Abstract
The precise adjustment of the timing of dormancy release according to final grain usage is still a challenge for many cereal crops. Grain sorghum [Sorghum bicolor (L.) Moench] shows wide intraspecific variability in dormancy level and susceptibility to pre-harvest sprouting (PHS). Both embryo sensitivity to abscisic acid (ABA) and gibberellin (GA) metabolism play an important role in the expression of dormancy of the developing sorghum grain. In previous works, it was shown that, simultaneously with a greater embryo sensitivity to ABA and higher expression of SbABA-INSENSITIVE 4 (SbABI4) and SbABA-INSENSITIVE 5 (SbABI5), dormant grains accumulate less active GA4 due to a more active GA catabolism. In this work, it is demonstrated that the ABA signalling components SbABI4 and SbABI5 interact in vitro with a fragment of the SbGA 2-OXIDASE 3 (SbGA2ox3) promoter containing an ABA-responsive complex (ABRC). Both transcription factors were able to bind the promoter, although not simultaneously, suggesting that they might compete for the same cis-acting regulatory sequences. A biological role for these interactions in the expression of dormancy of sorghum grains is proposed: either SbABI4 and/or SbABI5 activate transcription of the SbGA2ox3 gene in vivo and promote SbGA2ox3 protein accumulation; this would result in active degradation of GA4, thus preventing germination of dormant grains. A comparative analysis of the 5′-regulatory region of GA2oxs from both monocots and dicots is also presented; conservation of the ABRC in closely related GA2oxs from Brachypodium distachyon and rice suggest that these species might share the same regulatory mechanism as proposed for grain sorghum.
Key words: ABI4, ABI5, abscisic acid, GA 2-oxidase, germination, gibberellins, seed dormancy, Sorghum bicolor.
Introduction
Seed dormancy is considered as the failure of an intact viable seed to complete germination under conditions that are otherwise favourable for non-dormant seeds (Bewley, 1997). In wild plants from temperate habitats, seed dormancy improves their fitness as it avoids the occurrence of germination under unfavourable conditions, reduces intraspecific competition, and gives species the opportunity to survive natural catastrophes. On the other hand, the process of domestication has pushed towards a fast and uniform germination, followed by rapid seedling establishment and high crop yields (Finkelstein et al., 2008). This has been achieved through the selection of genotypes with a briefer dormancy. However, low dormancy levels before grain harvest can also lead to pre-harvest sprouting (PHS) if adequate temperature and humid conditions prevail during late maturation in the field. For these reasons, the precise adjustment of the timing of dormancy release according to grain usage requirements appears to be a trait of great importance for crop production.
It is widely documented that abscisic acid (ABA) and gibberellins (GAs) play an antagonistic role in the control of seed germination in species that display physiological dormancy (reviewed in Finch-Savage and Leubner-Metzger, 2006; Finkelstein et al., 2008; Nambara et al., 2010). Current knowledge on how GA and ABA control seed germination highlights the importance of inactivation of germination repressors for germination to take place. During dormancy release, ABA levels or sensitivity decline, while an enhanced response to GAs takes place (reviewed by Finch-Savage and Leubner-Metzger 2006). Moreover, it is precisely the ABA–GA balance which determines the expression of dormancy during seed imbibition in many species, including cereals (reviewed by Finch-Savage and Leubner-Metzger, 2006; Finkelstein et al., 2008; Nambara et al., 2010).
Grain sorghum [Sorghum bicolor (L.) Moench], like many other cereals, shows wide intraspecific variability in the pattern of dormancy release and PHS response, some genotypes being very susceptible and others resistant to PHS. Sorghum caryopses display coat-imposed physiological dormancy, and removal of the pericarp and endosperm leads to rapid embryo germination (Steinbach et al., 1995). Previous results have shown that embryonic ABA levels measured during grain incubation are not related to their dormancy level (Gualano et al., 2007; Rodríguez et al., 2009). In contrast, changes in embryo sensitivity to ABA correlated with the pattern of dormancy release of intact grains throughout development, for both sprouting-resistant and sprouting-susceptible genotypes (Steinbach et al., 1995; Gualano et al., 2007; Rodríguez et al., 2009). In agreement with these results, Rodríguez et al. (2009) reported that the transcription of the sorghum genes ABI3/VP1, ABI4, ABI5, and PKABA1 (positive regulators of ABA signalling) is stimulated during incubation of dormant grains of the sprouting-resistant inbred line (IS9530), but not in grains of the less dormant, sprouting-susceptible line RedlandB2. In particular, these authors pointed out that the expression of SbABA-INSENSITIVE 4 (SbABI4) and SbABI5 was transiently induced in IS9530 caryopses after 2–3 d of incubation, together with a similar increase in SbABI5 protein levels.
Genetic analysis of Arabidopsis aba-insensitive (abi) mutants revealed that the transcription factors ABI4 and ABI5 do not have an effect on dormancy induction or release but have a fundamental role in ABA-mediated inhibition of germination and seed maturation (Brocard-Gifford et al., 2003; Penfield et al., 2006; Piskurewicz et al., 2008; Daszkowska-Golec and Szareijo, 2013). ABI5 is a basic leucine zipper (bZIP) transcription factor, known to be involved in regulating germination in response to ABA and stress (Finkelstein and Rock, 2002). The leucine zipper domain of ABI5 is involved in protein dimerization, which is necessary for DNA binding to occur. Like other bZIP proteins, ABI5 binds ACGT core sequence elements, called ABA response elements or ABREs (Guiltinan et al., 1990; Mundy et al., 1990), found in many ABA-responsive promoters (reviewed by Shinozaki et al., 2007). On the other hand, ABI4 is a member of the APETALA2 domain family and it has been reported that the Arabidopsis abi4 mutant showed altered expression of ABA-responsive genes such as Em6 (Finkelstein, 1994). In vitro analysis of maize ABI4 binding sites revealed a CACCG consensus sequence, named coupling element 1 (CE1; Niu et al., 2002), and it has also been demonstrated that ABI4 binds to a sequence related to the S-box (CACYKSCA) (Bossi et al., 2009). However, Reeves et al. (2011) recently found that the promoters of many ABI4-regulated genes lacked these previously identified ABI4 binding sites, but were enriched for ABREs (bZIP binding sites), and they identified a group of genes that are synergistically co-regulated by ABI4 and bZIP proteins. Hence, it has been suggested that the minimal ABRC (ABA response complex) found in many promoters induced by ABA (such as HVA1 and HVA22) is composed of an ACGT core element (ABRE) and a coupling element (Shen and Ho, 1995; Shen et al., 1996). Similar ABRC configurations were found in the promoters of other ABA response genes such as Em genes (Guiltinan et al., 1990), Rab17 (Busk et al., 1997), and Rab28 (Busk and Pagés, 1998). Other elements that have been identified in ABA-inducible promoters include RY/Sph, MYC, and MYB elements (reviewed in Cutler et al., 2010). In particular, RY/Sph repeat elements have been found to be crucial for seed-specific promoters (Baumlein et al., 1992; Ezcurra et al., 1999).
As mentioned above, not only ABA but also GAs have a role in dormancy expression in the imbibed seed. In grain sorghum, Benech-Arnold et al. (2000) reported that embryo sensitivity to GA3 (measured as the ability of increasing GA3 concentrations to revert inhibition of embryo germination by 50 µM ABA) was not related to the contrasting levels of dormancy exhibited by sorghum lines RedlandB2 and IS9530. GA levels, in contrast, correlated with the expression of dormancy in sorghum grains. The embryo content of active GA4 in immature grains increased and reached a significantly higher value after a 4 d incubation period in the low dormancy line RedlandB2 as compared with the more dormant IS9530, and this increase occurred before embryo growth began (Perez-Flores et al., 2003; Rodríguez et al., 2012). It is precisely at this immature stage, before grain physiological maturity is achieved, that GA de novo synthesis contributes to germination of RedlandB2 seeds, as suggested by the results of Rodríguez et al. (2012).
Transcriptional analysis of several sorghum genes encoding putative GA synthesis enzymes (SbEKO, SbEKAH, SbGA20ox2, SbGA20ox3, and SbGA3ox1) showed a transient increase of these transcripts in dormant grains (IS9530) during the first 2–3 d of grain imbibition, but this did not occur in the less dormant genotype (RedlandB2) (Rodríguez et al., 2012). This evidence appears to be in contradiction to changes in GA4 levels in both lines. However, this GA synthesis ‘intention’ in the more dormant IS9530 occurred together with an evident promotion of the GA inactivation genes SbGA2ox1 and SbGA2ox3. This observation, together with a negative association between embryo content of active GA4 and its corresponding catabolite GA34, supported the idea that GA4 levels are kept low in dormant IS9530 grains as a result of a prominent catabolic activity by GA 2-oxidases (GA2oxs) which is not evident in RedlandB2 embryos. On the other hand, incubation of dormant grains in 100 μM GA3 promoted germination but did not reduce the expression of most key GA synthesis genes, ruling out the idea of a negative feedback regulatory mechanism driven by active GAs. In addition, and in contrast to other reports which show a feed-forward mechanism affecting expression of GA2ox genes by active GA levels in Arabidopsis (Ogawa et al., 2003; Rieu et al., 2008), expression of sorghum GA2ox1 and GA2ox3 was down-regulated by exogenously applied GA3 (Rodríguez et al., 2012). The temporal coincidence of expression patterns for SbABI4, SbABI5 (together with SbABI5 protein abundance), and SbGA2ox genes, coinciding also with impairment of GA4 accumulation and absence of germination, suggested ABA signalling as a candidate pathway for the regulation of the expression of SbGA2ox1 and SbGA2ox3 in dormant IS9530 grains (Supplementary Fig. S1 available at JXB online; Rodríguez et al., 2009, 2012).
Evidence of interactions between ABA signalling and GA metabolism had not been provided until recently. Lee et al. (2012) reported that AtABI5 is capable of binding AtGA3ox1 and AtGA3ox2 promoters in vivo and repressing their expression during the blocking of phyB-dependent germination. However, a cross-talk mechanism between ABA signalling and GA metabolism during dormancy expression has not been addressed for a species with agronomical importance such as grain sorghum. The in silico analysis of the SbGA2ox3 5′-regulatory region carried out by Rodríguez et al. (2012) revealed the presence of several cis-regulatory elements related to ABA and GA signalling. In particular, elements such as the RY repeat, CE, and ABRE were found to be located close to the TATA-box and with a spatial configuration similar to that which had been previously reported to be required for ABA induction of other promoters (Himmelbach et al., 2003; Shen et al., 2004). These findings suggested the possibility that the GA metabolism gene SbGA2ox3 could be regulated by some ABA signalling factors in immature dormant sorghum grains. In this regard, considering the synchrony of SbABI4, SbABI5, and SbGA2ox3 expression profiles and the SbABI5 abundance pattern in incubated dormant sorghum grains, together with the identification of a possible ABRC located on the SbGA2ox3 promoter, the possibility of a protein–DNA interaction between ABI4 and ABI5 proteins and the SbGA2ox3 5′-regulatory region was tested in vitro by performing electrophoretic mobility shift assays (EMSAs). The results show that both ABI5 and ABI4 can interact in vitro with a DNA fragment identical to a region of the SbGA2ox3 promoter containing an ABRE and CE, although they were not able to bind this DNA fragment simultaneously.
Although further evidence is needed to confirm the occurrence of this interaction in vivo, a model is proposed where SbGA2ox3 transcription could be enhanced by SbABI5 and/or SbABI4, increasing GA degradation, which would finally lead to the blocking of germination in dormant immature sorghum seeds. Furthermore, in order to explore whether this cross-talk scheme between ABA signalling and GA metabolism is conserved among different species, a phylogenetic analysis of GA2ox genes in monocot and dicot species was carried out, and the existence of conserved regulatory ‘complexes’ within their promoters was explored as evidence of probable functional importance. The ABRC first identified in the sorghum GA2ox3 promoter region emerged as a highly conserved module among several monocot species, which was absent in the analysed dicots.
Materials and methods
In silico analysis of the SbGA2ox3 promoter
The SbGA2ox3 gene KEGG ID code (Sb03g035000) was used at Phytozome to identify a genomic sequence of 2kb extending 5′ from the translation start site of the SbGA2ox3 gene. The 2kb sequence was considered a gene regulatory region or promoter as no introns were detected upstream of the translation start site of SbGA2ox3. The sequence was scanned for the presence of putative cis-regulatory elements at the PLACE database (http://www.dna.affrc.go.jp/htdocs/PLACE/).
Plant material, RNA extraction, and cloning of SbABI4 and SbABI5
IS9530 sorghum [S. bicolor (L.) Moench] inbred line was sown in the experimental field of the Institute for Agricultural Plant Physiology and Ecology (IFEVA) of the Faculty of Agronomy (Buenos Aires University). Anthesis date was recorded for each plant, and, at 30 d post-anthesis, seeds from eight plants were collected, pooled, and incubated in Petri dishes with distilled water at 25 °C. Embryos were dissected after 1, 2, and 3 d of incubation and stored in liquid N2. Samples of 30–45 embryos (70–100mg) were ground to powder using a mortar and pestle in liquid N2 and added to 600 µl of the RA1 extraction buffer included in the Nucleospin RNAII kit (Macherey-Nagel, Germany). PVP-40 (Sigma) was added to the RA1 buffer to a final concentration of 1% (wv). After 5min centrifugation, clear supernatant was used for the extraction protocol described in the kit’s manual. Reverse transcription was performed with M-MLV Reverse Transcriptase (Promega) and the resulting cDNA aliquots were pooled to use as PCR templates. SbABI4 and SbABI5 cDNA were cloned by PCR from these cDNAs with the addition of BamHI and XhoI restriction sites in the following primers: SbABI4, ACGGGATCCGAACCCAACAACAATCAG (forward) and GCGCTCGAGCTTGAGGAAGACATCAAACC (reverse); SbABI5, GAGAGGATCCAATTTCCCGGGAGGAAGCG (forward) and GAGACTCGAGCCACGGACCTGTCAATGTC (reverse). The resulting PCR products were purified with a NucleoSpin Gel and PCR Clean-up kit.
Expression and purification of recombinant SbABI4 and SbABI5 proteins
SbABI4 and SbABI5 purified cDNAs were ligated into the pGEM-T Easy vector (Promega) and used to transform DH5α Escherichia coli cells. Plasmids were purified by Wizard Plus Sv Minipreps DNA Purification Systems (Promega) and digested with BamHI and XhoI. The digestion product was subsequently ligated into pET24a containing a C-terminal histidine tag, and Rossetta pLys E. coli cells were transformed with the recombinant plasmids. Bacteria were grown in LB medium with kanamycin and chloramphenicol overnight, and 10ml were used to inoculate 1 litre of fresh LB medium. Cultures were grown until OD600=0.6 and induced for 16h with 0.05mM isopropyl-β-d-thiogalactopyranoside (IPTG), at 20 ºC. The resulting pellet was sonicated in lysis buffer (25mM Tris-HCl pH 7.4, 300mM NaCl, and 30mM imidazole) and loaded onto a His GraviTrap column (GE Healthcare). Elution of recombinant protein was achieved in buffer containing 25mM Tris-HCl, 300mM NaCl, and 350mM imidazole, according to the kit instructions. Purified protein was stored at –80 ºC with addition of 5% sucrose and 3mM 2-mercaptoethanol.
Electrophoretic mobility shift assays
SbGa2ox3 probe was obtained by PCR, with a 5′-biotinylated primer (5′BIOT GGGCGCCGTGGGAAAACTG3′) and a 3′-non-biotinylated primer (5′GGGCGGCACCTGGCTGGATG3′), using genomic DNA from the IS9530 grain sorghum line as template. The resulting fragment (242bp long) was purified with NucleoSpin Gel and PCR Clean Up (Macherey Nagel). The specific competitor fragment was obtained by PCR with the same primers as the probe, but without the biotinylated 5′ end. Alternative shorter biotinylated probes for the SbGA2ox3 promoter were generated by combining the same 5′-biotinylated primer and the following 3′-non-biotinylated primers: 5′GGGCGCGACGTGTCCGGACGCG3′ (131bp product, intact ABRE and CE); 5′GGGCGCGAATTGTC CGGACGCG3′ (131bp product, mutated ABRE), and 5′ACGCG ATCCACCGGAAGCAGG3′ (114bp product, mutated CE and no ABRE included). The non-specific competitor (183bp fragment) was amplified by PCR from the SbGAMyb gene. The AtEm6 promoter probe (186bp) was also generated by PCR on Arabidopsis genomic DNA as template and with the following primers: 5′BIOT AGTTAAAGAACACGCGGCGA3′ and 5′TCAATCCGGAGGGCGTTTTGG3′. Complete sequences for all probes are included in Supplementary Fig. S2 at JXB online.
Binding reactions were performed with binding buffer [0.5mM EDTA, 3mM MgCl2, 14mM 2-mercaptoethanol, 10% (v/v) glycerol, 0.05% (v/v) NP-40, 10 µg ml–1 bovine serum albumin (BSA), 25mM TRIS-HCl pH 7.4, 1mM dithiothreitol (DTT), and 30mM KCl], 50 µg ml–1 salmon sperm DNA, 40ng of biotinylated probe, a variable amount of purified recombinant protein SbABI5 or SbABI4, and specific or non-specific competitor fragments (5×, 10×, or 20×). Incubations were carried out for 30min at room temperature and loaded on an 8% (for SbABI4 assays) or 6% (for SbABI5 assays) polyacrylamide gel (29:1 acrylamide:bisacrylamide). For specific and non-specific (SbGAMyb) probe competition assays, proteins were incubated with competitor probes for 10min prior to the addition of biotinylated probe and incubation was continued for an extra 20min. For the non-specific protein assays, total protein extract was obtained from induced cultures of pET24a vector without insert and used for shift assays, incubating the reactions for 30min. In all cases, gels were electrophoresed at 120V in 0.5× TBE and transferred to a nylon membrane (Amersham Hybond-N+). The resulting blots were blocked with TTBS–5% low fat milk for 2h, and incubated overnight with High Sensitivity Streptavidin HRP Conjugate (Thermo). Detection of bound streptavidin protein was performed with ECL substrate and CL-X Posure Film (Thermo).
Identification of GA2ox sequences and phylogenetic analysis
In this work, the GA2ox full-length amino acid sequences of Arabidopsis thaliana (AtGA2ox), Oryza sativa (OsGA2ox), Sorghum bicolor (SbGA2ox), and Zea mays (ZmGA2ox) previously described by Rodríguez et al. (2012) were used. In order to expand the GA2ox family, the KEGG (http://www.genome.ad.jp/kegg/) nucleotide and protein sequence databases for fully sequenced genomes were scanned for GA2ox in the species Medicago truncatula (MtGA2ox), Vitis vinifera (VvGA2ox), Populus trichocarpa (PtGA2ox), and Brachypodium distachyon (BdGA2ox). Identification codes for sequences are listed in Supplementary Table S1 at JXB online. With these amino acid sequences, a set of GA2ox proteins represented in four monocot and four dicot species was completed. The alignment of the full-length amino acid sequences was performed in ClustalW (Thompson et al., 1997) using standard settings (Gonnet weight matrix, gap opening=10 and gap extension=0.2) and was adjusted by visual inspection. Bayesian phylogenetic analyses on aligned full-length sequences were performed with MrBayes v. 3.1.2 setting an MCMC algorithm (Ronquist and Huelsenbeck, 2003). Two independent runs were computed for 1 500 000 generations (Supplementary Fig. S3). In addition, using the Neighbor–Joining (NJ) and maximum parsimony (MP) methods, trees with similar topology were obtained (data not shown). All trees were visualized using the program MEGA 5.01 (Tamura et al., 2011). From the obtained tree (Supplementary Fig. S3), only the group including SbGA2ox3 (I) was selected and divided into subgroups D, M1, M2, and M3 for the following analysis. A 2kb fragment upstream of the ATG was identified for each gene as the putative regulatory region (promoter). The SbGA2ox4 promoter was not included in the search, as the SbGA2ox4 hypothetical protein is truncated and probably non-functional. A comparative analysis using the 2kb promoter sequences was performed within each of four subgroups (D, M1, M2, and M3) using the EARS tool (http://wsbc.warwick.ac.uk/ears/help.php). EARS software breaks sequences into small subsequences (windows) and carries out global alignments between each possible pair of windows, allowing the detection of conserved sequences. Four independent EARS runs were carried out, comparing the SbGA2ox3 promoter with D, M1, M2, and M3 GA2ox promoters, respectively. For all the runs, a windows size of 60bp and a cut off P-value of 0.0001 were used. The EARS result file for each run was analysed and, in the case of the M3 subgroup, the location of the significant peaks detected (1 and 2) within SbGA2ox3 and M3 GA2ox promoters was established. Finally, sequences corresponding to peaks 1 and 2 from BdGA2ox5, BdGA2ox8, OsGA2ox3, OsGA2ox4, and SbGA2ox3 promoters were run on MEME (Bailey et al., 2009) to identify the conserved motifs represented on those fragments.
Results
In silico analysis of the SbGA2ox3 5′-regulatory region
In previous work (Rodríguez et al., 2012), several regulatory motifs related to ABA signalling were found within the first 557bp upstream from the transcription initiation site of the SbGA2ox3 gene. In order to broaden this analysis, and taking into account that many authors have reported the occurrence of plant transcription factors binding cis-elements located at 2kb or 3kb upstream from the ATG (Kim, 2007; Oh et al., 2007; Lee et al., 2012), a genomic sequence of 2kb, considered to be the putative 5′-regulatory region (promoter), was cloned and sequenced for both sprouting-resistant (IS9530) and sprouting-susceptible (RedlandB2) genotypes. To assess the possibility of a differential sequence composition in the regulatory region between the sprouting-resistant and sprouting-susceptible lines, both sequences were analysed in the PLACE database to identify cis-acting regulatory elements. Several cis-acting regulatory sequences were identified, including previously reported ABA and GA signalling elements, with ABRE, CE, DRE (drought response element), RY repeat, MYB, E-box/MYC, and GA-down (similar to ABRE), being present at the highest density of these regulatory sequences found within the first 1000bp upstream from the ATG (Fig. 1). Among the ABA response group, an ABRE, a CE1, and a DRE motif were found to be located at positions –194, –218, and –225bp upstream of the TATA-box, respectively. This particular succession of regulatory elements has been previously reported by other authors to be present in the promoters of ABA-regulated genes (Himmelbach et al., 2003; Shen et al., 2004) and could be considered as an ‘ABA response complex’ (ABRC) on the SbGA2ox3 promoter. On the other hand, many RY and E-box/MYC elements were also found in the analysed region, and it has been reported that these motifs frequently occur in the 5′-regulatory sequence of genes that exhibit seed-specific expression (Stalberg et al., 1996; Ezcurra et al., 1999; Kim et al., 2007). In particular, the location of one of the RY sequences, at position –244bp upstream of the TATA box, and near the possible ABRC, suggested possible promoter seed-specific regulation by ABA for SbGA2ox3. These findings are in agreement with several studies that have shown that regulatory elements conferring seed-specific expression appear in the proximal region of the promoter, often within 500bp upstream of the transcriptional start (Wu et al., 2000; Chandrasekharan et al., 2003).
Fig. 1.
SbGA2ox3 putative 5′-regulatory region sequence, comprising 2000bp upstream from the ATG (in bold). Positions are given relative to the first base of the initiating methionine. Potential transcription binding motifs ABRE, CE, DRE, RY, MYB, and E-box/MYC are underlined and named below. The 242bp biotinylated fragment used as probe in electrophoretic mobility shift assay (EMSA) experiments is highlighted in grey. (This figure is available in colour at JXB online.)
The complete analysis of the 5′-regulatory region allowed the definition of a region within the promoter as a candidate sequence to be bound by SbABI4 and SbABI5. According to this analysis, the region located from –505bp to –263bp upstream from the ATG contained a putative ABRC (ABRE and CE1), a possible target for both transcription factors (ABI4 and ABI5). Therefore, this was precisely the fragment that it was decided to use as a probe in the first place for the subsequent EMSAs (Fig. 1).
When comparing the 2kb promoter region for SbGA2ox3 from both RedlandB2 and IS9530, some point mutations were detected, and only one base was different within the –505 to –236 sequence (probe region) which did not affect any of the ABRC elements (Supplementary Fig. S4 at JXB online). So far, considering cis-regulatory elements involved in ABA signalling, no differences were found in the SbGA2ox3 promoter region from both lines that could be related to the observed contrasting expression of this gene.
SbABI4 and SbABI5 bind to the SbGA2ox3 in vitro
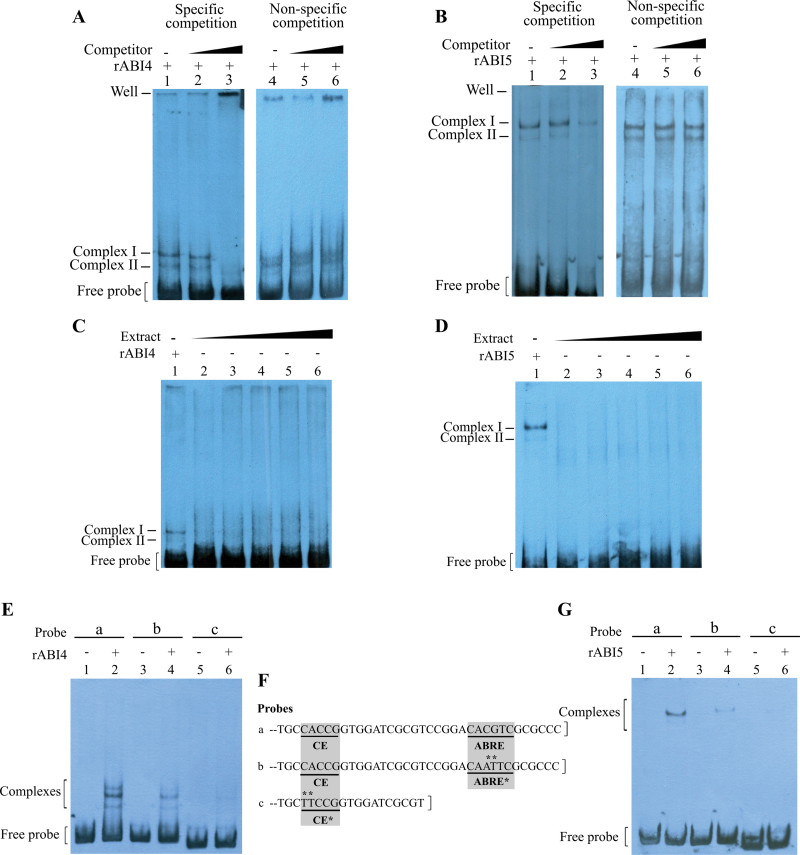
To test whether SbABI4 and SbABI5 can bind the SbGA2ox3 promoter region containing the putative ABRC, both S. bicolor cDNA fragments were cloned from IS9530 and both proteins were expressed with a C-terminal His-tag. The ability of recombinant ABI4 (rABI4) and rABI5 to bind the SbGA2ox3 promoter was analysed by EMSAs using the biotinylated –505bp to –263bp probe. Incubation of this probe with 0.135 μg of rABI4 led to the formation of two complexes (Fig. 2A), suggesting the presence of two binding sites for ABI4 in the SbGA2ox3 probe. Similar results were obtained when incubating the probe with purified rABI5: two major bands were observed using 0.75 μg of protein, indicating that the transcription factor can bind the probe at two sites (Fig. 2B). The slowest migrating complex I can be related to the transcription factor binding two cis-elements simultaneously, and the faster migrating complex II results from the probe being bound at only one of the cis-regulatory elements. For both transcription factors ABI4 and ABI5, complex I (double binding of the probe) showed a stronger signal than did complex II with all protein concentrations, indicating a lower probability of the single bound form in the in vitro conditions used here. When comparing EMSA results for ABI4 and ABI5 assays, the amount of protein that led to the formation of both complexes in each case was quite different. In this sense, a 5-fold higher amount of rABI5 was needed to detect the retarded complexes, as compared with the amount of rABI4. This can be ascribed in part to the fact that the SbABI5 functional form is a dimer of 82kDa while SbABI4 is 30kDa and binds as a monomer (Nakamura et al., 2001); hence, to obtain an equal number of ‘binding units’, a minimal 2.7-fold SbABI5 protein is required as compared with SbABI4, assuming a 100% efficiency of SbABI5 dimerization under the in vitro conditions used here.
Fig. 2.
DNA binding activity of recombinant S. bicolor ABI4 and ABI5 (rABI4 and rABI5). EMSA experiments were performed with 40ng of a 242bp (–505bp to –263bp) SbGA2ox3 promoter biotinylated probe and increasing amounts of recombinant proteins rABI4 and rABI5. Complexes detected, well position, and free probe are indicated. (A) Band shift pattern for rABI4. The protein amount in each reaction was as follows: lane 1, no protein; lane 2, 0.045 μg; lane 3, 0.075 μg; lane 4, 0.15 μg; lane 5, 0.225 μg; lane 6, 0.3 μg. (B) Band shift pattern for rABI5. The protein amount in each reaction was as follows: lane 1, no protein; lane 2, 0.03 μg; lane 3, 0.06 μg; lane 4, 0.15 μg; lane 5, 0.3 μg; lane 6, 0.375 μg; lane 7, 0.47 μg; lane 8, 0.6 μg. (This figure is available in colour at JXB online.)
To test the specificity of the binding of rABI4 and rABI5 to the SbGA2ox3 probe, several gel shift experiments were carried out using non-biotinylated specific and non-specific probes or non-specific protein extracts. Competition with increasing concentrations of the specific unlabelled probe of SbGA2ox3 completely displaced complexes I and II for both rABI4 and rABI5 incubations, whereas competition with the unlabelled fragment of non-specific probe (SbGAMyb) had no effect (Fig. 3A, B). To rule out a possible interaction between residual E. coli proteins and the probe used, incubation reactions were performed with variable amounts of crude protein extracts from E. coli transformed with empty pET24a; these incubations did not lead to the formation of any complex (Fig. 3C, D).
Fig. 3.
(A and B) Competition EMSAs for rABI4 and rABI5 performed with a 242bp SbGA2ox3 fragment (–505bp to –263bp) as biotinylated probe. For all lanes, 40ng of biotinylated probe and 0.225 μg of rABI4 (A) or 0.375 μg of rABI5 (B) were used. For specific competitions, the mass of unlabelled competitor DNA (SbGA2ox3 unlabelled fragment) in each reaction was as follows: lane 1, no competitor; lane 2, 200ng; lane 3, 600ng. For non-specific competitions, the mass of unlabelled competitor (SbGAMyb 183bp fragment) was as follows: lane 4, no competitor; lane 5, 200ng; lane 6, 600ng. (C and D) Control EMSAs for rABI4 (C) and rABI5 (D) performed with increasing concentrations of protein extract from E. coli cultures (transformed with empty pET24a) and 40ng of SbGA2ox3 biotinylated probe. For both C and D, the protein extract amount in each lane was as follows: lane 2, 10.74 μg; lane 3, 14.32 μg; lane 4, 17.9 μg; lane 5, 21.48 μg; lane 6, 25.06 μg. Lane 1 shows the positive control incubation reaction with 40ng of SbGA2ox3 biotinylated probe and 0.225 μg of rABI4 (C) and 0.375 μg of rABI5 (D). (E and G) EMSAs carried out with rABI4 (E) or rABI5 (G) and shorter SbGA2ox3 probes a, b, and c. For all lanes, 40ng of biotinylated probe was used. The protein amount was as follows: lanes 1, 3, and 5, no protein; lanes 2, 4, and 6, 0.225 μg of rABI4 (E) and 0.375 μg of rABI5 (G). (F) Sequences of probes a, b, and c with highlighted CEs and ABREs. Probe a, wild-type probe (intact ABRE and CE); probe b, mutated ABRE; probe c, mutated CE and no ABRE included. Asterisks in probes b and c indicate mutated bases and elements. Probe size was 131bp for a and b, and 114bp for c. (This figure is available in colour at JXB online.)
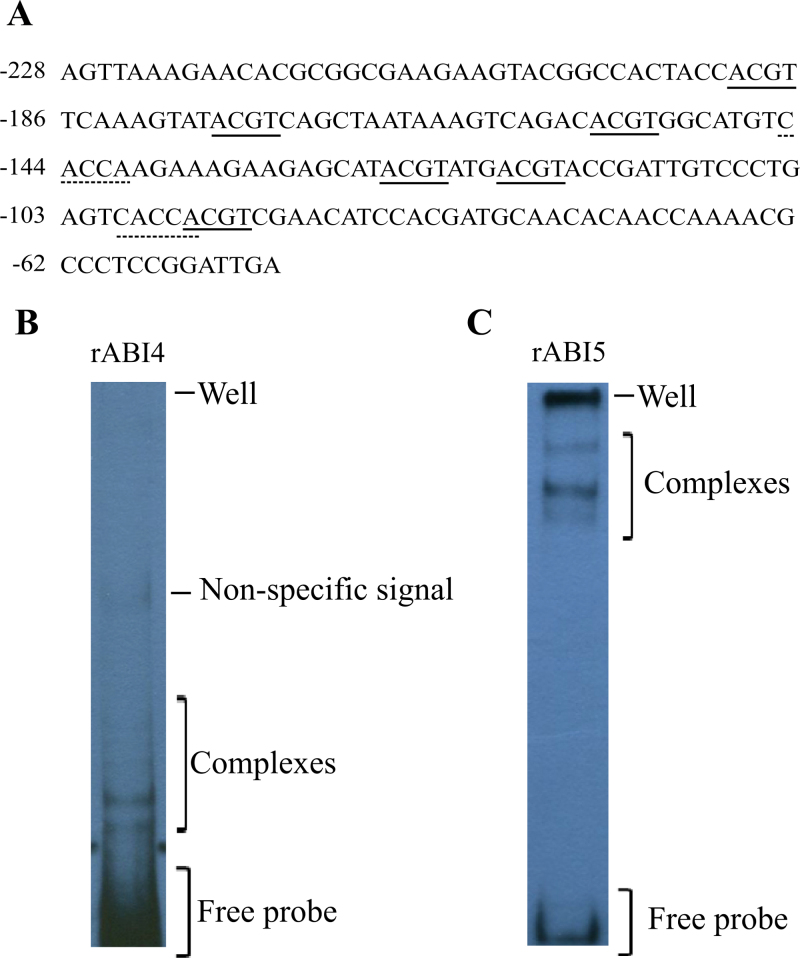
In order to examine the possibility that the ABRC included in the SbGA2ox3 5′-regulatory region was involved in the detected binding to both rABI4 and rABI5 and to discard possible artefacts due to the probe length used, shorter biotinylated probes were designed, including a wild-type sequence (with an intact ABRC), a mutated ABRE probe (with a mutated ABRE and intact CE), and a mutated CE probe (with a mutated CE and without the ABRE) (Fig. 3F). When EMSA experiments were performed with the wild-type sequence probe, both rABI4 and rABI5 produced similar complexes to those with the larger probe, with the appearance of an additional retarded complex in the case of rABI4. However, when rABI4 or rABI5 was incubated with the mutated ABRE probe, the intensity of all complexes decreased, indicating a lower affinity for that DNA fragment. Moreover, when the mutated CE probe was used (and no ABRE was included in this case), only traces of a retarded complex were detected, suggesting that the affinity for this probe was even weaker (Fig. 3E, G). On the other hand, positive control shift assays were performed incubating rABI4 or rABI5 with a biotinylated AtEm6 promoter probe, that contains six putative ACGT cores that could act like ABRE and two CE-like elements (Fig. 4A), and that had been previously demonstrated to be bound by AtABI5 (Carles et al., 2002). Both rABI4 and rABI5 were capable of binding the AtEm6 probe, giving rise to five and four retarded complexes, respectively (Fig. 4B, C). These results suggest that ABRE and CE have a decisive role in the binding of rABI4 and rABI5 to the SbGA2ox3 promoter. Taken together, the results of competition assays and positive and negative control experiments demonstrate that in vitro binding of rABI4 and rABI5 to the SbGA2ox3 probe occurs in a specific manner.
Fig. 4.
rABI4 and rABI5 binding affinity for an AtEm6 promoter biotinylated probe. (A) AtEm6 biotinylated probe sequence (186bp). Positions are given relative to the first base of the initiating methionine (ATG). ACGT cores for putative ABREs (solid lines) and CE-like (dashed lines) motifs are indicated. (B) Band shift pattern for rABI4. A 40ng aliquot of AtEm6 biotinylated probe was incubated with 0.225 μg of rABI4. (C) Band shift pattern for rABI5. A 40ng aliquot of AtEm6 biotinylated probe was incubated with 0.375 μg of rABI5. In all cases, complexes detected, well position, and free probe are indicated. (This figure is available in colour at JXB online.)
SbABI4 and SbABI5 do not bind simultaneously to the SbGA2ox3 in vitro
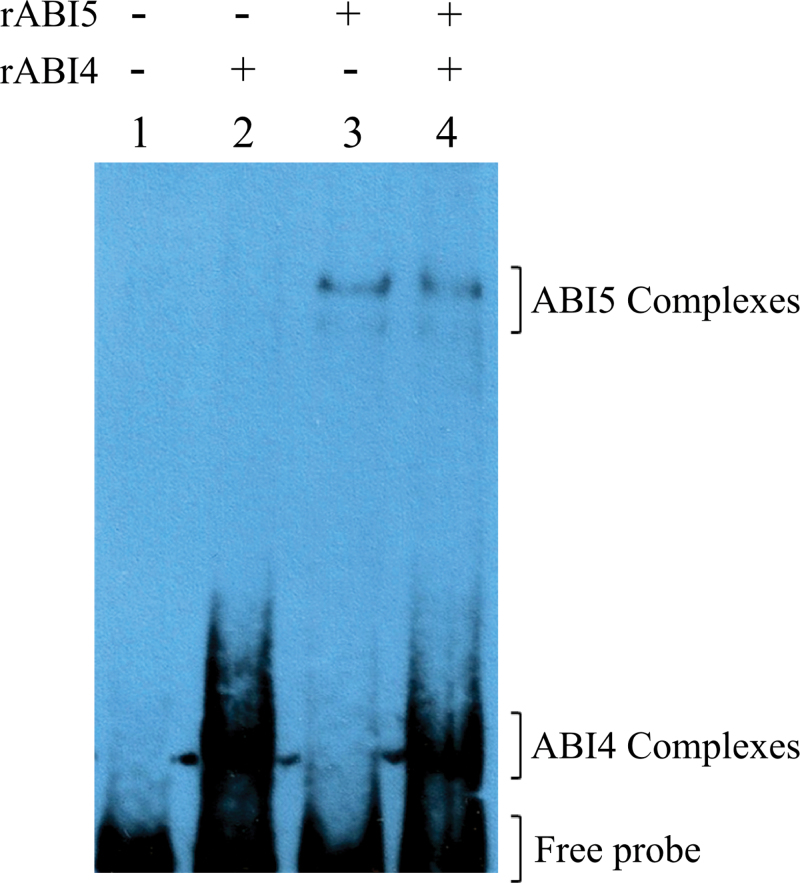
Considering that both SbABI4 and SbABI5 bound to the same SbGA2ox3 5′-regulatory region and that it has been recently reported that ABI4 and ABI5 could synergistically activate some plant promoters (Reeves et al., 2011), the possibility that ABI4 and ABI5 recombinant proteins could bind simultaneously to the same SbGA2ox3 promoter probe was examined. Contrary to this hypothesis, shift assays performed by co-incubating the SbGA2ox3 probe with both proteins did not generate any ternary complex, suggesting that SbABI4 and SbABI5 do not bind to the SbGA2ox3 promoter simultaneously (Fig. 5). Hence, it is possible that they have affinity for the same regulatory elements.
Fig. 5.
EMSA performed with rABI4 and rABI5 co-incubations with 40ng of SbGA2ox3 biotinylated probe. Protein amounts in the reactions were as follows: lane 1, no protein; lane 2, 0.225 μg of rABI4; lane 3, 0.375 μg of rABI5; lane 4, 0.225 μg of rABI4 and 0.375 μg of rABI5. Complexes and free probe are indicated. (This figure is available in colour at JXB online.)
ABRC is found in the promoters of other monocot GA2ox genes
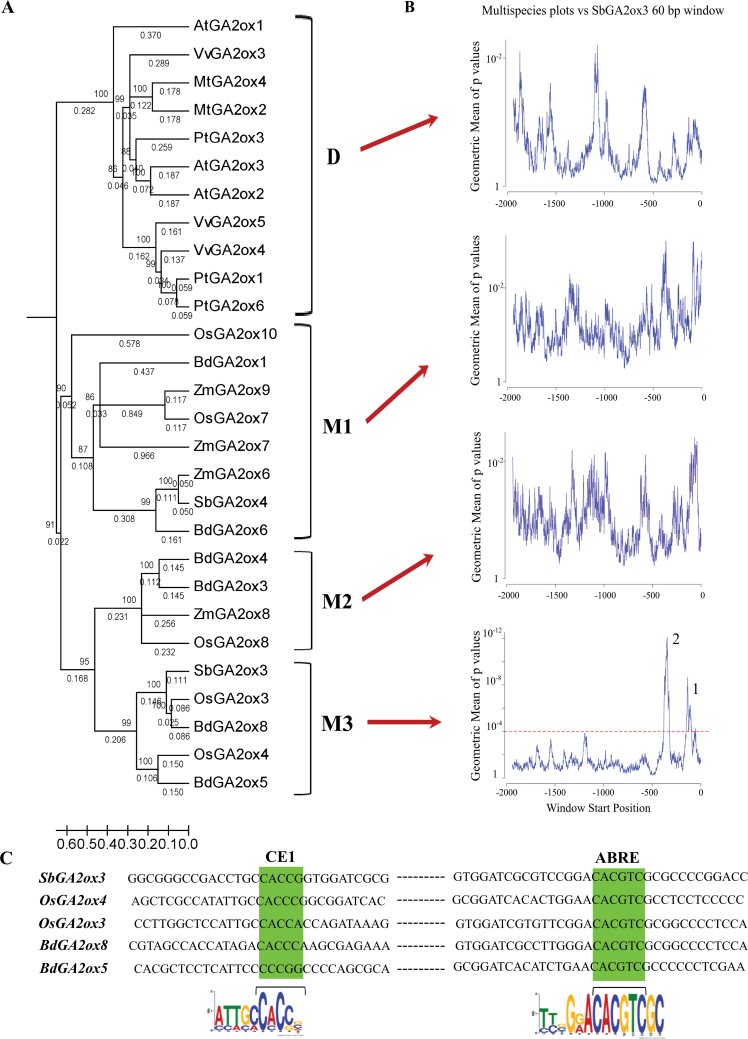
In view of the in vitro results obtained by EMSAs, the next objective was to investigate whether the interactions detected are part of a conserved regulatory mechanism of GA2ox genes in other species. Thus, the possibility that those GA2oxs with high sequence similarity to SbGA2ox3 would share the same regulatory elements in the 5′-regulatory region was tested. To address this question, a global phylogenetic analysis was first conducted using the whole set of described GA2oxs involved in GA metabolism from four dicot and four monocot species. The full-length GA2ox protein sequences from these eight species were completely aligned and a phylogenetic tree was obtained (Supplementary Fig. S3 at JXB online) using MrBayes (Huelsenbeck and Ronquist, 2001). Taking into account the tree topology structure, branch lengths, and clade support values, GA2ox proteins were classified into three groups: I, II, and III, corroborating the classification proposed by Lee and Zeevart (2005) and the specific results reported by Rodríguez et al. (2012). SbGA2ox1 and SbGA2ox2 were included in group II, while SbGA2ox3 and SbGA2ox4 were members of group I, and, as the main interest of this study was focused on SbGA2ox3, group I was selected for further analyses. A clear separation between monocot (M) and dicot (D) GA2oxs was observed within group I, and the M group was further divided into M1, M2, and M3 subgroups (Fig. 6A). When the motif composition of all monocot GA2oxs was examined (Supplementary Fig. S5), SbGA2ox4 appeared as a truncated and probably non-functional protein, almost certainly originating from a recent duplication of the SbGA2ox3 gene.
Fig. 6.
Comparative analysis of regulatory regions of GA2ox promoters. (A) Phylogenetic relationships for group I GA2oxs. The 28 GA2ox proteins were grouped into four subgroups: D, dicots; M1, monocots 1; M2, monocots 2; and M3, monocots 3. At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Mt, Medicago truncatula; Os, Oryza sativa; Pt, Populus trichocarpa; Sb, Sorghum bicolor; Vv, Vitis vinifera; Zm, Zea mays. Bootstrap support values are indicated and scale bars specify the number of changes per position for a unit branch length. Identification codes for sequences are listed in Supplementary Table S1 at JXB online. (B) Multispecies plots showing comparative analysis of the SbGA2ox3 promoter versus promoters of each subgroup (D, M1, M2, and M3), performed with the EARS tool. The red dashed line indicates the selected cut-off P-value (0.0001), suggesting that only in the case of M3 were two significant peaks detected. (C) Sequence alignments of significant peak sequences of M3 members (SbGA2ox3, OsGA2ox3, OsGA2ox4, BdGA2ox5, and BdGA2ox8). Conserved regions are highlighted in green: ABRE (CACGTC) and CE (CACCG). The MEME software (Bailey and Elkan, 1994) was used to find common motif logos in the promoters of subgroup M3 members. CE and ABRE motif logos are shown below each conserved sequence.
To test the hypothesis of a conserved regulatory mechanism involving the GA2ox gene promoter, a comparative analysis of 5′-regulatory sequences (2kb upstream of the ATG) of SbGA2ox3 against each subgroup promoter (i.e. D, M1, M2, and M3) was performed, with the aid of the EARS tool (Picot et al., 2010). Results obtained from these analyses showed that only promoters of GA2ox genes within the M3 subgroup share common regions with the 5′-regulatory sequence of SbGA2ox3. Two peaks with high significance levels were obtained, indicating the existence of two conserved regions within these promoters (Fig. 6B). Additionally, pairwise comparisons (i.e. SbGA2ox3 and each of the other GA2ox promoters within group M3) were performed, and it was found that the five M3 members shared the same two significant peaks (Supplementary Fig. S6 at JXB online). Then, with the aim of identifying the common motifs within these five GA2ox genes, the sequence composition of the two conserved regions was explored in detail. Peak 1 co-localized in all cases with the TATA-box, while peak 2 could be delimited between –460bp and –375bp upstream from the ATG of the SbGA2ox3 promoter. The sequence corresponding to that second peak comprised a portion of the probes that had been previously used in the EMSAs described above, and the ABRE and CE1 were represented in all cases. The CE1 sequence comprised point mutations for both Brachypodium GA2ox promoters, compared with the CE1 described in other species such as maize, which would lead to CE1-like elements (Niu et al., 2002). In all cases, the location of ABRE and CE1 with respect to the TATA-box was as had been previously described for ABRCs (Shen and Ho, 1995; Shen et al., 1996), supporting a possible transcriptional regulation by the ABA pathway for these genes (Fig. 6C). Moreover, although not included in the conserved regions detected in the present analysis (due to greater variation in the positioning of these elements), RY repeats were represented in all M3 subgroup promoters, indicating that the possible ABA induction of these genes is seed specific. Taken together, the results from the phylogenetic and comparative analyses indicate that M3 subgroup GA2oxs not only show a structural and probable functional similitude, but also might share a common transcriptional regulatory mechanism. Transcriptional factors ABI4 and ABI5 involved in ABA signalling emerge as strong candidates to be part of that mechanism.
Discussion
ABA signalling and GA metabolism are known to play a main role during dormancy expression in immature sorghum grains (Perez-Flores et al., 2003; Rodríguez et al., 2009, 2012). Previous results with the sorghum system used here (RedlandB2, low dormancy; and IS9530, high dormancy) indicated that embryo sensitivity to ABA was related to dormancy expression in these two genotypes (Steinbach et al., 1995; Gualano et al., 2007; Rodríguez et al., 2009). Similarly, Rodríguez et al. (2009) found that sorghum genes encoding putative ABI3/VP1, ABI4, ABI5, and PKABA1 (positive regulators of ABA signalling) were highly expressed during the incubation of IS9530 dormant immature grains, but not in the less dormant genotype RedlandB2. In addition, the content of active GA4 reached a significantly higher value in less dormant RedlandB2 embryos during day 4 of grain incubation as compared with the more dormant IS9530 (Perez-Flores et al., 2003; Rodríguez et al., 2012). A possible explanation for this differential GA4 accumulation was proposed by Rodríguez et al. (2012) who reported an increase in transcript levels for genes encoding putative GA synthesis enzymes in both dormant and non-dormant grains, but this was also accompanied by an evident promotion of the GA inactivation genes SbGA2ox1 and SbGA2ox3 only in dormant IS9530. A negative correlation between GA4 and the GA34 catabolite also supported an active role for GA catabolism in determining GA4 values during the expression of dormancy in these sorghum lines. Considering the coordinated expression of SbABI4, SbABI5, and SbGA2ox3 and the SbABI5 accumulation profile in incubated immature dormant sorghum grains (Supplementary Fig. S1 at JXB online; Rodríguez et al., 2009, 2012), together with the finding of a possible ABRC in the SbGA2ox3 promoter, an interaction between SbABI4, SbABI5, and the SbGA2ox3 5′-regulatory region was examined in vitro.
In this study, it is demonstrated that SbABI4 and SbABI5 recombinant proteins are both capable of interacting in vitro with a fragment of the SbGA2ox3 5′-regulatory region, suggesting a cross-talk between ABA signalling and GA metabolism during imbibition of immature, dormant sorghum seeds. Moreover, the results suggest that these interactions could be part of a transcriptional regulatory mechanism particular to a group of monocot GA2oxs, that has not been described previously.
In silico analysis of the SbGA2ox3 promoter revealed the presence of many cis-acting elements including several known to be related to ABA or GA signalling and seed-specific expression (Fig. 1). Of particular interest are the ABRE and CE1 motifs located close to the TATA-box that, together, constitute an ABRC. ABA-responsive elements including ABRE and CE have been previously identified in the promoters of the HVA1 Lea gene (Shen et al., 1996), ABA-induced HVA22 (Shen and Ho, 1995), wheat Em (Guiltinan et al., 1990), and rice genes Rab16b (Ono et al., 1996), Rab17 (Busk et al., 1997), Rab28 (Busk and Pagès, 1998), and OsEm (Hattori et al., 1995).
In this study it has been demonstrated that both transcription factors, SbABI4 and SbABI5, are capable of binding the SbGA2ox3 probe in a specific manner, generating two complexes; this indicates the existence of two binding sites within this region of the SbGA2ox3 promoter for each protein (Fig. 2). Although further experiments, such as DNA footprinting, are needed to elucidate the regulatory motifs that are bound by these proteins, shift assays performed with mutated ABRE or mutated CE probes demonstrated that both ABRE and CE elements are required for specific binding of SbABI4/SbABI5 to the SbGA2ox3 promoter, as the binding to the SbGA2ox3 probe was markedly reduced when ABRE and CE sequences were mutated. On this same theme, according to what had previously been reported in maize (Niu et al., 2002), a candidate element in the SbGA2ox3 promoter to be possibly bound by ABI4 would be the CE1, located –218bp upstream from the TATA-box. On the other hand, the ABRE (located –194bp upstream from the TATA-box) emerges as an additional candidate motif to be recognized by ABI4 as probe binding was reduced when the ABRE was mutated. This is also supported by the results of shift assays performed with the AtEm6 promoter probe containing six putative ABRE and two CE-like motifs, which gave rise to five complexes when incubated with rABI4. Moreover, Wind et al. (2012) proposed a model that predicts the binding of ABI4 to the G-box element, which is coincident with the strong ABRE sequence (CACGTG).
According to the existing information (reviewed in Shinozaki et al., 2007) and the present EMSA results obtained with the AtEm6 probe, ABI5 might bind the ABRE on the SbGA2ox3 regulatory region generating one of the complexes detected, and the CE appears as a possible candidate to be recognized by ABI5 and to give rise to the additional complex. In this sense, Casaretto et al. (2003) reported that the barley bZIP HvABI5 recognized both the ACGT-box and the CE3 of ABRC3, suggesting the possibility that SbABI5 binds both ABRE and CE. In the same vein, although both rABI4 and rABI5 were capable of binding the SbGA2ox3 promoter, they were not able to interact simultaneously with the biotinylated probe, suggesting that both proteins might compete for the same binding motifs. This is in accordance with results reported by Cassaretto et al. (2003) as described above and also with results of Reeves et al. (2011), who analysed the transcriptome of ABI4- and ABI5-overexpressing Arabidopsis plants and reported that the promoters of many target genes of ABI4 and/or ABI5 are enriched in ABRE but not in CE motifs. In this last work, Reeves et al. also reported binding of Arabidopsis ABI4 to a DNA probe containing only ABRE but no CE motifs.
The present results indicate that SbABI4 is required in smaller quantities than SbABI5 to detect retarded complexes. Part of this difference could be related to the fact that ABI5 is known to act as homodimer or heterodimer (Finkelstein and Lynch, 2000) and, on the other hand, it has been shown that phosphorylation stabilizes ABI5 and enhances its activity in vivo (Lopez-Molina et al., 2001; Piskurewicz et al., 2008), which would also lead to larger amounts of SbABI5 needed to detect in vitro binding. Taking into account that both SbABI4 and SbABI5 have the capability of binding the SbGA2ox3 promoter, both transcription factors could be part of a mechanism of fine-tuning the expression of SbGA2ox3 in imbibed dormant grains. Although SbABI4 expression has already been measured by Rodríguez et al. (2009), further experiments are needed to quantify SbABI4 protein abundance during the incubation of dormant grains in order to have a more accurate picture of SbABI4 accumulation during imbibition. Moreover, SbVP1 appears to be another strong candidate to regulate SbGA2ox3 expression, according to Himmelbach et al. (2003), whose work suggested that the transcriptional modulation of ABA-regulated genes might include the combined action of bZIP, AP2, and B3 domain transcription factors. In this sense, although ABI3 does not interact directly with ABREs or CEs, it is able to bind RY elements and, in combination with closely located ABREs or CEs, could act as a transcription enhancer of seed-specific ABA-regulated genes (reviewed by Holdswoth et al., 2008). In this context, the RY elements identified in the SbGA2ox3 promoter reinforce the possibility of SbAVP1/ABI3 acting as an accessory enhancer of SbGA2ox3 transcription in the seed.
Taken together, these results allow the proposal that during dormancy expression in dormant immature seeds, SbABI4 and/or SbABI5 (with the possible accessory action of SbVP1) might interact with the SbGA2ox3 promoter, enhance its transcription, and lead to SbGA2ox3 protein accumulation; this would result in subsequent active GA degradation, thus preventing germination of dormant grains. Conversely, ABA signalling components in non-dormant immature grains are scarcely accumulated during imbibition, and the SbGA2ox3 promoter could not be activated by SbABI5 and/or SbABI4; this would lead to the accumulation of active GA (due to weak inactivation) and, as a result, enhanced germination. Similarly, Lee et al. (2012) reasoned that during phyB-dependent inhibition of germination in Arabidopsis, AtABI5 interacts with AtGA3ox1 and AtGA3ox2, but suppresses their expression instead, which results in lower GA levels in the seed.
The phylogenetic and domain architecture analyses demonstrated that SbGA2ox3 is probably the only functional GA2ox within group I, as SbGA2ox4 appeared as a truncated duplication of SbGA2ox3. The probable lack of functionality of SbGA2ox4 highlights the central biological role of SbGA2ox3 in grain sorghum. In this same vein, the phylogenetic analysis and protein alignments performed showed that SbGA2ox3 shares not only a conserved structure/function with other M3 subgroup members, but also a common transcriptional regulation. No conserved regulatory complexes were observed for the remaining subgroups D, M1, and M2. The results demonstrate that the members of the M3 GA2ox subgroup have two conserved sequences, one of them including both the ABRE and CE (ABRC). The functionality of BdGA2ox5 the CE1-like element has not been tested in other promoters, and so it cannot be considered as a functional element until new evidence is reported, but the CE1-like element detected in the BdGA2ox8 promoter has been shown to be functional for ABI4 binding in maize ABI4, RAB28, and RAB17 and in barley HVA22 and HVA (Niu et al., 2002). These findings suggest that M3 GA2oxs could be ABA-regulated genes and that the ABA signalling and GA metabolism cross-talk proposed for grain sorghum could also operate in rice and Brachypodium, through the transcriptional regulation of OsGA2ox3, OsGA2ox4, BdGA2ox5, and BdGA2ox8. Furthermore, the finding of RY elements in the promoter of these four genes indicates that the regulation of ABA transcription could be also mediated by VP1/ABI3 in a seed-specific manner.
Along the same lines, a comparative analysis of ABA content on Brachypodium grains showed that although low dormant and high dormant genotype seeds that went through after-ripening had less ABA than dormant grains after 4 d of imbibition, ABA content in dry seeds from the less dormant genotype was higher than in seeds of the high dormant genotype. These results suggest that ABA levels by themselves cannot predict dormancy in different Brachypodium genotypes, and ABA sensitivity and other hormones such as GA could be playing an important role (Barrero et al., 2012). Studies of transcript levels of GA metabolism genes or embryo sensitivity to ABA have not been reported yet for Brachypodium, so a possible role for these components in dormancy expression cannot be ruled out. On the other hand, it has been proposed that ABA content during imbibition and seed development of weedy red rice (Oryza sativa) is not directly linked to the dormancy level. Instead, ABA sensitivity seems to be an important component for dormancy status (Gianinetti et al., 2007). Taken together, these results suggest that the ABA–GA cross-talk proposed for sorghum could also take place in rice and Brachypodium dormant seeds.
Although additional assays such as ChIP-PCR are necessary to confirm the in vivo occurrence of these interactions in the seed embryo, in vitro specific binding is the first proof required to clarify whether an interaction is possible or not. Current efforts are oriented towards studying the relevance of these interactions in vivo in sorghum immature, dormant grains, and identifying other potential transcriptional factors interacting with the SbGA2ox3 promoter region in one-hybrid yeast assays. As mentioned before, the first evidence of an interaction between ABA signalling and GA metabolism has recently been reported by Lee et al. (2012) in the model species A. thaliana, but similar interactions have not been described yet for any species of agronomic relevance.
To conclude, even though GA metabolism enzymes are highly conserved in their structure and probably in their function, the regulation of these genes across different species presents high variability. The identification of regulatory steps in species with agronomic relevance is crucial for the planning of breeding strategies. Sorghum lines IS9530 and RedlandB2 present an experimental system based on intraspecific variability for the level of dormancy and PHS response. The present work offers new insights into the regulation of a sorghum GA catabolism gene which in previous works emerged as a strong candidate to regulate active GA4 levels and the germination response. The contrasting expression pattern of the GA2ox3 gene in both lines is likely to rely on the differential activity of ABA pathway elements such as ABI4 and ABI5 rather than variability in the GA2ox3 promoter sequence. In this sense, activation of GA catabolism by ABA signalling factors could be interpreted as an additional factor that contributes to the inhibition of germination as the ABA/GA balance is pushed further towards the action of ABA. It would be interesting to find out if this mechanism is present in other monocots in which ABA sensitivity, rather than ABA metabolism, is in control of the level of dormancy. Further studies with TILLING mutants from sorghum, Brachypodium, or rice affected in the GA2ox genes that harbor the ABRC in their promoters might prove useful to understand the contribution of this gene to the expression of dormancy.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. (A) Embryonic transcript levels for SbABI4, SbABI5, and SbGA2ox3 genes during incubation at 20 ºC of immature grains of RedlandB2 and IS9530. (B) Evolution of germination percentage and embryonic content of GA4 during incubation of RedlandB2 and IS9530 immature seeds at 20 ºC.
Figure S2. Complete sequences of probes used in EMSA experiments.
Figure S3. Phylogenetic relationships of GA2oxs.
Figure S4. GA2ox3 probe sequence alignment for S. bicolor genotypes IS9530 and RedlandB2.
Figure S5. Domain architecture of group I monocot GA2ox proteins, showing the similarity in domain composition and location.
Figure S6. Pairwise comparison plots between SbGA2ox3 and each M3 promoter (OsGA2ox3, OsGA2ox4, BdGA2ox5, and BdGA2ox8) performed with the EARS tool.
Table S1. GA2ox protein sequence accessions, naming terminology, and database used for eight plant species.
Acknowledgements
This work was supported by a grant from the National Agency for Science and Technological Promotion (ANPCyT) PICT 2010 no. 2521 and a PhD grant from the Argentinean National Council of Scientific and Technical Research (CONICET). The authors would like to thank Paula Mut, Ana Distéfano, and Maximiliano Sánchez-Lamas for their skilful assistance with protein expression; Rodrigo Sieira for his expert help with the EMSA technique; Ángeles Zorreguieta for sharing her lab at Fundación Instituto Leloir; and Gabriela Auge for her collaboration in SbGA2ox3 promoter cloning.
References
- Bailey T, Bodén M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37, W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, 28–36 [PubMed] [Google Scholar]
- Barrero JM, Jacobsen JV, Talbot MJ, White RG, Swain SM, Garvin DF, Gubler F. 2012. Grain dormancy and light quality effects on germination in the model grass Brachypodium distachyon. New Phytologist 193, 376–386 [DOI] [PubMed] [Google Scholar]
- Baumlein H, Nagyt I, Villarroel R, Inzé D, Wobus U. 1992. Cis-analysis of a seed protein gene promoter: the conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. The Plant Journal 2, 233–239 [PubMed] [Google Scholar]
- Benech-Arnold RL, Enciso S, Sánchez RA, Carrari F, Perez-Flores L, Iusem N, Steinbach HS, Lijavetzky D, Bottini R. 2000. Involvement of ABA and GAs in the regulation of dormancy in developing sorghum seeds. In: Black M, Bradford KJ, Vázquez Ramos J, eds. Seed biology: advances and applications. Wallingford, UK, CAB International, 101–111 [Google Scholar]
- Bewley D. 1997. Seed germination and dormancy. The Plant Cell 9, 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi F, Cordoba E, Dupre P, Mendoza MS, Roman CS, Leon P. 2009. The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. The Plant Journal 59, 359–374 [DOI] [PubMed] [Google Scholar]
- Brocard-Gifford I, Lynch T, Finkelstein R. 2003. Regulatory networks in seeds integrating developmental, ABA, sugar and light signaling. Plant Physiology 131, 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk PK, Jensen AB, Pagés M. 1997. Regulatory elements in vivo in the promoter of the abscisic acid responsive gene rab17 from maize. The Plant Journal 11, 1285–1295 [DOI] [PubMed] [Google Scholar]
- Busk PK, Pagés M. 1998. Regulation of abscisic acid induced transcription. Plant Molecular Biology 37, 425–435 [DOI] [PubMed] [Google Scholar]
- Carles C, Bies-Etcheve N, Aspart L, León-Kloosterziel KM, Koorneef M, Echeverrria M, Delseny M. 2002. Regulation of Arabidopsis thaliana Em genes: role of ABI5. The Plant Journal 30, 373–383 [DOI] [PubMed] [Google Scholar]
- Casaretto J, Ho TH. 2003. The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. The Plant Cell 15, 271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Bishop KJ, Hall TC. 2003. Module-specific regulation of the betaphaseolin promoter during embryogenesis. The Plant Journal 33, 853–866 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. 2010. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology 61, 651–679 [DOI] [PubMed] [Google Scholar]
- Daszkowska-Golec A, Szareijo I. 2013. The molecular basis of ABA-mediated plant response to drought. In: Vahdati K, Leslie C, eds.Abiotic stress—plant responses and applications in agriculture. InTech, DOI: 10.5772/53128 [Google Scholar]
- Ezcurra I, Ellerström M, Wycliffe P, Stålberg K, Rask L. 1999. Interaction between composite elements in the napA promoter: both the B-box ABA-responsive complex and the RY/G complex are necessary for seed-specific expression. Plant Molecular Biology 40, 699–709 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. 2006. Seed dormancy and the control of germination. New Phytologist 171, 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein R. 1994. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. The Plant Journal 5, 765–771 [Google Scholar]
- Finkelstein R, Lynch T. 2000. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell 12, 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. 2008. Molecular aspects of seed dormancy. Annual Review of Plant Biology 59, 387–415 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Rock C. 2002. Abscisic acid biosynthesis and response. In: Somerville CR, Meyerowitz EM, eds. The Arabidopsis book. Rockville, MD: American Society of Plant Biologists, 1–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinetti A, Vernieri P. 2007. On the role of abscisic acid in seed dormancy of red rice. Journal of Experimental Botany 58, 3449–3462 [DOI] [PubMed] [Google Scholar]
- Gualano N, Carrari F, Rodriguez MV, Perez-Flores L, Sanchez RA, Iusem ND, Benech-Arnold R. 2007. Reduced embryo sensitivity to ABA in sprouting susceptible sorghum (Sorghum bicolor) variety is associated with an altered ABA signalling. Seed Science Research 17, 81–90 [Google Scholar]
- Guiltinan MJ, Marcotte WR, Jr, Quatrano RS. 1990. A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250, 267–271 [DOI] [PubMed] [Google Scholar]
- Hattori T, Terada T, Hamasuna S. 1995. Regulation of the Osem gene by abscisic acid and the transcriptional activator VP1: analysis of cis-acting promoter elements required for regulation by abscisic acid and VP1. The Plant Journal 7, 913–925 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E. 2003. Relay and control of abscisic acid signaling. Current Opinion in Plant Biology 6, 470–479 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJJ. 2008. Molecular networks regulating Arabidopsis seed maturation, afterripening, dormancy and germination. New Phytologist 179, 33–54 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Kim JK, Shin JS, Suh MC. 2007. The SebHLH transcription factor mediates trans-activation of the SeFAD2 gene promoter through binding to E- and G-box elements. Plant Molecular Biology 64, 453–466 [DOI] [PubMed] [Google Scholar]
- Lee KP, Piskurewicz U, Turecková V, Carat S, Chappuis R, Strnad M, Fankhauser C, Lopez-Molina L. 2012. Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes and Development 26, 1984–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DJ, Zeevaart JAD. 2005. Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris . Plant Physiology 138, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua N. 2001. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis . Proceedings of the National Academy of Sciences, USA 98, 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy J, Yamaguchi-Shinozaki K, Chua NH. 1990. Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proceedings of the National Academy of Sciences, USA 87, 1406–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR. 2001. Physical interactions between ABA response loci of Arabidopsis. The Plant Journal 26, 627–635 [DOI] [PubMed] [Google Scholar]
- Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y. 2010. Abscisic acid and the control of seed dormancy and germination. Seed Science Research 20, 55–67 [Google Scholar]
- Niu X, Helentjaris T, Bate NJ. 2002. Maize ABI4 binds coupling element1 in abscisic acid and sugar response genes. The Plant Cell 14, 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. 2003. Gibberellin biosynthesis and response during Arabidopsis seed germination. The Plant Cell 15, 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun T, Kamiya Y, Choi G. 2007. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. The Plant Cell 19, 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Izawa T, Chua NH, Shimamoto K. 1996. The rab16B promoter of rice contains two distinct abscisic acid-responsive elements. Plant Physiology 112, 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. 2006. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. The Plant Cell 18, 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Flores LJ, Carrari F, Osuna-Fernández HR, Enciso S, Stanelloni R, Sánchez RA, Bottini R, Iusem ND, Benech-Arnold RL. 2003. Expression analysis of a GA 20-oxidase in embryos from two sorghum lines with contrasting dormancy: possible participation of this gene in the hormonal control of germination. Journal of Experimental Botany 54, 2071–2079 [DOI] [PubMed] [Google Scholar]
- Picot E, Krusche P, Tiskin A, Carré I, Ott S. 2010. Evolutionary analysis of regulatory sequences (EARS) in plants. The Plant Journal 64, 165–176 [DOI] [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L. 2008. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. The Plant Cell 20, 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves WM, Lynch TJ, Mobin R, Finkelstein RR. 2011. Direct targets of the transcription factors ABA-insensitive (ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Molecular Biology 75, 347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, et al. 2008. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. The Plant Journal 53, 488–504 [DOI] [PubMed] [Google Scholar]
- Rodríguez MV, Mendiondo GM, Cantoro R, Auge GA, Luna V, Masciarelli O, Benech-Arnold RL. 2012. Expression of seed dormancy in grain sorghum lines with contrasting pre-harvest sprouting behavior involves differential regulation of gibberellin metabolism genes. Plant and Cell Physiology 53, 64–80 [DOI] [PubMed] [Google Scholar]
- Rodríguez MV, Mendiondo GM, Maskin L, Gudesblat GE, Iusem ND, Benech-Arnold RL. 2009. Expression of ABA signalling genes and ABI5 protein levels in imbibed Sorghum bicolor caryopses with contrasting dormancy and at different developmental stages. Annals of Botany 104, 975–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- Shen QJ, Casaretto JA, Zhang P, Ho TH. 2004. Functional definition of ABA-response complexes: the promoter units necessary and sufficient for ABA induction of gene expression in barley (Hordeum vulgare L.). Plant Molecular Biology 54, 111–124 [DOI] [PubMed] [Google Scholar]
- Shen Q, Ho THD. 1995. Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. The Plant Cell 7, 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Zhang P, Ho THD. 1996. Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. The Plant Cell 8, 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. 2007. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany 58, 221–227 [DOI] [PubMed] [Google Scholar]
- Stalberg K, Ellerstom M, Ezcurra I, Ablov S, Rask L. 1996. Disruption of an overlapping E-box/ABRE motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta 199, 515–519 [DOI] [PubMed] [Google Scholar]
- Steinbach HS, Benech-Arnold RL, Kristof G, Sánchez RA, Marcucci-Poltri S. 1995. Physiological basis of pre-harvest sprouting resistance in Sorghum bicolor (L.) Moench. ABA levels and sensitivity in developing embryos of sprouting-resistant and sprouting-susceptible varieties. Journal of Experimental Botany 46, 701–709 [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wind JJ, Peviani A, Snel B, Hanson J, Smeekens SC. 2013. ABI4: versatile activator and repressor. Trends in Plant Science 18, 125–132 [DOI] [PubMed] [Google Scholar]
- Wu C, Washida H, Onodera Y, Harada K, Takaiwa F. 2000. Quantitative nature of the prolamin-box, ACGT and AACA motifs in a rice glutelin gene promoter: minimal cis-element requirements for endosperm-specific gene expression. The Plant Journal 23, 415–421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.