is used to analyze data. ###P<0.001 versus UUO + control anti-miR. (C) Caspase-Glo 3/7 activity assay (Promega) is performed on protein lysates (n=6 per group) from C57bl/6 mice are subcutaneously injected with PBS, control anti-miR, or anti–miR-214 (as outlined in Concise Methods) and are subjected to UUO or sham surgery. Animals are euthanized 7 days after surgery. One-way ANOVA with Tukey’s post hoc test is used to analyze data. ###P<0.001 versus UUO + control anti-miR. (D) C57bl/6 mice are subcutaneously injected with PBS, control anti-miR, anti-miR-21, or anti-miR-214 (n=10 per group) as outlined in Concise Methods, and are subjected to UUO or sham surgery. Total RNA is extracted using a miRNeasy kit (Qiagen) and gene expression is assessed using specific probes for Psmd10 (Life Technologies) and normalized to mouse Gapdh. One-way ANOVA with Tukey’s post hoc test is used to analyze data. ###P<0.01 versus UUO + control anti-miR. DAPI, 4',6-diamidino-2-phenylindole; RQ, relative quantification. Scale bar, 20 µM in A and B.
To determine the expression of miR-214 in normal and diseased human kidneys, we performed in situ analysis (see the patient characteristics in Concise Methods). Histologic analysis of miR-214 expression revealed that miR-214 in the normal kidney was detectable in the distal convoluted tubules with a cytoplasmic and perinuclear staining pattern (Figure 8A). Perinuclear staining was also prominent in the nuclei of the proximal convoluted tubules (Figure 8A). Staining was also evident in the glomerular endothelial cell nuclei (and in other endothelial cells such as in arteries) (Figure 8A). In the cortical region from patients with CKD, tubular staining was more cytoplasmic and was localized predominantly, but not exclusively, to damaged tubules such as atrophic tubules (Figure 8B). Staining was also concentrated in the nucleus and cytoplasm of inflammatory cells (lymphocytes) (Figure 8B). Staining in the glomerulus was present in the epithelial cells (Figure 8B). Of interest was that distinct nucleolus staining was present for miR-214, which was previously observed for miR-206.33
Figure 8.
Evaluation of miR-214 expression in normal human kidneys and in CKD. In situ analysis of miR-214 expression is performed on human control renal tissue and biopsy tissue with H&E contiguous section. 5′,3′-digoxigenin-labeled LNA mercury probes (Exiqon) are used to detect miR expression. The asterisk indicates the nucleolus. H&E, hematoxylin and eosin. Scale bar, 100 µM.
Discussion
The major findings of our study are that in an in vivo experimental model of tubulointerstitial fibrosis, miR-21 and miR-214 regulation is independent of TGF-β canonical signaling and blockade of miR-214 expression through either genetic knockout or pharmacologic inhibition results in amelioration of fibrosis after UUO. The mechanism by which miR-214 exerts its effect is likely to be through a broad pleiotropic action because transcriptome analysis of the anti–miR-214 effect reveals significant gene expression changes. Evaluation of human kidney tissue reveals that miR-214 is expressed in the kidney and its expression is altered in CKD. In CKD, strong miR-214 staining is observed in damaged tubules and in inflammatory cells.
Tubulointerstitial fibrosis is the common end point of CKD. The molecular mechanisms underpinning fibrogenesis are still not fully understood. There is a clear need for a better understanding of this at the molecular and physiologic levels, which can then lead to the development of novel therapeutics. Current clinical treatments targeting renal fibrosis involve inhibition of the renin-angiotensin-aldosterone system.34,35 However, they are not always effective, especially when given in the later stages of CKD.36 Within the context of renal fibrosis, TGF-β is widely implicated in disease pathogenesis.3 This is also evidenced in our study by the efficacy of the ALK5 inhibitor SB525334, which blocked canonical TGF-β signaling and reduced fibrosis after UUO. Surprisingly, the increase in miR-21 and miR-214 after UUO was not altered by administration of SB525334. In contrast, levels of the miR-29 family (a known TGF-β–sensitive miRNA family21) normalized to sham-operated levels. The miR-29 family has been shown in the kidney and heart to regulate extracellular matrix, including collagens.8,21 This may account, at least in part, for some of the reduction in fibrosis observed after SB525334 treatment. However, deletion of both miR-21 and miR-214 independently ameliorated fibrosis after UUO. Therefore, the interplay of molecular mechanisms postinjury is complex and likely to involve many diverse pathways, encompassing multiple miRNA regulatory networks.
The kidney is an attractive organ for miRNA therapy due to the observed accumulation of inhibitors within it after systemic administration. However, when considering therapeutic applications, it will likely be important to target miR-214 therapies to the kidney, to avoid any potential disadvantages of modulating miR-214 in other tissues and organs such as the heart. In the heart, miR-214 genetic deletion coupled with ischemia reperfusion injury results in increased apoptosis and fibrosis.19 This was demonstrated to be through an increase in NCX-1, which caused Ca2+ induced toxicity within the cardiomyocyte.19 In the kidney, we found no increase in NCX-1 after injury induced by the UUO and observed that loss of miR-214 was protective against apoptosis and fibrosis. This illustrates the organ-selective effects of miRNAs and their response to different injurious stimuli. Thus, for translation, careful design of tissue selective delivery approaches may be needed.
Detection of miR-214 in the mouse and human kidney shows localization to key cell types (i.e., the tubular cells and the lymphocytes). In the kidney it is difficult to molecularly dissect whether the loss of miR-214 expression in one of these cell types is contributing more to the observed beneficial effect. Thus, future studies are required in this model system to assess whether targeted miR-214 knockdown in specific cell populations is effective.
Our microarray data indicated that loss of miR-214 leads to a substantial regulation of the transcriptome in the kidney, and that the cumulative effects of these alterations are likely responsible for the beneficial phenotypic changes observed. Further studies are required to fully elucidate the pathways central to this observed effect and to investigate the possible selective manipulation of those pathways.
In this study, we have shown a substantial therapeutic effect on renal fibrosis with an anti-miR against miR-214. These data suggest that administration of anti–miR-214 may be an attractive strategy to ameliorate progressive renal fibrosis.
Concise Methods
Ethical Statement
All animal experiments were approved by the University of Glasgow Animal Procedures and Ethics Committee and performed under UK Home Office license (PPL 60/4337; 60/3654) in strict accordance with UK Home Office guidelines.
All human samples were provided with full ethical approval. Patient biopsy tissue was from the Health Sciences Scotland South-East Scotland Bioresource after approval from the Local Scientific Review Committee and the National Health Service Greater Glasgow & Clyde Bio-repository Local Ethics Review Committee (reference numbers 10/S1402/33 and 06/MRE00/93, respectively). Biopsy tissue was from male patients aged 37–73 years, with stage 4 CKD due to diabetic nephropathy. Normal renal tissue was from the Glasgow Bio-repository and was confirmed to have no interstitial fibrosis.
Model of Tubulointerstitial Fibrosis: UUO
Eight-week-old male C57/BL6 mice, wild-type (miR-21+214+/21+214+), miR-21−/−,37 and miR-214−/− mice19 weighing approximately 21–23 g were used. The miRNA-21−/− mice37 and miR-214−/−mice19 were previously described (kindly provided by the laboratory of Eric Olson, University of Texas Southwestern Medical Center, Dallas, TX). Briefly, the lines were generated by a targeting strategy, which introduced a neomycin resistance cassette and loxP sites that flanked the pre-miR. Targeted embryonic stem cells carrying the disrupted allele were identified by Southern blot analysis and PCR. The miR-21 or miR-214 targeted embryonic stem clones were used for blastocyst injection. The resulting chimeras were bred to C57BL/6 mice to obtain germline transmission of the mutant allele (conditional) and CAG-Cre mice to directly obtain the global miR knockout. Littermates were randomly assigned to either UUO (n=6) or sham (n=5) surgery. Animals were anesthetized by isoflurane inhalation and subjected to midline laparotomy, and the right ureter was exposed by blunt dissection. Complete ureteral obstruction was carried out by tying two 4–0 silk sutures around the isolated ureter. Control groups were subjected to sham operations including laparotomy and ureter handling but not ligation. Animals were euthanized 7 days postoperatively.
SB525334 is an orally bioavailable and selective small molecule inhibitor that blocks ALK5 serine/threonine kinase activity and prevents phosphorylation of the R-smads 2 and 3 and their subsequent nuclear translocalization and gene activation. To assess the effect of TGF-β inhibition in vivo, SB525334 (Tocris/R&D Systems, Abingdon, UK) in 0.1% v/v DMSO in PBS was administered daily at 10 mg/kg per day body weight by standard oral gavage in a final volume of 100 μl. Animals were euthanized at day 7.
For anti-miR studies, four groups were used. Eight-week-old C57/BL6 mice were purchased (n=40) from Charles River Laboratories, were randomly assigned into one of four groups (n=10 mice per group), and UUO or sham surgery was performed. Anti-miR was obtained from miRagen Therapeutics (Boulder, CO) and administered subcutaneously at −15, −8, and −1 days at a total dose of 10 mg/kg in 200 µl PBS, with animals receiving 5 mg/kg of anti–miR-214 and 5 mg/kg control anti-miR. UUO or sham surgery was performed on day 0 and animals were euthanized 7 days postsurgery.
Northern Blot Analyses
Total RNA was extracted from mouse kidneys using the miRNeasy Mini Kit (Qiagen, Crawley, UK). We resolved 2–10 μg of total RNA in a 15% Tris Borate-EDTA-urea denaturing gel (Life Technologies, Paisley, UK), transferred onto Hyband-NX membrane (GE Healthcare Little Chalfont, Buckinghamshire, UK). After 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride cross-linking, membranes were hybridized with 5′-digoxigenin-labeled LNA mercury probes (Exiqon, Vedbaek, Denmark) at 50°C (miR-21 probe), 60°C (miR-214 probe), or 55°C (U6) overnight. After posthybridization washes, membranes were incubated with anti-digoxigenin-AP antibody (1:5000; Roche, Burgess Hill, UK) in 1% blocking reagents (Roche) and signals were detected using CDP-Star (Sigma-Aldrich, Dorset, UK) according to the manufacturer’s instructions.
Analysis of miRNA and Gene Expression
Quantitative real-time PCR was performed. The amplification step was carried out using specific TaqMan miRNA probe or for gene expression, inventoried gene expression specific primers (Life Technologies) on the Applied Biosystems 7900 HT real-time PCR system following the manufacturer’s instructions. U6 was used as an endogenous control for the miRNA expression and mouse Gapdh was used as an endogenous control for gene expression. Results are shown as the relative quantification ± the maximum relative quantification, calculated with the SEM.
Biodistribution Assay to Assess Anti-miR-214 Biodistribution
A sandwich hybridization assay was used to quantify anti-miR-214 in tissue samples as previously described with modifications for miR-214.38 Briefly, probes for the hybridization assay were synthesized with 2’OMe modified nucleotides, including 5′mC.mA.mC.mA.mG.mA.mC.mA.bTEG-sup-3′ (capture probe) and 5′-6FAM.mC.mA.mG.mC.mA.mG.mG-sup-3′ (detection probe). Detection was accomplished using antifluorescence–peroxidase, Fab fragments (Roche), and a 3,3′,5,5′-tetramethylbenzidine peroxidase substrate (KPL). Standard curves were generated with nonlinear logistic regression analysis with four parameters (4-PL). The working concentration range of the assay was 1–536 ng/ml. Tissue samples were prepared at 100 mg/ml by homogenizing in 3 mol/L guanidinium thiocyanate buffer (3 mol/L guanidine isothiocyanate, 0.5 mol/L NaCl, 0.1 mol/L Tris, pH 7.5, and 10 mmol/L EDTA) 2 times for 30 seconds with an MP FastPre-24 at a speed setting of 6.0. Tissue homogenates were diluted in 1 mol/L guanidinium thiocyanate buffer (1 mol/L guanidine isothiocyanate, 0.5 mol/L NaCl, 0.1 mol/L Tris, pH 7.5, and 10 mmol/L EDTA) for testing.
Histologic Stains
Formalin fixed and paraffin embedded (FFPE) 3-µM sections were deparaffinized and rehydrated. For picrosirius red staining, sections were stained with Weigert’s hematoxylin (Sigma-Aldrich, Poole, UK) and Solution B (Sigma-Aldrich) for 10 minutes at room temperature. Slides were washed and incubated in the dark with Sirius red F3B (0.1% w/v) (Sigma-Aldrich), washed in acidified water, dehydrated, and mounted. Quantification of collagen staining was performed using a background subtraction method using Image Pro Plus (Media Cybernetics, Basingstoke, UK). Briefly, six nonoverlapping bright field images of renal cortex were captured and a single pixel representing positive red staining was selected. Pixel values representing the number and intensity across the entire captured image were transformed to OD units and normalized to the total OD. Quantification (n=8–10 per group) is presented as the percentage of the ratio of OD of positive staining to the entire spectrum of a given image. For Masson’s trichrome staining, 3-µM FFPE sections were deparaffinized and rehydrated, the trichrome (Masson) stain kit (Sigma-Aldrich) was then used as per the manufacturer’s instructions, and slides were dehydrated and mounted. For hematoxylin and eosin staining, sections were incubated with hematoxylin solution for 5 minutes and then washed in running tap water, rinsed in 95% alcohol and counterstained in eosin Y solution, and then dehydrated and mounted.
TUNEL Assay
FFPE mouse kidney 3-µM sections (n=3;) were digested with 5 µg/ml (w/v) proteinase K (Promega, Southampton, UK) in 1× Tris-EDTA pH 7.4 at room temperature for 15 minutes. Cells were washed and treated with a prewarmed reaction mixture containing 0.01 mM (w/v) dATP, 0.01 mM (w/v) biotin-16-dUTP, 33.3 U/ml terminal dUTP transferase, and 1× reaction buffer (Promega) for 60 minutes at room temperature. Slides were washed and incubated with goat anti-rabbit Alexa-488 (1:400; Life Technologies) for 2 hours in the dark and then treated with 0.1% v/v Sudan Black (Sigma-Aldrich) in methanol for 10 minutes in the dark. Cells were mounted in ProLong Gold with 4',6-diamidino-2-phenylindole (DAPI; Life Technologies), with additional DAPI (100 µg/ml) and cured overnight. Images were taken with a Zeiss confocal imaging system (LSM500). To quantify the number of TUNEL-positive cells, six nonoverlapping fields of view were captured and the total number of nuclei and the number of TUNEL-positive cells were counted in the field of view.
Immunofluorescence
FFPE sections (3-µM) were deparaffinized, rehydrated and antigen unmasked using citrate buffer. Sections were blocked in blocking buffer (15% goat serum; Vector Laboratories, Peterborough, UK), 0.05% (v/v) Tween-20 (Sigma) in Tris-buffered saline for 30 minutes at room temperature. Primary anti-rabbit Smad 2/3 (1:100, sc-11769R; Santa Cruz) or negative control rabbit IgG (Dako, Cambridgeshire, UK) at a matched concentration were incubated overnight at 4°C in blocking buffer. Slides were washed and then incubated with anti-rabbit Alexa 488 (1:400; Life Technologies) used. Detection was as with the TUNEL assay. To quantify positive nuclei, six nonoverlapping fields of view were captured and the total number of nuclei and the number of p-Smad 2/3–positive nuclei counted in the field of view.
Western Blot Analyses and Densitometry
Protein lysate was fractionated on a SDS polyacrylamide gel. Protein was transferred to Hybond-P membrane (GE Life Sciences, Buckingham, UK) and blocked in 10% BSA in Tris-buffered saline and Tween 20. Membranes were incubated with rabbit anti-p-Smad 2 (1:500; Cell Signaling Technologies, New England Biolabs, Hitchin, UK), rabbit anti-p-Smad 3 (1:500; Cell Signaling Technologies), mouse anti-NCX-1 (1:500; Swant, Switzerland), or rabbit anti-Gapdh (1:1000; Cell Signaling Technologies). Membranes were washed and incubated with goat-anti-rabbit horseradish peroxidase (1:1000; Dako) or goat-anti-mouse horseradish peroxidase (1:1000) for 1 hour. Amersham enhanced chemiluminescence detection reagent was used and films were developed by a Kodak X-Omat 1000 developer. Films were scanned by the Bio-Rad Fluro-S Multi-Imager and bands quantified using Quantity One software.
Caspase-Glo 3/7 Activity Assay
Protein lysates were prepared from kidneys of C57bl/6 mice that were subcutaneously injected with PBS and had sham surgery, or were subcutaneously injected with control anti-miR or anti-miR-214 and underwent UUO surgery (n=6 per group). Then 25 µl of protein sample was mixed with 25 µl Caspase-Glo substrate (Promega) in a white plate and incubated for 1 hour. Luminescence was determined using a Wallac Victor 2 (PerkinElmer, Buckingham, UK) and readings normalized to protein concentration determined by bicinchoninic acid assay (ThermoFisher Scientific, Loughborough, UK).
Microarray
Total RNA was extracted using miRNeasy kit (Qiagen) from kidneys of animals treated with control anti-miR (n=4) or anti–miR-214 (n=4); in vitro transcription was performed using the Ambion Illumina TotalPrep RNA Amplification Kit (Life Technologies). The microarray was performed using the Illumina MouseWG-6 v2.0 Expression BeadChip Kit (Illumina, Braintree, UK) and data were generated using GenomeStudio (Illumina) (n=4 per group,1 sample per animal). To assess the statistical significance of pairwise intergroup differences, RP39 was initially used. RP has been specifically developed for the analysis of microarray experiments,40,41 and has been shown to perform very well with small sample sizes like that used in this study (n=4 per group).41 Significance was assessed using the false discovery rate (FDR) multiple testing correction method42 with a FDR cut-off of 5%. A secondary analysis was conducted using linear models for microarray data,43 and the significance cut-off was a FDR adjusted P value of <0.05. Data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession #GSE48194). Data were analyzed using the IPA (Ingenuity Systems, www.ingenuity.com), DAVID,44 and miRWalk databases.45
Statistical Analyses
From previous studies, we assumed a within-group SD of 1.5 in order to achieve 90% power at 5% significance level to determine group size. Littermates and purchased animals were randomly assigned into surgical groups. The t test (2-group studies) and one-way ANOVA with a Tukey’s multiple comparison post hoc test (>2-group studies) were used. In all cases, a 5% statistical significance cut-off was used.
Disclosures
E.v.R. is a cofounder of miRagen Therapeutics, a company focused on developing miRNA-based therapies for cardiovascular disease. B.D. is an employee of miRagen Therapeutics.
Supplementary Material
Acknowledgments
We thank Eric Olson (The University of Texas Southwestern Medical Center) for generously supplying knockout mice and Xuan T. Beatty (miRagen Therapeutics) for tissue handling and hybridization assays. We also thank Nicola Britton and Gregor Aitchison for technical assistance and Dr. Martin McBride (University of Glasgow) for assistance in microarray analysis.
This work was supported by a personal Kidney Research UK Fellowship (PDF6/2012) to L.D., a British Heart Foundation Chair of Translational Cardiovascular Science to A.H.B., a Medical Research Council Doctoral Training Grant to A.H.B. and L.D., and a Capacity Building Award in Integrative Mammalian Biology funded by the BBSRC, British Pharmacological Society, Knowledge Transfer Network, Medical Research Council, and Scottish Funding Council (BB/E527071/1) to L.D.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013010072/-/DCSupplemental.
References
- 1.Bohle A, Mackensen-Haen S, von Gise H: Significance of tubulointerstitial changes in the renal cortex for the excretory function and concentration ability of the kidney: A morphometric contribution. Am J Nephrol 7: 421–433, 1987 [DOI] [PubMed] [Google Scholar]
- 2.Liu Y: Renal fibrosis: New insights into the pathogenesis and therapeutics. Kidney Int 69: 213–217, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Eddy AA, Neilson EG: Chronic kidney disease progression. J Am Soc Nephrol 17: 2964–2966, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Sharma K, Jin Y, Guo J, Ziyadeh FN: Neutralization of TGF-beta by anti-TGF-beta antibody attenuates kidney hypertrophy and the enhanced extracellular matrix gene expression in STZ-induced diabetic mice. Diabetes 45: 522–530, 1996 [DOI] [PubMed] [Google Scholar]
- 5.Grygielko ET, Martin WM, Tweed C, Thornton P, Harling J, Brooks DP, Laping NJ: Inhibition of gene markers of fibrosis with a novel inhibitor of transforming growth factor-β type I receptor kinase in puromycin-induced nephritis. J Pharmacol Exp Ther 313: 943–951, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Zeisberg M, Hanai J-i, Sugimoto H, Mammoto T, Charytan D, Strutz F, Kalluri R: BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med 9: 964–968, 2003 [DOI] [PubMed] [Google Scholar]
- 7.Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, Qi X, Gautron L, Elmquist JK, Bassel-Duby R, Olson EN: A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell 149: 671–683, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN: Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A 105: 13027–13032, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V: The functions of animal microRNAs. Nature 431: 350–355, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Ma L, Teruya-Feldstein J, Weinberg RA: Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449: 682–688, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Denby L, Ramdas V, McBride MW, Wang J, Robinson H, McClure J, Crawford W, Lu R, Hillyard DZ, Khanin R, Agami R, Dominiczak AF, Sharpe CC, Baker AH: miR-21 and miR-214 are consistently modulated during renal injury in rodent models. Am J Pathol 179: 661–672, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chau BN, Xin C, Hartner J, Ren S, Castano AP, Linn G, Li J, Tran PT, Kaimal V, Huang X, Chang AN, Li S, Kalra A, Grafals M, Portilla D, MacKenna DA, Orkin SH, Duffield JS: MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med 4: 121ra118, 2012 [DOI] [PMC free article] [PubMed]
- 13.Zarjou A, Yang S, Abraham E, Agarwal A, Liu G: Identification of a microRNA signature in renal fibrosis: Role of miR-21. Am J Physiol Renal Physiol 301: F793–F801, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godwin JG, Ge X, Stephan K, Jurisch A, Tullius SG, Iacomini J: Identification of a microRNA signature of renal ischemia reperfusion injury. Proc Natl Acad Sci U S A 107: 14339–14344, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zavadil J, Narasimhan M, Blumenberg M, Schneider RJ: Transforming growth factor-beta and microRNA:mRNA regulatory networks in epithelial plasticity. Cells Tissues Organs 185: 157–161, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Zhong X, Chung AC, Chen HY, Meng XM, Lan HY: Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J Am Soc Nephrol 22: 1668–1681, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L-M, Hou D-X, Guo Y-L, Yang J-W, Liu Y, Zhang C-Y, Zen K: Role of microRNA-214-targeting phosphatase and tensin homolog in advanced glycation end product-induced apoptosis delay in monocytes. J Immunol 186: 2552–2560, 2011 [DOI] [PubMed] [Google Scholar]
- 18.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN: A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A 103: 18255–18260, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aurora AB, Mahmoud AI, Luo X, Johnson BA, van Rooij E, Matsuzaki S, Humphries KM, Hill JA, Bassel-Duby R, Sadek HA, Olson EN: MicroRNA-214 protects the mouse heart from ischemic injury by controlling Ca²⁺ overload and cell death. J Clin Invest 122: 1222–1232, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chevalier RL, Forbes MS, Thornhill BA: Ureteral obstruction as a model of renal interstitial fibrosis and obstructive nephropathy. Kidney Int 75: 1145–1152, 2009 [DOI] [PubMed] [Google Scholar]
- 21.Qin W, Chung ACK, Huang XR, Meng X-M, Hui DSC, Yu C-M, Sung JJY, Lan HY: TGF-β/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J Am Soc Nephrol 22: 1462–1474, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamashita J, Kita S, Iwamoto T, Ogata M, Takaoka M, Tazawa N, Nishikawa M, Wakimoto K, Shigekawa M, Komuro I, Matsumura Y: Attenuation of ischemia/reperfusion-induced renal injury in mice deficient in Na+/Ca2+ exchanger. J Pharmacol Exp Ther 304: 284–293, 2003 [DOI] [PubMed] [Google Scholar]
- 23.Wakimoto K, Kobayashi K, Kuro-O M, Yao A, Iwamoto T, Yanaka N, Kita S, Nishida A, Azuma S, Toyoda Y, Omori K, Imahie H, Oka T, Kudoh S, Kohmoto O, Yazaki Y, Shigekawa M, Imai Y, Nabeshima Y, Komuro I: Targeted disruption of Na+/Ca2+ exchanger gene leads to cardiomyocyte apoptosis and defects in heartbeat. J Biol Chem 275: 36991–36998, 2000 [DOI] [PubMed] [Google Scholar]
- 24.Cho CH, Kim SS, Jeong MJ, Lee CO, Shin HS: The Na+ -Ca2+ exchanger is essential for embryonic heart development in mice. Mol Cells 10: 712–722, 2000 [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A, Li D, Hou Z, Lv K, Kan G, Cao H, Wu H, Song J, Pan X, Sun Q, Ling S, Li Y, Zhu M, Zhang P, Peng S, Xie X, Tang T, Hong A, Bian Z, Bai Y, Lu A, Li Y, He F, Zhang G, Li Y: miR-214 targets ATF4 to inhibit bone formation. Nat Med 19: 93–100, 2013 [DOI] [PubMed] [Google Scholar]
- 26.Tan R, Zhang X, Yang J, Li Y, Liu Y: Molecular basis for the cell type specific induction of SnoN expression by hepatocyte growth factor. J Am Soc Nephrol 18: 2340–2349, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Prunier C, Pessah M, Ferrand N, Seo SR, Howe P, Atfi A: The oncoprotein Ski acts as an antagonist of transforming growth factor-β signaling by suppressing Smad2 phosphorylation. J Biol Chem 278: 26249–26257, 2003 [DOI] [PubMed] [Google Scholar]
- 28.Docherty NG, O’Sullivan OE, Healy DA, Fitzpatrick JM, Watson RW: Evidence that inhibition of tubular cell apoptosis protects against renal damage and development of fibrosis following ureteric obstruction. Am J Physiol Renal Physiol 290: F4–F13, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Thomas GL, Yang B, Wagner BE, Savill J, El Nahas AM: Cellular apoptosis and proliferation in experimental renal fibrosis. Nephrol Dial Transplant 13: 2216–2226, 1998 [DOI] [PubMed] [Google Scholar]
- 30.Misiewicz-Krzeminska I, Sarasquete ME, Quwaider D, Krzeminski P, Ticona FV, Paíno T, Delgado M, Aires A, Ocio EM, García-Sanz R, San Miguel JF, Gutiérrez NC: Restoration of microRNA-214 expression reduces growth of myeloma cells through positive regulation of P53 and inhibition of DNA replication. Haematologica 98: 640–648, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higashitsuji H, Higashitsuji H, Itoh K, Sakurai T, Nagao T, Sumitomo Y, Masuda T, Dawson S, Shimada Y, Mayer RJ, Fujita J: The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell 8: 75–87, 2005 [DOI] [PubMed] [Google Scholar]
- 32.Small EM, Olson EN: Pervasive roles of microRNAs in cardiovascular biology. Nature 469: 336–342, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Politz JC, Zhang F, Pederson T: MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci U S A 103: 18957–18962, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brenner BM, Cooper ME, de Zeeuw D, Keane WF, Mitch WE, Parving H-H, Remuzzi G, Snapinn SM, Zhang Z, Shahinfar S, RENAAL Study Investigators : Effects of losartan on renal and cardiovascular outcomes in patients with type 2 diabetes and nephropathy. N Engl J Med 345: 861–869, 2001 [DOI] [PubMed] [Google Scholar]
- 35.Balakumar P, Arora MK, Ganti SS, Reddy J, Singh M: Recent advances in pharmacotherapy for diabetic nephropathy: Current perspectives and future directions. Pharmacol Res 60: 24–32, 2009 [DOI] [PubMed] [Google Scholar]
- 36.Ruggenenti P, Cravedi P, Remuzzi G: Mechanisms and treatment of CKD. J Am Soc Nephrol 23: 1917–1928, 2012 [DOI] [PubMed] [Google Scholar]
- 37.Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN: Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest 120: 3912–3916, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Efler SM, Zhang L, Noll BO, Uhlmann E, Davis HL: Quantification of oligodeoxynucleotides in human plasma with a novel hybridization assay offers greatly enhanced sensitivity over capillary gel electrophoresis. Oligonucleotides 15: 119–131, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Breitling R, Armengaud P, Amtmann A, Herzyk P: Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92, 2004 [DOI] [PubMed] [Google Scholar]
- 40.Breitling R, Herzyk P: Rank-based methods as a non-parametric alternative of the T-statistic for the analysis of biological microarray data. J Bioinform Comput Biol 3: 1171–1189, 2005 [DOI] [PubMed] [Google Scholar]
- 41.Jeffery IB, Higgins DG, Culhane AC: Comparison and evaluation of methods for generating differentially expressed gene lists from microarray data. BMC Bioinformatics 7: 359, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y, Hochberg Y: Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300, 1995 [Google Scholar]
- 43.Smyth GK: Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3, 2004 [DOI] [PubMed]
- 44.Huang W, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2009 [DOI] [PubMed] [Google Scholar]
- 45.Dweep H, Sticht C, Pandey P, Gretz N: miRWalk—database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44: 839–847, 2011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.