Abstract
CLAVATA3 (CLV3), a stem cell marker in Arabidopsis thaliana, encodes a secreted peptide that maintains the stem cell population within the shoot apical meristem. This work investigated the CLV3 orthologue in a major legume crop, soybean (GmCLV3). Instead of being expressed in the three outermost layers of the meristem as in Arabidopsis, GmCLV3 was expressed deeper in the central zone beneath the fourth layer (L4) of the meristem, overlapping with the expression of soybean WUSCHEL. Subsequent investigation using an alternative stem cell marker (GmLOG1) revealed its expression within layers L2–L4, indicating that GmCLV3 is not a stem cell marker. Overexpression studies of GmCLV3 in Arabidopsis and complementation of clv3-2 mutant suggest similar functional capacity to that of Arabidopsis CLV3. The expression of soybean CLV1, which encodes a receptor for CLV3 in Arabidopsis, was not detectable in the central zone of the meristem via reverse-transcription PCR analysis of amplified RNA from laser-microdissected samples or in situ, implicating a diverged pathway in soybean. This study also reports the novel expression of GmLOG1 in initials of axillary meristem in the boundary region between the SAM and developing leaf primordia, before the expression of GmWUS or GmCLV3, indicating cytokinin as one of the earliest signals in initiating and specifying the stem cell population.
Key words: CLAVATA3, cytokinin, shoot apical meristem, soybean, stem cell.
Introduction
Plant peptide ligands are signalling molecules that play vital roles in various aspects of plant growth and development. Among them is the CLAVATA3/embryo-surrounding region (CLE) family of peptide, which is characterized by an N-terminal signal peptide and a conserved 14 residues domain (CLE motif) at the C-terminus (Cock and McCormick, 2001). CLAVATA3 (CLV3) from Arabidopsis thaliana represents the best-understood member of the CLE family, with the CLV3 ligand known to function in maintaining the stem cell population within the shoot apical meristem (SAM).
Stem cells residing in the SAM are the source of new cells for the development of all aboveground organs. They can self-maintain as well as give rise to daughter cells that leave the stem cell niche to develop into specialized cell type. The SAM can be divided into two distinct regions: the surface tunica layer where cells divide anticlinally resulting in an expansion of the surface area and the underlying corpus consisting of cells that divide in all planes increasing the volume of the apex (reviewed by Steeves, 2006). The tunica comprises of one to five clonal layers (L1–L5) with one layer found in monocot. In addition to this layered organization, the SAM can also be divided into three zones of distinct functions: (i) the central zone that contains stem cells; (ii) the surrounding peripheral zone; and (iii) the underlying rib zone where the initiation of lateral organs and the central stem tissue take place. The rate of cell division in the central zone is much lower than that in the peripheral zone, and stem cells can be distinguished morphologically by a large nucleus with dense cytoplasm and the lack of a large central vacuole.
In Arabidopsis, CLV3 is known to be expressed in the three outermost layers of the central zone (L1–L3) acting in a feedback loop involving CLV1 and a homeobox transcription factor, WUSCHEL (WUS), to regulate the dynamic balance between the two activities of stem cells, proliferation and differentiation. The CLV3 gene acts as a negative regulator for stem cell proliferation (Clark et al., 1995; Fletcher et al., 1999) while WUS is expressed in the underlying region in the organizing centre promotes stem cell activity (Mayer et al., 1998). CLV3 encodes a small extracellular protein that is processed into a ligand of 13 amino acids (Kondo et al., 2006; Ohyama et al., 2009) and CLV1 encodes a transmembrane receptor kinase expressed primarily in the L3 layer of the SAM (Clark et al., 1997; Jeong et al., 1999). The CLV3 ligand has been demonstrated to bind CLV1 (Ogawa et al., 2008) leading to the downregulation of WUS expression. Mutation in CLV1 or CLV3 loci thus result in an overproliferation of stems cells in the central zone. Recent study has highlighted the dynamic of the feedback loop as WUS not only activates CLV3 expression (Schoof et al., 2000), it can also directly repress the expression of CLV1 and hence negatively modulates the CLV signalling pathway (Busch et al., 2010).
The phytohormone cytokinin has also been shown to be essential in the regulation of stem cell activities (reviewed by Sablowski, 2011). Mutation in LONELY GUY (LOG) that encodes an enzyme involved in the conversion of inactive cytokinin to its biologically active form in rice resulted in plants with defective SAM (Kurakawa et al., 2007). There is evidence implicating cytokinin in the positioning of the stem cell niche (Leibfried et al., 2005; Gordon et al., 2009). In fact, cytokinin has been reported to activate WUS expression while WUS directly represses the transcription of several two-component type-A ARABIDOPSIS RESPONSE REGULATOR (ARR) genes, which function as negative regulators for cytokinin signalling (Leibfried et al., 2005; Gordon et al., 2009).
The activity at the SAM largely determines the general architecture of a plant. This has implication on the total amount of sunlight intercepted by a plant, which is critical for biomass accumulation. Later in plant development, the reproductive SAM gives rise to flowers that form seeds following fertilization. Having a better understanding of shoot meristem biology in crops therefore offers potential strategies in improving yield to meet the increasing global food demand. Soybean is the largest legume crop in the world responsible for more than 50% of worldwide oilseed production. While Arabidopsis is an excellent model system for studying regulatory network governing SAM function, much remains to be uncovered for that of soybean meristem. Furthermore, the translation of fundamental knowledge obtained using the model plant Arabidopsis to corresponding processes in legume crop remains a challenge due to obvious vegetative and floral developmental differences.
This study isolated the soybean orthologue of Arabidopsis CLV3 and characterized its expression in relation to the spatial expression of GmWUS and GmLOG1. GmCLV3 functional characteristics were further examined through ectopic expression in Arabidopsis and clv3-2 mutant complementation. This study implies a diverged CLV pathway in soybean and also reveals evidence that supports cytokinin as one of the earliest signals in initiating and specifying the stem cell population.
Materials and methods
Plant materials and growth conditions
Soybean plants (Glycine max L. Merr. cv. Bragg) were grown from seeds in a greenhouse located at the University of Melbourne, Victoria, Australia while Arabidopsis thaliana (Col0) and the clv3-2 mutant line were obtained from the Arabidopsis Biological Resource Centre and maintained under long-day conditions (16/8 light/dark 22 °C) in growth chambers.
RNA extraction and reverse-transcription PCR analysis
Total RNA from different soybean tissues of 10-day-old plants were isolated using Qiagen RNeasy Mini Kit with on-column DNAse digestion (Qiagen). Subsequent reverse-transcription PCR (RT-PCR) was carried out using a one-step RT-PCR kit (Qiagen) according to manufacturer’s instructions with 20ng total RNA as template. The PCR amplification step was routinely carried out for 30 cycles.
Plant vectors and transformation
The full-length GmCLV3a gene was amplified by PCR using cDNA derived from SAMs and primers (GmCLV3aBamH1F: 5′-CAGGATCCGATCTTCACCACACAACATTAC-3′; GmCLV3aXho1R: 5′-TAGCTCGAGGCCATAAGCTGGTAGAT GTTC-3′). The amplified fragment was subcloned into pENTR1A vector (Invitrogen) at the BamH1 and Xho1 site. Following verification of the cloned gene via sequencing, the fragment was transferred into pB7WG2 vector via LR-mediated recombination reaction (Invitrogen). The construct was then transferred into Arabidopsis via Agrobacterium tumefaciens AGL1-mediated floral-dip transformation method (Clough and Bent, 1998). Primary transformants were screened on soil saturated with 40 µg l–1 of the herbicide glufosinate ammonium.
Laser microdissection of central zone
Dissected soybean shoot apexes were fixed in Farmer’s solution (ethanol/acetic acid 75:25) for 4h on ice. Tissues were then dehydrated through ethanol series starting with 75% ethanol and processed according to Wong et al. (2012). Paraffin blocks were sectioned into 10-µm thickness before being stretched on prewarmed diethyl pyrocarbonate-treated H2O bath (37 °C) and placed on PALM membrane slides (1mm PEN, P.A.L.M., Bernried, Germany). Slides were then dried for 3h at 37 °C and stored at 4 °C until laser microdissection. Immediately before laser microdissection, slides were deparaffinized in Histochoice (Sigma). The PALM Robot Microbeam laser microdissection system was used for collecting cells from the central zone of meristem. Cells were first selected using the graphics tools of the P.A.L.M. RoboSoftware and then isolated by the laser microbeam. The triangular region presumed to be the central zone was then collected by laser pressure catapulting into the lid of a 0.5-ml microfuge tube placed in a holder closely above the slide. RNA was then extracted and amplified using PicoPure kit and AmbionMessageAmp III RNA amplification kit, respectively, according to manufacturer’s instructions.
In situ hybridization analysis
The soybean shoot apices from 10-day-old plants were dissected and fixed with 4% paraformaldehyde in phosphate-buffered saline overnight at 4 °C following vacuum infiltration. The tissue was then dehydrated through a series of ethanol solution and embedded in paraplast (Sigma, St Louis, MO, USA) following standard methods. The in situ hybridization was carried out according to modified protocols from Jackson (1991) as described in Haerizadeh et al. (2009). Digoxigenin-labelled antisense RNA probes were transcribed from T7 or SP6 promoter of pGEMT-Easy vector (Promega, Madison, WI, USA) using the DIG RNA Labeling Kit (Roche Diagnostics, Indianapolis, IN, USA) according to manufacturer’s instructions. All hybridization results were observed and photographed using a BX60 microscope and DP70 digital camera (Olympus, Centre Valley, PA, USA).
Results
Identification of soybean CLV3-like genes
Though soybean genome has recently been sequenced, no CLV3-like orthologues could be identified in the predicted Glyma1.0 gene set (Schmutz et al., 2010). When searches were performed using existing CLV3 sequences against the soybean genome sequence, two matching regions were identified where no gene was annotated. These regions were subjected to further gene prediction analysis at FGENESH (http://www.softberrry.com) and two potential CLV3-like transcripts were identified (Gm12:34,928,570..34,929,240 and Gm13:39,688,331..39,688,917). The presence of two CLV3-like transcripts in soybean genome is a result of the more recent whole genome duplication event (Schmutz et al., 2010).
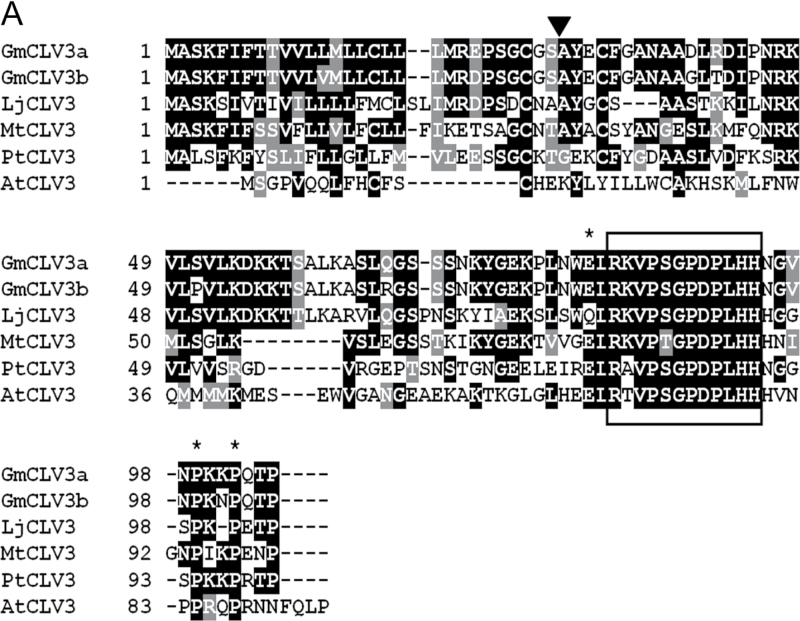
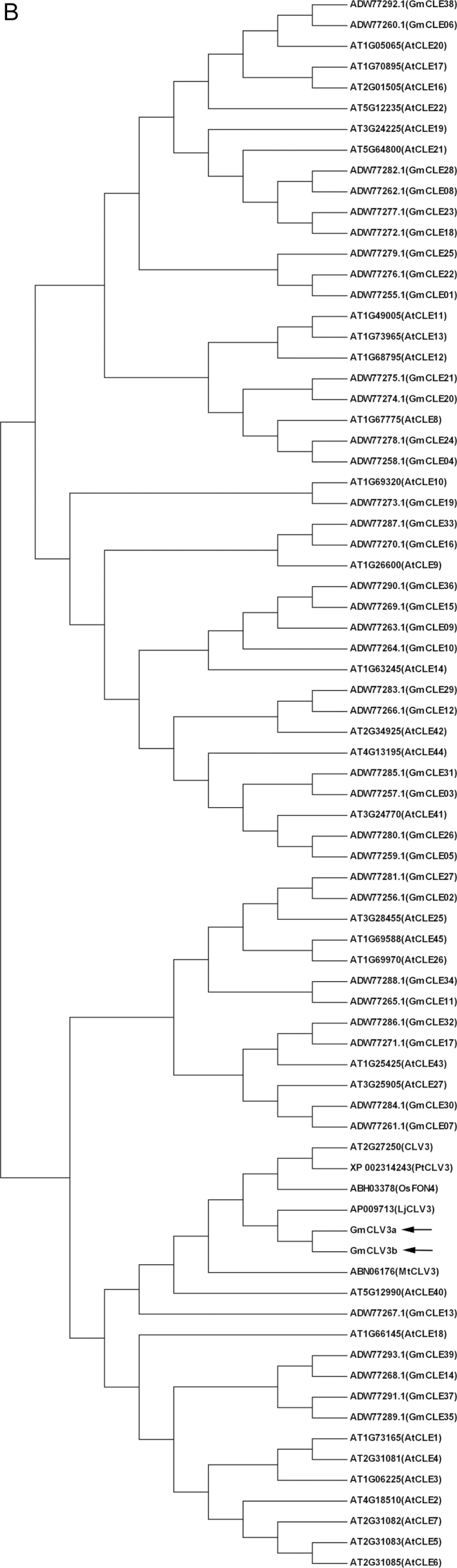
As with Arabidopsis CLV3, both GmCLV3 genes also consist of three exons. They encode small proteins of 105 amino acids in length with a signal peptide predicted at the N-terminal region between amino acids 29 and 30 (Dyrløv Bendtsen et al., 2004). A recent study has divided the CLE family into 13 groups based on sequence similarity at the C-terminal region (12–18 amino acids) encompassing the CLE domain (Oelkers et al., 2008). Arabidopsis CLV3 clustered in group 3 and, in addition to the CLE motif, members of this cluster also contains a conserved secondary motif adjacent to the CLE residues (Oelkers et al., 2008). When the virtually translated amino acid sequences of the gene pair were aligned with known full-length dicot CLV3 protein sequences, the highly conserved CLE motif as well as the secondary motif specific to group 3 can be identified (Fig. 1A). In fact, the GmCLV3 homologue only differs at one residue (residue 2) in the CLE domain from that of Arabidopsis (Fig. 1A). This study also compared the CLE domains of GmCLV3 with those encoded by all members of CLE family identified in soybean and Arabidopsis. As shown in Fig. 1B, GmCLV3a and GmCLV3b form a superclade together with CLV3. These features suggest that this homologous pair is the orthologous sequence of CLV3 in soybean. Hereafter, these are referred to as GmCLV3a (Gm12:34,928,570..34,929,240) and its homologue GmCLV3b (Gm13:39,688,331..39,688,917).
Fig. 1.
Sequence alignment analysis of GmCLV3. (A) Predicted protein sequences for GmCLV3a and GmCLV3b were aligned with other existing CLV3 orthologues using CLUSTALW (Larkin et al., 2007) and the output was displayed with Box Shade (http://www.ch.embnet.org/software/BOX_form.html). The predicted cleavage site for the signal peptide of GmCLV3s is indicated with a triangle. The 12-amino acid CLE motif is boxed. The three amino acid residues that are part of the secondary conserved motif adjacent to the CLE motif specific to the CLV3 group (Oelkers et al., 2008) are marked with asterisks. AtCLV3, Arabidopsis thaliana CLV3 (At2g27250); LiCLV3, Lotus japonicus CLV3 (AP009713); MtCLV3, Medicago truncatula CLV3 (ABN06176); PtCLV3, Populus trichocarpa CLV3 (XP_002314243). (B) Alignment analysis of the CLE domain encoded by all CLE members reported to date for soybean and Arabidopsis indicating GmCLV3a and GmCLV3b (marked with arrowheads) are orthologous to Arabidopsis CLV3. (Figure is overleaf).
Expression of GmCLV3 and GmCLV1
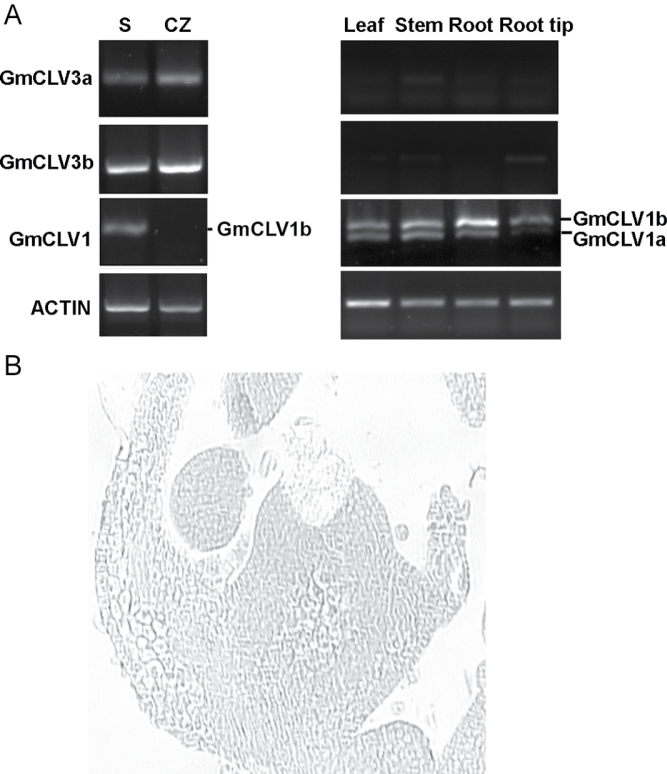
This study investigated the expression pattern of the GmCLV3 gene pair in different soybean tissues using RT-PCR analysis with primers specific to each of the homologue. To assess their expression in the SAM, laser-microdissection technology was used to obtain amplified RNA from the central zone of the meristem. As expected, both genes were expressed only in the central zone of the SAM but not in other vegetative tissues examined (Fig. 2A).
Fig. 2.
Expression profiles of GmCLV3 and GmCLV1 in vegetative soybean tissues. (A) Reverse-transcription PCR was carried out with amplified RNA (left panel) or total RNA (right panel) as templates. S is a sample consisting of amplified RNA derived from one section of a complete shoot apex scratched from the glass slide and CZ is from the central zone. For GmCLV1 amplification, the lower band (403 bp) corresponds to GmCLV1a whereas the upper band (457 bp) represents GmCLV1b. ACTIN was used as an internal control. (B) One section of the SAM shoot apex after central zone was laser microdissected.
CLV3 is known to function as a ligand that binds to CLV1 receptor to regulate stem cells’ function (Ogawa 2008). Thus the expression pattern of GmCLV1 was investigated. The soybean counterparts of CLV1 are annotated in the Glyma1.0 gene set as Glyma11g12190 (GmCLV1a) and Glyma12g04390 (GmCLV1b) and they are the same GmCLV1 sequences isolated by Yamamoto et al. (2000). While GmCLV1a function is unknown, GmCLV1b corresponds to GmNARK that has been reported to play a role in nodulation (Searle et al., 2003) and likely in partnership with members of the CLE family (Reid et al., 2013). The current study proceeded to examine the expression of the GmCLV1 homologue via RT-PCR analysis (Fig. 2A). GmCLV1a transcript was detected throughout vegetative tissues but not in the root tip and central zone of SAM. The expression of GmCLV1b is similar to its homologue except its transcripts are also found in the root tip. This expression pattern of GmCLVb is largely consistent with that reported by Nontachaiyapoom et al. (2007).
Spatial expression pattern of SAM regulatory genes
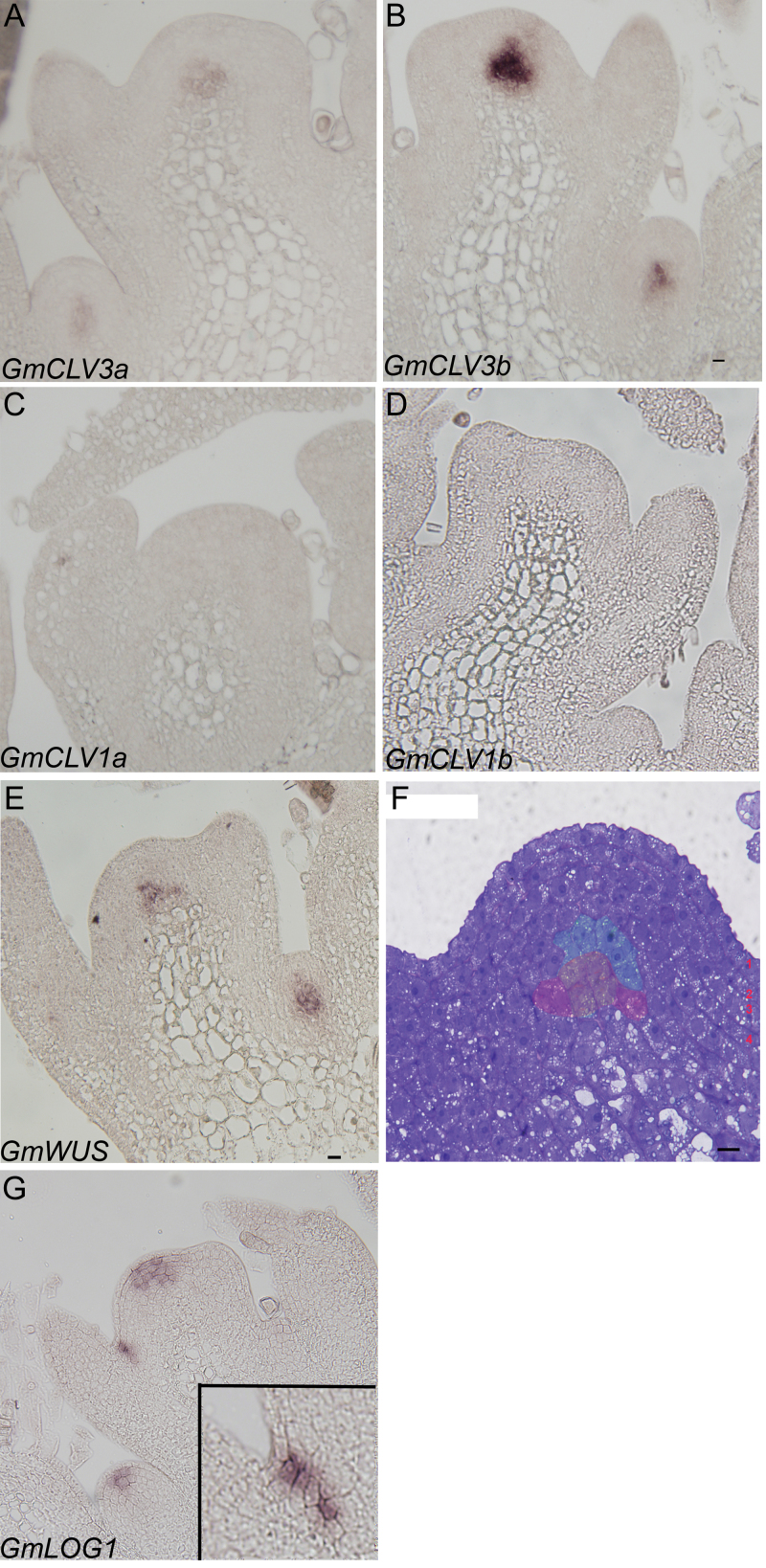
When this work examined the spatial localization of GmCLV3 in the soybean shoot apex, GmCLV3 was found remarkably to be expressed deeper in the central zone and, more specifically, GmCLV3 is expressed beneath the fourth layer of the meristem (Fig. 3A and B). This is in stark contrast to CLV3 expression in Arabidopsis whereby CLV3 is known to be expressed in the first three layers of the meristem, L1, L2, and L3. The expression of GmCLV3 deeper in the axillary meristem was also observed (Fig. 3A and B). This work also examined the expression of the GmCLV1 homologous pair in the shoot apex but no signal was observed for either of the gene (Fig. 3C and D) likely due to the absence or low expression level of the gene that is beyond the detection limit of the technique.
Fig. 3.
RNA in situ hybridization analysis. (A–E and G) Longitudinal sections of soybean shoot apexes hybridized with antisense digoxigenin-labelled RNA as indicated. (A and B) Spatial expression of GmCLV3a and GmCLV3b are very similar, beneath the fourth layer of the meristem albeit a weaker expression for GmCLV3a. (C and D) No signals were detected for either GmCLV1a or GmCLV1b. (E) GmWUS is expressed beneath the fifth outermost layer of the meristem. (F) A longitudinal section of soybean shoot apical meristem stained with toluidine blue with the four distinct zonal layers indicated; the expression domain of GmCLV3 is false-coloured green (approximately 9 cells in total) while that of GmWUS red and the overlapping region yellow. (G) GmLOG1 transcripts are found with the L2–L4 layer. The inset shows a close up of its expression in the boundary region between the SAM and the developing leaf primordia. Bar, 0 μm (this figure is available in colour at JXB online).
WUS is known to be expressed in the organizing centre of the SAM. To determine whether the region where the GmCLV3 was expressed corresponded to the organizing centre and hence the expression of GmWUS, the spatial expression pattern for GmWUS in the SAM was also examined. Consistent with Wong et al. (2011), the expression of GmWUS was observed in similar area in the central zone but beneath the fifth layer of the meristem (Fig. 3E). In fact, the GmWUS expression domain overlaps with that of GmCLV3. When compared to a toluidine-blue-stained section of soybean SAM, GmCLV3- or GmWUS-expressing cells contain large nuclei with dense cytoplasm and small vacuoles (Fig. 3F). It is also obvious from the section that the tunica consists of four layers indicating soybean SAM has an additional L4 layer, unlike in Arabidopsis.
Recently, alternative stem cell markers with CLV3-like expression pattern have been reported in Arabidopsis and they are APUM-10 (AT1G35750), MCT2 (AT5G07930), and AtLOG1 (AT5G06300) (Aggarwal et al., 2010). To identify location of stem cells in soybean SAM, the spatial expression of corresponding soybean orthologues was interrogated. While this work failed to detect the expression of GmAPUM-10 (Glyma08g19140) and GmMCT2 (Glyma18g01600) in the SAM (data not shown), it was successful with GmLOG1 (Glyma10g09480; Fig. 3G). Based on the expression of GmLOG1 (Fig. 3G), it is clear that the expression of GmLOG1 is at least partly confined to the four outermost layer of the meristem except the L1. The absence of GmLOG1 from the L1 layer can also be seen in the axillary meristem. Intriguingly, GmLOG1 is also expressed in the boundary region between the SAM and developing leaf primordia (Fig. 3G, inset), likely marking the cells that are competent to initiate axillary meristem in the axils of the primary shoot subsequently. However, no GmWUS or GmCLV3 transcripts were detected in these initials.
Constitutive expression of GmCLV3 promotes consumption of the SAM
This study next examined the functional characteristics of GmCLV3. As both GmCLV3 copies are identical in the CLE motif region and since they are expressed only in the SAM during the vegetative development, they likely perform the same functions. Thus, the further analysis focused on the ectopic expression of GmCLV3a. The full length GmCLV3a cDNA was cloned to be driven by the constitutively active cauliflower mosaic virus 35S promoter and the transgene was introduced into Arabidopsis via Agrobacterium-mediated floral dip transformation (Clough and Bent, 1998).
The independent Arabidopsis transgenic lines successfully transformed with 35S::GmCLV3a displayed significant developmental defects (Fig. 4). The SAM stopped initiating organ after the appearance of first leaves (Fig. 4B and C). While strong 35S::GmCLV3 lines died prematurely (35 out of 47 independent lines), moderate to weak transgenic lines (12 out of 47 independent lines) overcame the initial meristem arrest and subsequently produced misshapen leaves with irregular phyllotaxis (Fig. 4D). The mortality observed is in line with the reported ectopic expression analysis of CLV3 and several members of the CLE family (Brand et al., 2000; Strabala et al., 2006). The ability of several 35S::GmCLV3a lines to overcome the initial meristem arrest is consistent with reports that long-term expression of high levels of CLV3 can result in the reactivation of WUS expression (Muller et al., 2006). Surviving plants displayed significant developmental timing delays as they remained vegetative for close to 2 months after germination (Fig. 4D) while first floral bud was visible with wild-type plants 5 weeks after germination under these growth conditions (data not shown). Bolting was eventually initiated almost simultaneously from primary as well as axillary meristem (Fig. 4E). As these plants developed further into the reproductive stage, they produced flowers with missing carpel and fewer anthers than normal. As a result, no siliques were formed from any of these lines (Fig. 4E). These phenotypes are reminiscent of the gain-of-function analysis with Arabidopsis CLV3 (Brand et al., 2000).
Fig. 4.
Phenotypes of 35S::GmCLV3 Arabidopsis plants. (A–C) One-week-old seedlings of wild-type (A) and 35S::GmCLV3 (B and C) displaying a temporary arrest on leaf initiation for plant B while plant C never recovered from the defect. (D) Two-month-old 35S::GmCLV3 plant with leaves formed at irregular phyllotaxis resulting in a ‘bushy’ appearance. (E) 35S::GmCLV3 plant with bolts initiated from primary as well as axillary meristems; no siliques were formed from the plant. (F) Wild-type flower. (G) Defective flower of 35S::GmCLV3 with missing carpel. (H) Phenotypes of 35S GmCLV3 in clv3-2 background with normal shaped buds shown in the inset. (I) Three individual clv3-2 plants with enlarged floral buds (inset). (J) Siliques of 35S::GmCLV3a in clv3-2 background (left) and club-shaped silique of clv3-2 (right). Bar, 1 mm (A–C, F, G, J), 1 cm (D, E, H, I) (this figure is available in colour at JXB online).
Ectopic expression of GmCLV3 overrides Arabidopsis clv3 mutant phenotypes
The clv3-2 mutant is characterized by enlarged vegetative meristem and floral buds, and with short, club-shaped siliques (Clark et al., 1995). To further confirm a conserved function of the GmCLV3 with its Arabidopsis counterpart, the construct (35S::GmCLV3a) was transformed into clv3-2 plants (Clark et al., 1995). The overexpression of GmCLV3a in clv3-2 mutant lines rescued the phenotype (Fig. 4H–J) and most notably resulted in the generation of normally shaped siliques (Fig. 4J).
Discussion
This study has identified the Arabidopsis CLV3 counterpart in a major legume crop, soybean. Intriguingly, unlike in Arabidopsis whereby CLV3 is expressed in stem cells of all three outermost layers (L1–L3), GmCLV3 is expressed deeper in the meristem beneath the fourth layer and coinciding with GmWUS expression in the organizing centre (Fig. 3). Subsequent investigation with an alternative stem cell marker (GmLOG1) reveals a different expression pattern of GmLOG1 to that of GmCLV3. Based on the expression of GmLOG1, the location of soybean stem cells is likely to be within L2 to L4 layers of the central zone (Fig. 3D) and hence indicating GmCLV3 is not a stem cell marker in soybean.
The markedly different expression of GmCLV3 implicates its signalling mechanisms must have diverged from that of Arabidopsis especially since GmCLV1 is not detectable in the central zone of the SAM via RT-PCR analysis. The absence of GmCLV1 in the SAM is further corroborated with previous microarray dataset derived from studies examining the expression profile of dissected soybean SAM as the expression of GmCLV1 was identified as absent in the SAM microarray dataset (Haerizadeh et al., 2009). This study also failed to localize the expression of GmCLV1 using in situ hybridization analysis. To date, two other pathways involving CLV2/CORYNE heterodimer or a receptor-like protein kinase TOAD2 homomer have been found to act as receptors for CLV3 ligand in parallel to the CLV1 pathway in Arabidopsis to negatively regulate the expression of WUS (Muller et al., 2008; Kinoshita et al., 2010). GmCLV3 could thus act through similar CLV2/CORYNE and/or TOAD2-associated pathways in soybean.
Studies in Arabidopsis have demonstrated the movement of the CLE peptide of CLV3 and WUS in the SAM, although the relevance of this movement is unclear (Nimchuk et al., 2011; Yadav et al., 2011). It is likely that the overlapping expression of GmCLV3 and GmWUS in soybean reflects the situation in many plants. Nevertheless, future studies examining the receptor systems of GmCLV3 shall shade light on the biological relevance of this novel spatial expression pattern of GmCLV3.
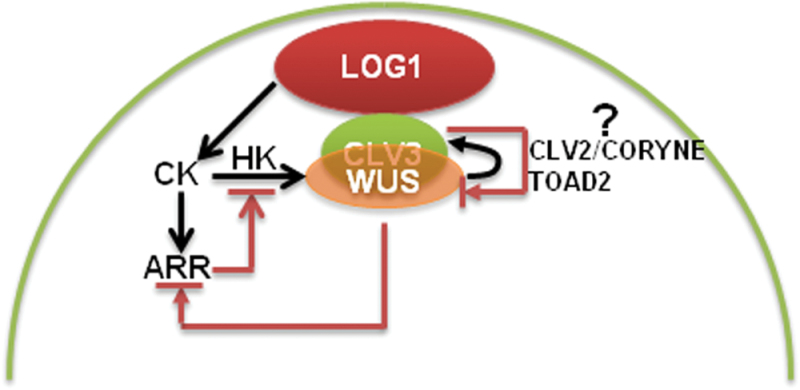
Stem cells in the SAM as well as axillary meristem in soybean are associated with the expression of GmLOG1. In fact, GmLOG1 expression marks the initials for axillary meristem at the leaf axil and, as far as is known, this spatial expression pattern of GmLOG1 has not been reported before (Kurakawa et al., 2007; Aggarwal et al., 2010). The current study thus implicates cytokinin in early axillary meristem development, possibly by inhibiting cell growth or conferring meristematic competency to axillary initials. Furthermore, given that neither GmWUS nor GmCLV3 expression is detected in these axillary initials or the surrounding neighbouring cells but only in axillary meristems, it is conceivable that cytokinin may be the primary signal that initiates and sustains an environment promoting stem cell organization. In Arabidopsis, cytokinin has been implicated in specifying a spatial WUS-expressing domain which then promotes stem cell number in the overlaying region acting in concert with cytokinin receptors and response regulators (Gordon et al., 2009). We have recently carried out transcriptome sequencing of total RNA derived from micro-dissected SAM (Wong et al., 2013). An examination of the resulting dataset reveals the expression of several putative cytokinin receptors or regulators in the SAM (Supplemental Table 1). Similar histidine kinases and response regulators could function as receptors or regulators for cytokinin. They could thus act in a similar signalling feedback loops that operate and may take precedence over GmCLVs in the control of stem cell number within soybean SAM (Fig. 5).
Fig. 5.
Regulatory networks in the soybean shoot apical meristem. A hypothetical model showing the action of cytokinin, WUS, and CLV3 in concert with other factors (described in the text) in regulating the stem cell population within the shoot apical meristem in soybean. Histidine kinase (HK) functions as a receptor for cytokinin (CK) while type-A ARABIDOPSIS RESPONSE REGULATOR (ARR) acts as a negative regulator for cytokinin signalling (Leibfried et al., 2005; Gordon et al., 2009) (this figure is available in colour at JXB online)
Meanwhile, the absence of GmLOG1 from the outermost layer of the soybean meristem is noteworthy. It is possible that true stem cell marker is yet to be uncovered and the expression of GmLOG1 in stems cells located in the L2–L4 layer may serve as a sufficient source of cytokinin for it to exert its roles in the SAM.
Despite the novel expression pattern, GmCLV3 seems to have the capacity to function as CLV3 when ectopically expressed in Arabidopsis and clv3-2 mutant. This implies that the GmCLV3 ligand could interact with the Arabidopsis CLV3 receptor(s) and, hence, consistent with previous studies reporting the capacity of CLE ligands for functional redundancy (Wang et al., 2005; Strabala et al., 2006).
In summary, this work successfully isolated the soybean CLV3 orthologue (GmCLV3) and showed its markedly different spatial expression pattern from its Arabidopsis counterpart. Though ectopic expression studies of GmCLV3 in Arabidopsis and mutant complementation work suggest largely similar functional characteristics, the novel expression pattern of GmCLV3 and the lack of GmCLV1 expression in the central zone implicate divergence in the CLV pathway in soybean. Moreover, this spatial localization study of GmLOG1 implicates a primary role of cyokinin in initiating and sustaining stem cell population within the meristem.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. 1. Alignment analysis of GmCLV3a and GmCLV3b
Supplementary Table 1. Expression levels of putative cytokinin receptors and response regulators in soybean shoot apical meristem
Acknowledgements
The authors wish to thank Associate Professor Bernie Carroll for soybean seeds and Miss Soo Yee Khor for her technical assistance in floral-dipping, screening, and maintaining Arabidopsis plants. C.E. Wong gratefully acknowledges financial support from the University of Melbourne Early Career Researcher Grant scheme. Financial support from the Australian Research Council in the form of the ARC Centre of Excellence for Integrative Legume Research (CE0348212 and ARC DP0988972) is also gratefully acknowledged.
References
- Aggarwal P, Yadav RK, Reddy GV. 2010. Identification of novel markers for stem-cell niche of Arabidopsis shoot apex. Gene Expression Patterns 10, 259–264 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. 2000. Dependence of stem cell fate in Arabidopsis an a feedback loop regulated by CLV3 activity. Science 289, 617–619 [DOI] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, et al. 2010. Transcriptional control of a plant stem cell niche. Developmental Cell 18, 849–861 [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. 1995. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067 [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis . Cell 89, 575–585 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Cock JM, McCormick S. 2001. A large family of genes that share homology with CLAVATA3. Plant Physiology 126, 939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrløv Bendtsen J, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology 340, 783–795 [DOI] [PubMed] [Google Scholar]
- Fletcher LC, Brand U, Running MP, Simon R, Meyerowitz EM. 1999. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914 [DOI] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. 2009. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proceedings of the National Academy of Sciences, USA 106, 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerizadeh F, Wong CE, Singh MB, Bhalla PL. 2009. Genome-wide analysis of gene expression in soybean shoot apical meristem. Plant Molecular Biology 69, 711–727 [DOI] [PubMed] [Google Scholar]
- Jackson D. 1991. In situ hybridization in plants. In: Gurr SJ, McPherson MJ, Bowles DJ, editors, Molecular plant pathology: a practical approach : Oxford University Press, Oxford, 163–174 [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE. 1999. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. The Plant Cell 11, 1925–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S. 2010. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis . Development 137, 3911–3920 [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. 2006. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848 [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. 2007. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- Leibfried A, To JPC, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU. 2005. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175 [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815 [DOI] [PubMed] [Google Scholar]
- Muller R, Bleckmann A, Simon R. 2008. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. The Plant Cell 20, 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R, Borghi L, Kwiatkowska D, Laufs P, Simon R. 2006. Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. The Plant Cell 18, 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM. 2011. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Current Biology 21, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nontachaiyapoom S, Scott PT, Men AE, Kinkema M, Schenk PM, Gresshoff PM. 2007. Promoters of orthologous Glycine max and Lotus japonicus nodulation autoregulation genes interchangeably drive phloem-specific expression in transgenic plants. Molecular Plant–Microbe Interactions 20, 769–780 [DOI] [PubMed] [Google Scholar]
- Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T. 2008. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biology 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. 2008. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294–294 [DOI] [PubMed] [Google Scholar]
- Ohyama K, Matsubayashi Y, Shinohara H, Ogawa-Ohnishi M. 2009. A glycopeptide regulating stem cell fate in Arabidopsis thaliana . Nature Chemical Biology 5, 578–580 [DOI] [PubMed] [Google Scholar]
- Reid DE, Li D, Ferguson BJ, Gresshoff PM. 2013. Structure–function analysis of the GmRIC1 signal peptide and CLE domain required for nodulation control in soybean. Journal of Experimental Botany 64, 1575–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R. 2011. Plant stem cell niches: from signalling to execution. Current Opinion in Plant Biology 14, 4–9 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, et al. 2010. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jurgens G, Laux T. 2000. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644 [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM. 2003. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299, 109–112 [DOI] [PubMed] [Google Scholar]
- Steeves TA. 2006. The shoot apical meristem: an historical perspective. Canadian Journal of Botany—Revue Canadienne De Botanique 84, 1629–1633 [Google Scholar]
- Strabala TJ, O’Donnell PJ, Smit A-M, Ampomah-Dwamena C, Martin EJ, Netzler N, Nieuwenhuizen NJ, Quinn BD, Foote HCC, Hudson KR. 2006. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiology 140, 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Mitchum MG, Gao BL, Li CY, Diab H, Baum TJ, Hussey RS, Davis EL. 2005. A parasitism gene from a plant-parasitic nematode with function similar to CLAVATA3/ESR. Molecular Plant Pathology 6, 187–191 [DOI] [PubMed] [Google Scholar]
- Wong C, Khor S, Bhalla P, Singh M. 2011. Novel spatial expression of soybean WUSCHEL in the incipient floral primordia. Planta 233: 553–560 [DOI] [PubMed] [Google Scholar]
- Wong CE, Singh MB, Bhalla PL. 2012. Sample preparation for laser-microdissection of soybean shoot apical meristem. International Journal of Plant Biology 3, e3 [Google Scholar]
- Wong CE, Singh MB, Bhalla PL. 2013. The dynamics of soybean leaf and shoot apical meristem transcriptome undergoing floral initiation process. PLoS One 8, e65319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV. 2011. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes and Development 25, 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E, Karakaya HC, Knap HT. 2000. Molecular characterization of two soybean homologs of Arabidopsis thaliana CLAVATA1 from the wild type and fasciation mutant. Biochimica et Biophysica Acta 1491, 333–340 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.