Abstract
The central catalyst in eukaryotic ATP-dependent homologous recombination consists of RAD51 proteins, polymerized around single-stranded DNA. This nucleoprotein filament recognizes a homologous duplex DNA segment and invades it1,2. After strand exchange, the nucleoprotein filament should disassemble in order for the recombination process to complete3. The molecular mechanism of RAD51 filament disassembly is poorly understood. Here, we have combined optical tweezers with single-molecule fluorescence microscopy and microfluidics4,5 to reveal that disassembly results from the interplay between ATP hydrolysis and release of the tension stored in the nucleoprotein filament. Applying external tension to the DNA, we found that disassembly slows down and can even be stalled. We quantified the fluorescence of RAD51 patches and found that disassembly occurs in bursts interspersed by long pauses. Upon relaxation of a stalled complex, pauses were suppressed resulting in a large burst. These results imply that tension-dependent disassembly takes place only from filament ends, after tension-independent ATP hydrolysis. This integrative single-molecule approach allowed us to dissect the mechanism of this key homologous recombination reaction step, which in turn clarifies how disassembly can be influenced by accessory proteins.
Homologous recombination is a vital mechanism that maintains genome integrity by repairing double-strand breaks in DNA, and generates genetic diversity by exchanging DNA between chromosomes during meiosis. The central process in homologous recombination is the strand exchange between homologous DNA segments. Recombinase proteins such as RecA and RAD51 catalyze this process by forming an ATP-dependent helical filament around single-stranded DNA (ssDNA)1. This filament finds a homologous segment of double-stranded DNA (dsDNA), invades it and catalyzes strand exchange to generate a joint molecule. This resulting structure is further processed in multiple steps by additional proteins, finally yielding two intact, homologous dsDNAs1,2. For these steps to proceed properly, it is essential that RAD51 filaments disassemble. Hydrolysis of ATP bound at the interface between adjacent monomers is a prerequisite for filament disassembly6-8. RecA and RAD51 not only form ATP-dependent filaments on ssDNA, but also on dsDNA4,9-11. These dsDNA nucleoprotein filaments may have deleterious effects in vivo, e.g., by sequestering these recombinases in nonfunctional complexes that could obstruct other DNA transactions. RAD51 filament disassembly can be aided by auxiliary proteins3. To understand recombinase removal, it is essential to elucidate the molecular mechanism of the intrinsic RAD51 disassembly reaction.
To follow this process under controlled conditions, we developed an instrument that combines fluorescence microscopy with force-measuring dual optical traps and a custom-built multi-channel microfluidic flow cell (Supplementary Fig. S1)11-13. This instrument enabled us to control and trigger biochemical reactions while mechanically manipulating individual DNA molecules. At the same time, it allowed us to image and quantify the fluorescence from functional human RAD51 variants with a single surface-exposed cysteine, labeled with Alexa Fluor 5554,14.
Figure 1a depicts our experimental assay, in which we moved single Ca2+-stabilized RAD51-dsDNA complexes4,15,16 to a Mg2+-containing buffer by swiftly shifting the microscope stage between parallel flow channels. This buffer exchange activates ATP hydrolysis. Figure 1b shows a kymograph4 of fluorescently labeled RAD51 polymerized onto a dsDNA molecule, held from one side by a single optically trapped bead and stretched by buffer flow (Supplementary Video 1). The triggered ATP hydrolysis results in filament disassembly, evidenced by a steady decrease of intensity and a marked shrinkage of the complex. This shrinkage, caused by relief of RAD51-induced DNA extension, immediately excludes photobleaching as the cause of intensity decrease4. Some patches appear to shrink from their ends (e.g. the one marked with an asterisk, Fig. 1b), suggesting that disassembly occurs from filament ends, as reported for RecA17-19. With a more sophisticated analysis, we will address this question in more detail below.
Figure 1. Assay for triggering of RAD51 disassembly.
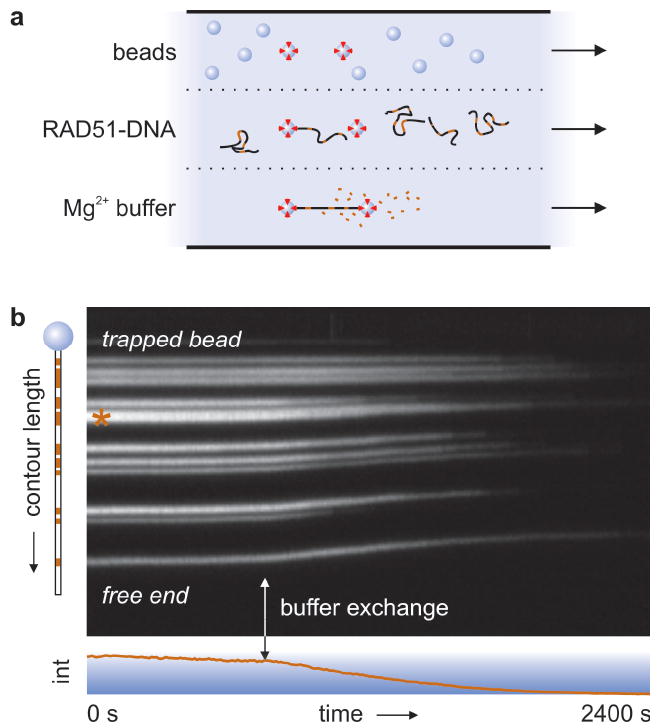
a, Schematic of the multi-channel flow cell. After capturing RAD51-bound dsDNA molecules by one or both ends using optical traps, ATP hydrolysis is triggered by moving the complex to a Mg2+-containing channel, setting off disassembly. b, Kymograph of a RAD51-coated dsDNA molecule held from one end, stretched by flow. RAD51 patches are marked red in the cartoon on the left. Mg2+-induced filament disassembly (vertical arrow) is evidenced by simultaneous DNA contraction and a steady decrease in fluorescence (bottom graph).
RAD51 forms helical filaments on dsDNA that extend the DNA by about 50% compared to B-form4,7,10,14. Could the tension thus stored be a driving force for the disassembly process20? To test this hypothesis, we captured RAD51–DNA complexes from both ends between two optically trapped beads4,13. Figure 2a shows the time course of fluorescence intensity and tension for a complex undergoing disassembly while being held at fixed end-to-end distance (Supplementary Video 2). Due to the shrinking contour length, the DNA pulls itself taut, after which tension gradually builds up. We observed that disassembly slowed down with increasing tension and even stalled at a tension of 48 ± 3 pN (s.e.m.; n = 7). To test that this slowing down is an actual characteristic of RAD51 and not due to the decreasing number of monomers left to dissociate, we examined the effect of a sudden tension release, induced by instantaneously moving the optical traps closer together. Indeed, Fig. 2b shows that disassembly immediately reinitiates after tension release, confirming the stabilizing effect of tension on RAD51 filaments.
Figure 2. RAD51 disassembly rate is reversibly reduced by DNA tension.
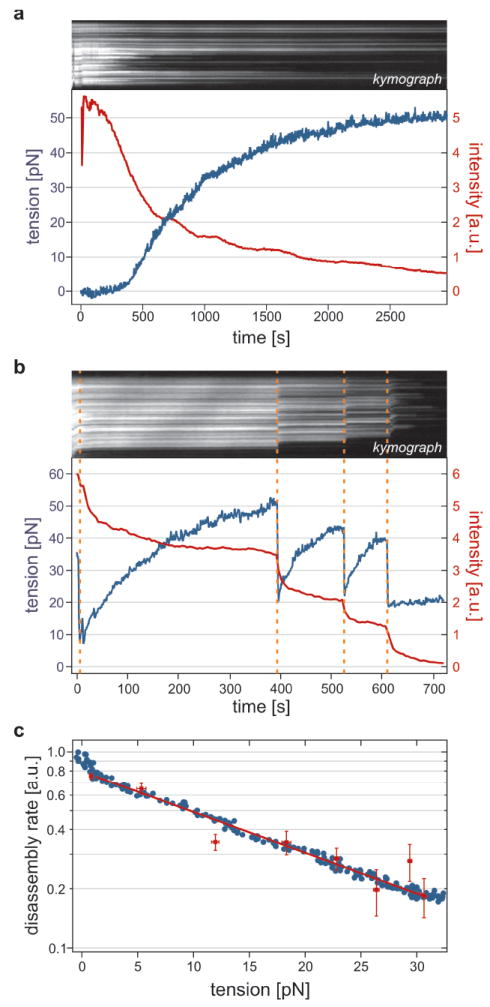
a, Kymograph and intensity trace (red) of a RAD51–dsDNA complex, held at fixed length, triggered to disassemble at t = 0. Tension (blue) increases due to disassembly-induced DNA contraction, leveling off to stall around 50 pN. Intensity decrease slows down accordingly. b, Tension-stalled disassembly is reinitiated by tension release (orange dashes). c, Disassembly rates decrease exponentially with tension. Directly differentiated intensity trace (red symbols) and smoothed trace (blue) are fitted by Arrhenius’ equation, yielding the same x†-value (0.20 ± 0.01 nm for this complex). See Supplementary Information for calculation.
Figure 2c shows how the disassembly rate, calculated as the time derivative of the fluorescence intensity, decreased with tension. Apparently, the energy barrier of disassembly is raised by a tension increase, stabilizing the RAD51-bound state. The rate decrease is well fit by a single exponential, suggesting a dependence according to Arrhenius’ law: k(F) = k(0) exp[-x†F / kBT], where kBT is the thermal energy, F the tension and x† the distance to the transition state along the relevant reaction coordinate21. This transition state is intermediate between the initial state with RAD51 bound to extended DNA, and the final one with relaxed DNA without RAD51. We determined x† to be 0.27 ± 0.04 nm (s.e.m., n = 9). One RAD51 monomer covers 3 basepairs and holds them in an extended conformation of about 1.5 nm in length (compared to 1 nm in canonical B-form DNA)4,7,10,16. Therefore, upon disassembly of a single monomer the DNA shrinks at most half a nanometer. Hence, our value for the location of the transition state, x†, is consistent with filaments disassembling one monomer at a time.
Can we demonstrate more directly that RAD51 filaments disassemble as monomers from filament ends and can we extract kinetic rates? To address these questions, we calibrated the fluorescence intensity to numbers of RAD51 monomers using single-molecule photobleaching steps in Ca2+-stabilized, optically trapped RAD51–DNA complexes (Supplementary Figs. S2 and S3). Figure 3a shows a kymograph and corresponding disassembly traces of four isolated RAD51 patches. A striking feature emerges: the intensity decrease is not continuous, but occurs in bursts of varying size, interspersed with pauses on the order of minutes. We fitted many such isolated disassembling filaments using a step fitting algorithm (blue lines in Fig. 3a; see Supplementary Fig. S2)22. Using the fitted steps, the kinetics of the disassembly can be analyzed from distributions of pause durations and burst sizes (Fig. 3b and Supplementary Figs. S4 and S6). Pause durations are exponentially distributed with a time constant of 152 ± 9 s, suggesting that the pauses were caused by a single Poisson waiting step in the reaction.
Figure 3. RAD51 disassembly occurs in bursts interspersed with pauses.
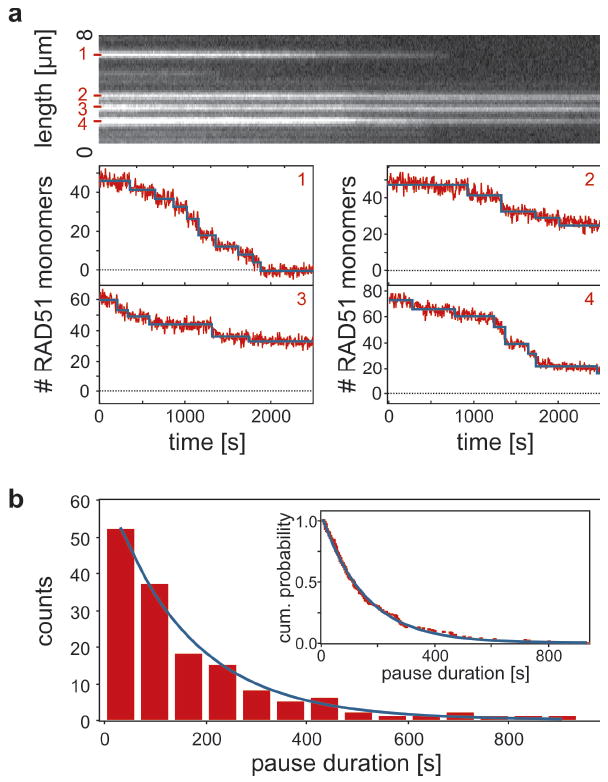
a, Calibrated intensity traces of isolated, short RAD51 patches show a burst pattern of disassembly activity interspersed by pauses on the order of minutes. These staircases are well fitted by a step fitting routine (blue lines)22. b, Pause durations are exponentially distributed, both when binned into a histogram (decay constant 152 ± 9 s) and when plotted as a cumulative probability distribution (inset, decay constant 152 ± 1 s).
This burst-wise disassembly can be understood with a model in which monomers dissociate exclusively from filament ends after ATP hydrolysis (graphically depicted in Fig. 4a). Assuming ATP hydrolysis to take place uniformly along the nucleoprotein filament (like RecA23), we interpret pauses as events in which filament disassembly transiently halts because the terminal monomer has ATP bound. Once that ATP is hydrolyzed, the terminal monomer loses contact with the DNA and dissociates, as do the neighbors that have already hydrolyzed their ATP. This burst of disassembly stops once an ATP-bound monomer is encountered. An inference of this model is that the ATPase rate of RAD51 bound to dsDNA is the reciprocal of the average pause duration. To accurately determine this rate we need to take into account that the fit residuals in the pause plateaus show a small but nonzero average slope (-0.01 monomers/s). This indicates that small steps (1–3 monomers) are hidden in the noise in our single-patch intensity traces (Fig. 3a) under the applied illumination conditions. From this we could determine that on average one short disassembly event per fitted pause was not detected and that we thus overestimated the average pause duration by a factor 2. Taking this into account, we determined that the ATP hydrolysis rate, khydr, is 0.6–1.3 · 10-2 s-1. This value is similar to that measured with bulk chemical kinetics assays24, which confirms that the observed pauses are governed by ATP hydrolysis. Interestingly, burst-wise disassembly was still observed in the presence of 2 mM ATP (khydr = 1.0 · 10-2 s-1, Supplementary Fig. S4). This implies that ATP renewal along the filaments takes place at a significantly slower rate than hydrolysis, if occurring at all, suggesting that ADP release is slower than disassembly. This is consistent with ATPase assays that showed that ADP release is the rate-limiting step in the ATPase cycle24-26. Our model also predicts the shape of the intensity curves such as shown in Fig. 2a (see Supplementary Fig. S5), yielding an estimate for the average filament length of 10–50 monomers, in agreement with earlier results16. Moreover, we can predict that on average 5–10 monomers are involved in a burst, which is in agreement with our measurements (cf. the total numbers of bursts in Figure 3a and Supplementary Fig. S6).
Figure 4. RAD51 disassembly pathway.
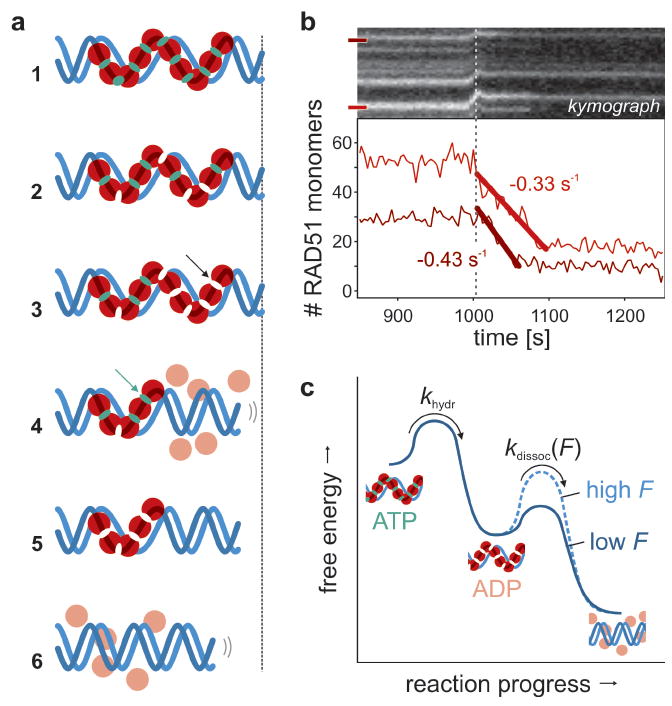
a, [1] All RAD51 (red) starts out ATP (green) and DNA (blue) bound. [2] ATP hydrolysis is triggered; filaments remain stable as long as terminal RAD51s have ATP bound. [3] ATP hydrolysis at terminal monomer. [4] Monomers dissociate until next ATP-bound monomer is terminal (arrow). [5] Disassembly pauses until terminal ATP hydrolyzes. [6] Disassembly relaxes DNA. b, Stalled disassembly reinitiated by tension release (dotted line). The slopes of the prolonged bursts yield the intrinsic dissociation rate. c, Free energy diagram of RAD51 disassembly. After ATP hydrolysis by a terminal monomer (with tension-independent rate khydr), ADP-bound monomers dissociate with rate kdissoc(F) that exponentially decreases with tension. Tension either increases the energy of the DNA-bound ADP-state, or the energy barrier to the dissociated state (as depicted).
In our disassembly model, ATP hydrolysis precedes dissociation of monomers. Which of these two causes the reaction to stall, when we apply tension to the DNA (Fig. 2)? In case only dissociation would depend on tension, ATP hydrolysis would proceed even while disassembly is stalled with high tension. When such a stalled complex is relaxed, an extended disassembly burst without pauses is expected. Indeed, Fig. 4b shows that isolated stalled patches exhibit such extended disassembly, confirming that ATP hydrolysis continues even at high DNA tension. Apparently, DNA tension changes the rate-limiting reaction step: ATP hydrolysis on filament ends at no tension and monomer dissociation at high tension. The extended bursts in Fig. 4b allowed us to directly determine the intrinsic dissociation rate of RAD51 monomers at low tension from the slope of the intensity decrease: kdissoc = 0.51 ± 0.14 s-1 (s.e.m., n = 5). Figure 4c summarizes the complete mechanokinetic model for ATP hydrolysis and tension-dependent RAD51 disassembly in a free-energy diagram. The extension of DNA that RAD51 imposes acts as a loaded spring that, in part, drives the disassembly reaction. For this to occur, the RAD51–DNA complex must shorten by x† in order to reach the transition state through thermal fluctuations. The amplitudes of thermal fluctuations shortening the DNA are reduced by external tension, kinetically disfavoring disassembly. In contrast, ATP hydrolysis seems to be independent of DNA tension, suggesting that hydrolysis does not alter the extended conformation of the DNA.
Our data and model illustrate how RAD51 filament stability crucially depends on the nucleotide state of the filament terminus. When the terminal RAD51 is in the ATP-bound state, the filament end is stably attached to the DNA. After hydrolysis, the terminal RAD51 loses its DNA affinity and detaches. Within a filament, RAD51 monomers appear to be locked on the DNA by their neighbors, independent of nucleotide state. Tight coupling between the nucleotide state, interactions between recombinase monomers and DNA binding have been observed in RecA-DNA co-crystals27. Notably, with our data and model it now becomes possible to shed new light on the way accessory proteins can catalyze filament disassembly. First, our data show that RAD51 nucleoprotein filament ends dominate the disassembly process, clarifying why proteins such as RAD54 interact with filament termini to stimulate disassembly28. If RAD54 destabilizes the interaction of the terminal RAD51 with DNA, this would result in accelerated disassembly by decreasing the duration of pauses that we show to occur during unaided RAD51 disassembly. Second, it explains why stable RAD51 filaments in the presence of ADP can only form when there is at least some ATP present29: the end caps need to contain ATP to stabilize a filament. This, in turn, clarifies why the rate of RAD54-assisted filament disassembly is higher in the presence of ADP30. These ADP-containing filaments would give rise to longer disassembly bursts and RAD54 would less often need to remove ATP-stabilized end caps. To conclude, concurrent visualization, quantification, manipulation and triggering of the dynamics of RAD51 filaments provided a comprehensive understanding of spontaneous RAD51 nucleoprotein filament disassembly, which in turn allowed new insights in the disassembly assisted by accessory proteins. We expect that integrative single-molecule approaches, such as those used here, will also be of great value in dissecting many other complex biological reactions.
Methods Summary
Biotinylated λ-DNA and fluorescently labeled human RAD51 (isoform Q313) were prepared as described elsewhere12-14. RAD51-dsDNA nucleoproteins were pre-assembled in Ca2+-stabilized conditions as used before4,14. ATP hydrolysis was triggered in the flow cell by exposing the trapped RAD51-dsDNA complex to a buffer containing 10 mM Mg2+ and 10 mM EGTA.
Descriptions of the combined dual optical tweezers and fluorescence microscope4 as well as data analysis procedures are provided in the Supplementary Information and Supplementary Figs. S1–S3.
Supplementary Material
Acknowledgments
The authors thank Bram van den Broek and Remus Dame for discussions and a critical reading of the manuscript and Jacob Kerssemakers for kindly providing his step fitting algorithm. This work was supported by the Biomolecular Physics program of the Dutch organization for Fundamental Research of Matter (FOM) (RK, CW, EJGP and GJLW), and grants from the Dutch Cancer Society (KWF), the Netherlands Organization for Scientific Research (NWO), the Netherlands Genomics Initiative/NWO, the Association for International Cancer Research and the European Commission Integrated Projects Molecular Imaging and DNA Repair and a National Cancer Institute-NIH USA program project (CW and RK). EJGP, GJLW and CW are recipients of Vidi grants and a Vici grant from NWO, respectively.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 2.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 Recombinase and Recombination Mediators. J Biol Chem. 2003;278(44):42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 3.Symington LS, Heyer WD. Some disassembly required: role of DNA translocases in the disruption of recombination intermediates and dead-end complexes. Genes Dev. 2006;20(18):2479–2486. doi: 10.1101/gad.1477106. [DOI] [PubMed] [Google Scholar]
- 4.van Mameren J, et al. Dissecting elastic heterogeneity along DNA molecules coated partly with Rad51 using concurrent fluorescence microscopy and optical tweezers. Biophys J. 2006;91(8):L78–L80. doi: 10.1529/biophysj.106.089466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brau RR, et al. Interlaced optical force-fluorescence measurements for single molecule biophysics. Biophys J. 2006;91(3):1069–1077. doi: 10.1529/biophysj.106.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kowalczykowski SC, Eggleston AK. Homologous Pairing and DNA Strand-Exchange Proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 7.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13(23):5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi P, et al. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair. 2006;5(3):381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Hegner M, Smith SB, Bustamante C. Polymerization and mechanical properties of single RecA-DNA filaments. Proc Natl Acad Sci USA. 1999;96(18):10109–10114. doi: 10.1073/pnas.96.18.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ristic D, et al. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33(10):3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443(7113):875. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 12.Noom MC, van den Broek B, van Mameren J, Wuite GJL. Visualizing single DNA-bound proteins using DNA as a scanning probe. Nat Methods. 2007;4(12):1031–1036. doi: 10.1038/nmeth1126. [DOI] [PubMed] [Google Scholar]
- 13.van den Broek B, Noom MC, Wuite GJ. DNA-tension dependence of restriction enzyme activity reveals mechanochemical properties of the reaction pathway. Nucleic Acids Res. 2005;33(8):2676–2684. doi: 10.1093/nar/gki565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Modesti M, et al. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15(5):599. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci USA. 2004;101(27):9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Heijden T, et al. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35(17):5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindsley JE, Cox MM. Assembly and disassembly of RecA protein filaments occur at opposite filament ends. Relationship to DNA strand exchange. J Biol Chem. 1990;265(16):9043–9054. [PubMed] [Google Scholar]
- 18.Arenson TA, Tsodikov OV, Cox MM. Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J Mol Biol. 1999;288(3):391–401. doi: 10.1006/jmbi.1999.2705. [DOI] [PubMed] [Google Scholar]
- 19.Joo C, et al. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126(3):515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 20.Wyman C. Monomer networking activates recombinases. Structure. 2006;14(6):949–950. doi: 10.1016/j.str.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Evans E. Probing the relation between force - lifetime - and chemistry in single molecular bonds. Annu Rev Biophys Biomol Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 22.Kerssemakers JWJ, et al. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442(7103):709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 23.Brenner SL, et al. RecA protein-promoted ATP hydrolysis occurs throughout recA nucleoprotein filaments. J Biol Chem. 1987;262(9):4011–4016. [PubMed] [Google Scholar]
- 24.Tombline G, Fishel R. Biochemical characterization of the human RAD51 protein. I. ATP hydrolysis. J Biol Chem. 2002;277(17):14417–14425. doi: 10.1074/jbc.M109915200. [DOI] [PubMed] [Google Scholar]
- 25.Tombline G, Shim KS, Fishel R. Biochemical characterization of the human RAD51 protein. II. Adenosine nucleotide binding and competition. J Biol Chem. 2002;277(17):14426–14433. doi: 10.1074/jbc.M109916200. [DOI] [PubMed] [Google Scholar]
- 26.Shim KS, et al. Magnesium influences the discrimination and release of ADP by human RAD51. DNA Repair. 2006;5(6):704–717. doi: 10.1016/j.dnarep.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Chen Zhucheng, Yang Haijuan, Pavletich Nikola P. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453(7194):489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 28.Kiianitsa K, Solinger JA, Heyer WD. Terminal association of Rad54 protein with the Rad51-dsDNA filament. Proc Natl Acad Sci USA. 2006;103(26):9767–9772. doi: 10.1073/pnas.0604240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaitseva EM, Zaitsev EN, Kowalczykowski SC. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1999;274(5):2907–2915. doi: 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
- 30.Li X, et al. Rad51 and Rad54 ATPase activities are both required to modulate Rad51-dsDNA filament dynamics. Nucleic Acids Res. 2007;35(12):4124–4140. doi: 10.1093/nar/gkm412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


