Abstract
The breakdown of organic nitrogen in soil is a potential rate-limiting step in nitrogen cycling. Arbuscular mycorrhizal (AM) fungi are root symbionts that might improve the ability of plants to compete for organic nitrogen products against other decomposer microbes. However, AM uptake of organic nitrogen, especially in natural systems, has traditionally been difficult to test. We developed a novel quantitative nanotechnological technique to determine in situ that organic nitrogen uptake by AM fungi can occur to a greater extent than has previously been assumed. Specifically, we found that AM fungi acquired recalcitrant and labile forms of organic nitrogen. Moreover, N enrichment of soil reduced plot-scale uptake of these compounds. Since most plants host AM fungi, AM use of organic nitrogen could widely influence plant productivity, especially where N availability is relatively low.
Keywords: Arbuscular mycorrhizal fungi, Boreal forest, Chitosan, Emission fingerprinting, Glycine, Organic nitrogen, Quantitative confocal microscopy, Quantum dots, Raster image correlation spectroscopy
1. Introduction
Arbuscular mycorrhizal fungi are common in most terrestrial ecosystems and form relationships with the majority of plant families (Newman and Reddell, 1987; Smith and Read, 2008; Treseder and Cross, 2006). Specifically, they colonize the interiors of plant roots and transfer nutrients to their host plants in exchange for photosynthate-C. Arbuscular mycorrhizal fungi have traditionally been thought to specialize primarily on inorganic nutrients (Smith and Read, 2008). Recent microcosm studies have challenged this notion by demonstrating that AM fungi can access N derived from organic nitrogen sources (Hodge et al., 2001; Whiteside et al., 2009; Hodge and Fitter, 2010). Nevertheless, evidence of direct uptake of organic nitrogen by AM fungi remains sparse. In addition, in the field, Näsholm et al. (1998) demonstrated that AM host plants can take up amino acids from boreal forest soil. However, they could not determine whether AM fungi contributed to the uptake (Näsholm et al., 1998).
Indeed, microbial processes in natural systems have traditionally been difficult to study. For example, it has been challenging to follow the fate of organically-derived nutrients from the soil to mycorrhizal fungi. One approach has been to apply isotopically-labeled compounds to soil as nutrient tracers (e.g., Näsholm et al., 1998). However, under field conditions it is extremely difficult to determine the contribution of mycorrhizal fungi versus direct uptake by plant roots. In a recent greenhouse based study to test the uptake of organically-derived N by AM fungi, Hodge and Fitter (2010) applied 15N labeled plant litter to split microcosms where AM extramatrical hyphae were in separate compartments from their plant hosts. The authors found that as much as 31% of the N obtained by compartmented AM hyphae was derived from decomposing organic matter, but it is unclear whether the organic material was broken down by AM fungi or saprotrophs present in the soil. In natural soils, it may be difficult to reliably remove mycorrhizal fungi from soil patches while leaving the remainder of the soil community intact. Hobbie and Hobbie (2006) were among the first investigators to estimate uptake of N by mycorrhizal fungi in the field; they used natural abundance of 15N in plants and fungal fruit bodies to approximate the contribution of ectomycorrhizal fungi to plant N and found that ectomycorrhizal fungi are responsible for 61–86% of N uptake by plants in arctic tundra. We developed an additional approach toward estimating the contribution of arbuscular mycorrhizal fungi (and microbes in general) to biogeochemical processes such as N-cycling: quantum dots (QDs).
Quantum dots are semiconducting nanoparticles that are often composed of cadmium selenide or cadmium telluride cores, encased in zinc sulfide shells (Peng and Peng, 2000; Lin et al., 2004). When exposed to UV light, QDs fluoresce in bright, pure, and precise colors of almost any wavelength of visible light (Bruchez et al., 1998; Chan and Nie, 1998; Bailey and Nie, 2003). They can be covered with protective polymer coatings and embedded with receptor molecules (often carboxyl or amino groups) that allow the QD to be conjugated with other compounds, including antibodies. In this way, QDs can be used to track the movement of diverse compounds at minute spatial scales within microbes and soil. In a recent laboratory study, Whiteside et al. (2009) completed a qualitative proof-of-method that demonstrates fungi and plants actively take up and translocate QD-labeled organic nitrogen. Here, we developed a quantitative method (raster image correlation spectroscopy, RICS) for measuring the amount of QDs present within a sample. This advance allows QDs to be used in ways similar to quantitative isotopic techniques, such as in tests for environmental controls over nutrient fluxes.
We applied this quantitative QD technique in a boreal forest with low baseline N availability to test hypotheses related to organic nitrogen acquisition by AM fungi in response to N addition. This is the first in situ use of QDs to test an ecological question. Specifically, wehypothesized that specific uptake of organically-derived N by AM fungi will decline with increasing N availability. As with all enzymes, those required for acquisition of organic nitrogen require N and C to construct. Thus, AM fungi could allocate resources more efficiently by investing more in organic nitrogen uptake when N is relatively unavailable (Allison et al., 2010). Consequently, AM use of organic nitrogen per unit biomass (i.e., specific uptake) should decrease under N enrichment. In addition, we hypothesized that plants should reduce their investment in AM fungi if N availability increases, because mycorrhizal relationships would become less cost-effective. As a result, AM abundance should decline, as has often been observed in other ecosystems (Treseder, 2004). A decrease in specific uptake rates of organic nitrogen by AM fungi and/or reductions in AM biomass could ultimately lead to declines in organic nitrogen uptake via AM fungi per m2 of soil.
We used glycine—the smallest of the amino acids—for labile organic nitrogen and chitosan for recalcitrant organic nitrogen. For our purposes, we define recalcitrant organic nitrogen as polymeric, structurally complex, relatively large organic nitrogen compounds. Chitosan is deacetylated chitin; it is larger (average molar weight: 1526 g mol−1) and more complex (Bertz complexity index: 2630) than glycine (molar weight: 75.06 g mol−1, complexity: 42.9). Generally, larger and more complex compounds turn over more slowly (Muir and Howard, 2006). Chitosan is thought to degrade on the same timescale as chitin, which has residence times of approximately one year in the environment (Gooday, 1990). Chitosan is relatively abundant in the biosphere. It is directly produced by zygomycete and ascomycete fungi as well as arthropods (Gooday, 1994b). In addition, chitosan is formed as one of the first steps of chitin degradation, and chitin is one of the most abundant polymers on Earth, with annual production rates of 1013–1014 kg (Gooday, 1990). A number of studies have demonstrated that ectomycorrhizal and ericoid fungi can use chitin (or the products of chitin breakdown catalyzed by chitinases) as a sole C or N source when they are grown in pure culture (Bajwa and Read, 1986; Leake and Read, 1990; Kerley and Read, 1995; Read and Perez-Moreno, 2003).
2. Methods
2.1. Sites
Our study site was an upland boreal forest near Delta Junction, Alaska (63°55′ N, 145°44′ W). It had burned in a severe fire in 1999. At the time of sample collection, an AM graminoid (Festuca altaica Trin.), an AM forb (Epilobium angustifolium Lam.), ericoid evergreen shrubs (Ledum palustre L. and Vaccinium vitisidaea L.), and an ericoid deciduous shrub (Vaccinium uliginosum L.) were common (Mack et al., 2008; Treseder et al., 2004). Populus tremuloides Michx. and species of Salix were also abundant; they are deciduous and can each be colonized by AM as well as ectomycorrhizal fungi. Approximately 80% of annual NPP was contributed by plants that form relationships with AM fungi only, and 9% by those that can host both AM and ectomycorrhizal fungi (Mack et al., 2008). Permafrost is discontinuous in this area and was not present in the site. The local climate was cold and dry, with a mean annual temperature of −2 °C and a precipitation rate of 303 mm y−1 (http://weather.noaa.gov/). The site consisted of a block design described in Treseder et al. (2007). Each block contained an N-fertilized plot and a control plot. The N-fertilized plots were originally dosed with 200 kg N ha−1 y−1 in June 2002, and thereafter received 100 kg N ha−1 y−1 in June of each year. We sampled in three of these blocks (n = 3), chosen at random. Our replicate number was limited owing to the immediate availability of QDs at the field site.
2.2. Preparation of quantum dots
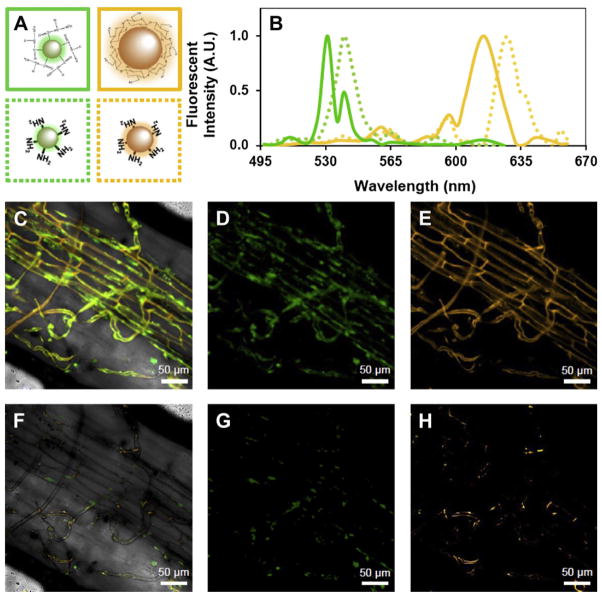
To conjugate QDs to organic nitrogen substrates, we used the same methods and concentrations as described by Whiteside et al. (2009). Briefly, commercial green (535 nm emission) and orange (620 nm emission) carboxyl QDs were purchased from Vive Nano (Ontario, Canada). Green QDs were bound to the amino groups of glycine (MP Biomedical, Solon, Ohio, USA), and orange QDs were bound to the amino groups of chitosan (Fig. 1; MP Biomedical). Quantum dot size (i.e., the average hydrodynamic diameter) was estimated using RICS and the Stokes–Einstein equation (Digman et al., 2005; Uhlenbeck and Ornstein, 1930). Diameters of the green QDs averaged ~3 nm; orange QDs ~8 nm. Approximately five glycine molecules were conjugated to each green QD, and one chitosan molecule to each orange QD.
Fig. 1.
Emission spectra of QD-glycine and QD-chitosan within intraradical AM hyphae. (A) Conceptual diagram of QD-glycine (solid green box), QD-chitosan (solid orange box), and amino terminated QDs representing the N mineralized form of each (dashed boxes of corresponding color). (B) Reference spectra of QD-glycine (solid green line); “N mineralized” QD-glycine (dotted green line); QD-chitosan (solid orange line); and “N mineralized” QD-chitosan (dotted orange line). For both QD-glycine and QD-chitosan, substrate mineralization resulted in a slight red shift in the emission spectra. (C) Superimposed laser scans of a plant root (from an N-fertilized plot) colonized by AM hyphae: where (D) green represents intact QD-glycine and (E) orange represents intact QD-chitosan. (F) Superimposed scans of “N mineralized” QD-glycine (G) and mineralized (H). Sample was taken from an N fertilized plot. Emission fingerprints were scanned from 494 to 660 nm (4.74 nm increments) using a Zeiss 710 (Quasar spectral detection unit) confocal microscope, on a 40× oil objective. Scale bars are 50 μm. Conceptual diagram (A) is not drawn to scale. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Amino terminated QDs were prepared in the same manner, except using low molecular weight poly(allylamine hydrochloride) in place of glycine or chitosan (Jin and Gao, 2009). These QDs were solely used to represent mineralized forms of QD-glycine and QD-chitosan during spectral analysis (see below). They were not included as field injections.
2.3. Field incubations
To assess uptake of organic nitrogen by AM fungi in the field, on September 9, 2008 we injected two QD cocktails into each of the N fertilized and control plots (Table S1). The first cocktail contained an equal (0.1 μM) mixture of green-labeled glycine and orange-labeled chitosan. The second cocktail contained an equal mixture of orange and green QD controls, which consisted of unbound QDs subjected to the same conditions as the labeled conjugates, but lacking the binding reagent. Twenty ml of each (0.1 μM) QD cocktail were injected ~1 cm into the soil. Each plot received either two injections of QD controls plus two injections of glycine-bound QDs mixed with chitosan-bound QDs, or three injections of each. Twenty-four hours following injection, 2 cm diameter × 10 cm deep soil cores were taken from each injection point. In addition, in each plot, one “uninjected” core was collected from a random area of no injection, for a total of 34 cores. QD-glycine and QD-chitosan were stored separately and mixed immediately before injection. The soil corer was cleaned with sodium hypochlorite between samples. Cores were initially frozen at −4 °C and stored at −80 °C.
2.4. Arbuscular mycorrhizal colonization of roots
We used wet sieving through 1 mm mesh to extract all fine (<2 mm diameter) roots from each core. Standard techniques were used on a subset of randomly-selected 1 cm root segments to determine percent root length colonized by intraradical AM hyphae (Koske and Gemma, 1989; McGonigle et al., 1990). Colonization by AM fungi was assessed based on the presence of arbuscules and of characteristic intraradical hyphae with irregular walls, angular branching, and lack of septa (Bonfante-Fasolo, 1986). All calculations were performed per core and averaged per plot.
2.5. Uptake of organic nitrogen by AM fungi in the field
To quantify QDs in AM fungi, ten fine root segments were chosen at random and placed on a microscope slide with 250 μL of bicarbonate buffer (pH 7.4–8.0) and a coverslip. These were not the same segments that were stained to quantify AM abundance. We used the same morphological criteria to identify intraradical AM hyphae here as for the stained samples (above).
Standard features of confocal microscopy allowed us to confirm that we were only quantifying QD-glycine or QD-chitosan within the interior of fungal cells (i.e., not attached externally to cell walls). Confocal microscopes eliminate light signals from all but the precise focal plane of interest. Thus, we could move the focal plane of the image vertically through the fungal cell and ensure that we were examining space enveloped by, but not including, cell walls.
2.6. Quantification of specific uptake of QDs by AM fungi
We developed a RICS protocol to quantify the uptake of QDs by AM fungi. RICS can be used to calculate the concentrations of particular fluorophores (like QDs) within specimens, and to date it has primarily been used in cell biology (Digman et al., 2005; Rossow et al., 2010). In this procedure, any laser scanning microscope can be used to document the spatial distribution and diffusion of fluorophores in a sample. Generally, fluorophores with similar sizes and chemical compositions diffuse at similar rates within a cell. RICS analysis can be employed to identify individual fluorophore particles that diffuse at similar rates, and group them accordingly. Quantum dots, for example, are manufactured with relatively little variation in diameter and composition, so they tend to diffuse at comparable speeds. In contrast, the diffusion rates of QDs are often detectably different from other cellular constituents such as proteins. In this case, RICS can identify and count QD particles within mycorrhizal tissue because they diffuse at expected, consistent rates. At the same time, this method can be used to discount background fluorescence, because other fluorescent molecules will likely not diffuse at the same rate as QDs. In theory, RICS can detect as little as one fluorophore per biovolume sample. In practice, the RICS technique detected as few as four fluorescent QDs per biovolume sample in our study. This approach is broadly accessible to researchers, since RICS software can be used in conjunction with commercially available laser scanning microscopes (Rossow et al., 2010).
Raster image correlation spectroscopy analysis applies the principles of spatial autocorrelation to confocal raster-scan images to extract the movement of a particle from pixel to pixel across an image. These autocorrelations provide a statistical function that can be used to determine the diffusion and concentration of fluorescent particles. In order to correlate molecules that are diffusing along adjacent pixels via Brownian motion, the scanned pixel size must be four to five times smaller than the optical focal volume characterized by the confocal system (Rossow et al., 2010). Likewise, the speed at which the laser dwells in each pixel location during the raster scan (image acquisition) must be faster than the movement of the molecule across the volume of excitation. The pixel dwell time τp was selected according to Eq. (1):
| (1) |
where ω0 is the radial waist of the focal spot and D is the diffusion coefficient of the particles. Images were scanned at 33.8× zoom yielding an individual pixel size of 20 nm. We used the approximate diffusion rates of QD-chitosan and QD-glycine suspended in borate buffer to select an optimal pixel dwell time of 25.21 μs pixel−1; yielding a total line speed of 6.454 ms (in the x direction). Preliminary trials confirmed that these parameter settings could detect QDs bound to both glycine and chitosan.
To remove background due to immobilized structures in the sample, including intracellular autofluorescence, we performed moving average background subtractions before calculating image autocorrelations (Rossow et al., 2010). This procedure, which in effect acts as a high pass filter, also compensates for motion of the sample in times longer than the frame time (2 s). We used a high pass filter setting of 4 (corresponding to 4 frames as the moving average). We found that the amount of immobilized QD was negligible.
After background subtraction, image autocorrelation functions were calculated as:
| (2) |
where i(x, y) is the intensity at each pixel of the image, ξ and ψ are the x and y spatial correlation shifts, δ(i) = i − 〈i〉 is the intensity fluctuation and 〈…〉x,y indicates that the values are averaged over the image (Rossow et al., 2010).
In order to extract the number of QD particles (P) in the volume of excitation we fitted the autocorrelation function (Eq. (2)) to the RICS equations for a model of particles diffusing in 3 dimensions (Rossow et al., 2010):
| (3) |
S(ξ, ψ) describes the spatial part of the autocorrelation function:
| (4) |
and G(ξ, ψ) describes the temporal part of the correlation function:
| (5) |
where γ is a numerical factor (=0.3536), D is the diffusion coefficient, τp is the pixel dwell time, τl is the time between lines, δr is the distance between pixels, and ω0 is the beam waist of excitation in the radial direction and ωz in the axial direction. The RICS auto-correlation function was calculated according to Eq. (2) and fitted using Eq. (5) to extract the number of particles (P) in the excitation volume. Raster images were analyzed using Zen 2009 software (Carl Zeiss, Jena, Germany), and SimFCS was used for data fit (Laboratory for Fluorescence Dynamics, University of California, Irvine, USA).
2.7. Calculation of specific uptake of QDs by AM fungi
Twenty-five random AM intraradical hyphae were selected for each fine root slide, without regard for the presence or absence of QDs. Fifty images were collected per mycorrhizal location. Specifically, raster-scan images (256 × 256 pixels) were acquired using a Zeiss LSM 710 (Quasar spectral detection unit) confocal microscope equipped with a Pecon XL3 incubator to provide thermal stability to both the microscope and samples during time-lapse imaging (Carl Zeiss, Jena, Germany). A 40× water immersion objective was used for all samples. QDs were excited with an Argon ion laser (488 nm) at 0.5% power. For the best signal to noise ratio, QDs were detected at 535 ± 4 nm and 620 ± 4 nm at 1 airy unit (AU). For the wavelength used, 0.300 μm was obtained as the radial waist of the excitation beam (ωo) for our system. Finally, RICS analysis was used to extract the number of particles (P) in the excitation volume. Specific uptake of organic nitrogen (QD particles absorbed μm−3 tissue AM) was estimated from the number of QD particles present per biovolume tissue visible in each confocal frame. Arbuscular mycorrhizal fungi showed no uptake of QD controls.
2.8. Calculation of plot-level QD uptake by AM fungi
Plot-level uptake (nmol QD m−2 ground area) of QD-organic nitrogen was calculated as the product of standing AM biomass and specific uptake by intraradical AM hyphae. For intraradical AM hyphae, we calculated biovolumes of hyphae and then assumed a fresh tissue density of 1.1 g cm−3 and a dry weight content of 40% (Paul and Clark, 1996). The biovolume of AM hyphae within roots (VAM) was approximated as , where rr is fine root radius; LAM, AM root length; and KAM, the fraction of colonized root volume that is fungal (Toth et al., 1991). We assumed a value of 0.06 for KAM (Toth et al., 1991).
To determine the radius of fine roots in the study plots (rr), we extracted all fine roots from each soil core. Roots were extracted by hand-sieving, washed three times in deionized water, and digitally scanned. The radius of each root was measured manually with WinRHIZO Tron MF software (Regent_Instruments, 2003).
To calculate AM root length (LAM), we took the product of percent root length colonized by intraradical AM hyphae and standing fine root length. Standing fine root length was measured on September 9, 2008 by using minirhizotrons exactly as described in Treseder et al. (2007), and by assuming that the viewing depth of the minirhizotron microscope was 2 mm (The same plots and tubes were used in this study as in that of Treseder et al., 2007.) All calculations (for root and QD data) were performed separately for each core and averaged per plot.
2.9. Mineralization of QD-organic nitrogen
We used an emission fingerprinting technique to determine the extent to which chitosan and glycine were degraded before being accessed by AM fungi. Emission fingerprinting is a method of analyzing multiple fluorescent molecules which may have similar or even widely overlapping emission spectra (Zimmermann et al., 2003). Because emission fingerprinting identifies individual molecules by their specific spectral profile rather than fluorescence intensity alone, it can be used to analyze fluorescent particles with varying amounts of bound substrates (Lin et al., 2004). We created reference spectra for QD-glycine, QD-chitosan, and the mineralized forms of each (represented by N-terminated green and orange QDs; Fig. 1A and B). Emission spectra were used to separate mineralized from non-mineralized substrates. Spectral analyses were performed using Zen 2009 software (Carl Zeiss, Jena, Germany; Fig. 1).
To estimate the extent that QD-organic nitrogen was mineralized before uptake by AM fungi, fluorescence intensity profiles were analyzed in eight samples chosen at random, of which half were selected from N enriched plots, and half from controls; with an equal number of QD-glycine and QD-chitosan per treatment. These samples were the same as those obtained from the field incubation for quantification of QD-organic nitrogen uptake. Percent mineralization was calculated as the quotient of mineralized fluorescent intensity divided by mineralized + unmineralized fluorescence intensity multiplied by 100. Calculations were performed individually and averaged across all samples (n = 8).
2.10. Statistics
We performed analyses of variance (ANOVAs) to test for N effects (JMP, Version 7, SAS Institute Inc., Cary, NC, USA). Specifically, we used two-tailed ANOVAs with specific uptake of QD-glycine or QD-chitosan by AM fungi, AM biomass, or plot-level uptake of QD-glycine or QD-chitosan by AM fungi as dependent variables. Block effects were not significant, and were omitted from final analyses. Because we were unable to transform our data to meet assumptions of normality, we performed our analyses on ranked data. Differences were considered significant when P < 0.05. For all comparisons, n = 3, with plot as the unit of replication.
3. Results
Arbuscular mycorrhizal fungi took up QD-glycine and QD-chitosan (Fig. 1, Table 1). Uptake was documented in internal AM hyphae. Based on spectral analyses, we estimated that few organic nitrogen substrates were mineralized before being taken up by AM fungi (3.0 ± 1.0%; Fig. 1). Nitrogen additions significantly decreased AM specific uptake (QD particles μm−3 fungal tissue) of QD-glycine (P = 0.006) but not QD-chitosan (Table 1).
Table 1.
Organic N uptake by intraradical hyphae of arbuscular mycorrhizal fungi under N fertilization for the top 10 cm of soil (as means ± SEM, n = 3).
| Parameter | Control | N-fertilized | P-value |
|---|---|---|---|
| Specific uptake of chitosan (QDs μm−3 hyphae) | 2360 ± 750 | 1390 ± 740 | 0.524 |
| Specific uptake of glycine (QDs μm−3 hyphae) | 3260 ± 1010 | 805 ± 257 | 0.006 |
| Plot-level uptake of chitosan (nmol QDs m−2 soil) | 0.176 ± 0.056 | 0.140 ± 0.074 | 0.711 |
| Plot-level uptake of glycine (nmol QDs m−2 soil) | 0.243 ± 0.075 | 0.080 ± 0.026 | 0.001 |
The biomass of intraradical AM hyphae did not respond significantly to N fertilization (Table 2). Although standing root length was significantly higher in N-fertilized plots compared to control plots (P = 0.034), this increase was offset by a trend toward decreases in root diameter (Table 2). Moreover, percent root length colonized by AM fungi did not differ significantly among treatments (Table 2).
Table 2.
Root and arbuscular mycorrhizal (AM) parameters under N fertilization for the top 10 cm of soil (as means ± SEM, n = 3).
| Parameter | Control | N-fertilized | P-value |
|---|---|---|---|
| Standing root length (km m−2) | 7.1 ± 2.3 | 14.5 ± 1.1 | 0.034 |
| Root diameter (mm) | 0.048 ± 0.005 | 0.034 ± 0.007 | 0.148 |
| Root colonization by intraradical AM hyphae (% root length) | 27.3 ± 3.6 | 28.3 ± 2.0 | 0.832 |
| Biomass of intraradical AM hyphae (g m−2) | 0.088 ± 0.030 | 0.157 ± 0.097 | 0.533 |
At the plot-level, uptake of QD-glycine by AM fungi (nmol QD m−2 ground area) significantly declined under N fertilization (P = 0.001). In comparison, N fertilization had no significant effect on plot-scale QD-chitosan uptake by AM fungi (Table 1).
4. Discussion
It has traditionally been assumed that AM fungi do not exploit organic nitrogen—especially recalcitrant organic nitrogen—in soils (Smith and Read, 2008). Our findings of QD-chitosan and QD-glycine uptake by AM fungi in the field challenge this view and suggest that AM fungi may play a greater role in organic nitrogen use than previously thought. Nonetheless, our results are consistent with others who have recently observed that AM fungi can access (but not necessarily break down) N from organic material in laboratory settings (Hodge et al., 2001; Hodge and Fitter, 2010; Whiteside et al., 2009). In addition, Cappellazzo et al. (2008) documented gene expression of amino acid permeases in Glomus mosseae. Furthermore, AM species can produce chitinases, which are commonly used to defend against root pathogens (Azcón-Aguilar and Barea, 1997; Gianinazzi-Pearson, 1996). In a previous laboratory study, cultures of AM fungi were able to access QD-chitosan (Whiteside et al., 2009). Since AM fungi are associated with almost 75% of plant species worldwide (Newman and Reddell, 1987), this process may influence organic nitrogen cycling in many terrestrial ecosystems.
Arbuscular mycorrhizal fungi displayed significantly lower specific uptake rates (i.e., per unit biovolume) of labile organic nitrogen under N fertilization (Table 1). This result suggests that AM fungi adjust uptake rates of these substrates potentially to curtail costs of N acquisition. Since membrane transporters and extracellular enzymes can require up to 17% of C and 6% of N resources for microbes and constitute as much as 50% of cell membrane mass (Frankena et al., 1988; Gooday, 1994a), then microbes should allocate these resources elsewhere when N is abundant (Allison et al., 2010). This process could be regulated genetically. In a recent study, Cappellazzo et al. (2008) found that the AM fungus G. mosseae expressed fewer amino-acid transporters when inorganic N was abundant. It has not been previously tested under field conditions. Alternately, the shift in specific uptake could have resulted from any shifts in AM fungal composition under N fertilization. The decrease in specific uptake rates of labile organic nitrogen by AM fungi led to a decline in plot-scale uptake rates, even though AM abundance did not change significantly (Tables 1 and 2).
We restricted our analyses to comparisons of relative uptake values of organic nitrogen within QD size classes across N treatments. To analyze labile and recalcitrant organic nitrogen simultaneously required two colors—and therefore two sizes—of QDs. As a consequence, we could not compare labile organic nitrogen uptake to recalcitrant organic nitrogen uptake. This constraint need not influence future studies, however, since recently-developed QDs can emit light at different pulse rates even when similarly-sized (Xiao-Wei et al., 2010). Although QDs are nanoscale, with sizes comparable to those of small peptides (3–8 nm), we expect that their mass influenced movement of the QD-conjugated organic nitrogen to some extent. In addition, it is possible that the conjugation of organic nitrogen to QDs could have facilitated organic nitrogen uptake by AM fungi through an as-yet-unknown mechanism. In this case, uptake rates from this study may overestimate actual uptake rates of unconjugated glycine or chitosan.
Although a small portion (3.0 ± 1.0%) of the organic nitrogen compounds bound to QDs may have been mineralized by soil microbes prior to uptake by the AM fungi, these compounds likely contained at least one carboxyl group bound to one amino group. The QDs used in this study were wrapped in carboxyl terminating polymers. These carboxyl polymers were covalently bound to amino groups of glycine or chitosan. In addition, control QDs, which contained unbound carboxyl polymers, were not taken up detectably by AM fungi. These results suggest that if the amino group (and the remainder of the organic nitrogen molecule) were removed from the glycine- or chitosan-bound QDs, the QDs would probably not have been taken up by the AM fungi.
Our study is one of the first to confirm AM uptake of recalcitrant (i.e., relatively large and complex) organic nitrogen in situ. Classically, mineralization of organic nitrogen to ammonium-N or nitrate-N by microbes was thought to determine levels of N available for plant uptake (Aber and Melillo, 2001; Russell, 1912), and, ultimately, net primary productivity (NPP) (Reich et al., 1997). However, findings that plants can take up organic nitrogen such as amino acids (e.g., Näsholm et al., 1998) contributed to the development of an alternative paradigm in which the depolymerization of relatively large organic nitrogen compounds in the soil by extracellular enzymes is the major rate-limiting step in the N cycle (Chapin et al., 2002; Schimel and Bennett, 2004). Our observations support the mycorrhizal component of this alternate paradigm (Schimel and Bennett, 2004), and provide N conditions under which the paradigm may operate. For instance, AM uptake of organic nitrogen is apparent but less prevalent under N enrichment compared to baseline conditions. Our results suggest that shifts in specific uptake of organic nitrogen by AM fungi may be a mechanism that drives this pattern. Thus, if mycorrhizal fungi process significant amounts of organic nitrogen in ecosystems—as has been suggested (Lundeberg, 1970; Näsholm et al., 2009; Talbot et al., 2008)—this pathway could be a crucial step in plant N acquisition and ultimately productivity; increasingly so in areas that contain lower levels of available N. Nevertheless, further research is needed to assess how much of this N is transferred to plants. In addition, we did not address the extent to which AM fungi depolymerize proteins prior to uptake of their constituent amino acids.
We used RICS as a novel approach for QD quantification. We were able to quantify the number of QD-organic nitrogen particles independently of the level of autofluorescence. In addition, we were able to determine that the majority of these substrates were taken up by AM fungi as organic nitrogen rather than inorganic form (Fig. 1). Moreover, we could detect as few as four QDs per biovolume sample Thus, QDs would be especially useful in micro-scale studies where sample sizes or fluxes are too small for isotopic analysis. For instance, it can be time-consuming and challenging (but not impossible; see Hodge and Fitter, 2010; Staddon et al., 2003) to isolate enough uncontaminated microbial biomass from the soil to perform isotopic measurements. In contrast, QDs can provide quantitative, real-time, spatially-explicit information within microscopic structures. Indeed, the applicability of QDs to ecosystem research may extend beyond the approaches outlined here. For example, QDs could be employed to examine a variety of ecological processes, including competition between bacteria and fungi for organic substrates, nutrient allocation and transport within plants, or herbivory or predation rates. In addition, they could be conjugated with a broad array of organic compounds to measure total contributions of mycorrhizal fungi to C and N cycling within ecosystems.
Supplementary Material
Acknowledgments
We thank R. Aicher, S. Allison, S. Kivlin, H. McGray, K. McGuire, K. Mooney, M. Spasojevic, K. Suding, and J. Talbot for intellectual contributions; M. Mack for collaboration in establishing and maintaining the field site; Ft. Greely and the U.S. Army for access to the field site; the Optical Biology Core facility of the UCI Developmental Biology Center (supported in part by a Cancer Center Support Grant and a Center for Complex Biological Systems Support Grant) for facility use; and the U.S. National Science Foundation and the Kearney Foundation for funding. M.D. acknowledges support from the National Institutes of Health (PHS 5 P41-RR003155, U54 GM064346 Cell Migration Consortium and P50-GM076516 grants).
Appendix A. Supplementary material
Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.soilbio.2012.06.001.
References
- Aber J, Melillo J. Terrestrial Ecosystems. 2. Harcourt Academic Press; San Diego, California, USA: 2001. [Google Scholar]
- Allison SD, Weintraub MN, Gartner TB, Waldrop MP. Evolutionary-economic principles as regulators of soil enzyme production and ecosystem function. In: Shukla GC, Varma A, editors. Soil Enzymology. Springer; Heidelberg: 2010. pp. 245–258. [Google Scholar]
- Azcón-Aguilar C, Barea JM. Arbuscular mycorrhizas and biological control of soil-borne plant pathogens – an overview of the mechanisms involved. Mycorrhiza. 1997;6:457–464. [Google Scholar]
- Bailey RE, Nie S. Alloyed semiconductor quantum dots: tuning the optical properties without changing the particle size. Journal of the American Chemical Society. 2003;125:7100–7106. doi: 10.1021/ja035000o. [DOI] [PubMed] [Google Scholar]
- Bajwa R, Read DJ. Utilization of mineral and amino N sources by the ericoid mycorrhizal endophyte Hymenoscyphus ericae and by mycorrhizal and non-mycorrhizal seedlings of Vaccinium. Transactions of the British Mycological Society. 1986;87:269–277. [Google Scholar]
- Bonfante-Fasolo P. Anatomy and morphology of VA mycorrhizae. In: Powell C, Bagyaraj D, editors. VA Mycorrhiza. CRC Press; Boca Raton, FL: 1986. pp. 2–33. [Google Scholar]
- Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- Cappellazzo G, Lanfranco L, Fitz M, Wipf D, Bonfante P. Characterization of an amino acid permease from the endomycorrhizal fungus Glomus mosseae. Plant Physiology. 2008;147:429–437. doi: 10.1104/pp.108.117820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WCW, Nie SM. Quantum dot bioconjugates for ultrasensitive non-isotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- Chapin FS, Matson PA, Mooney HA. Principles of Terrestrial Ecosystem Ecology. Springer; New York: 2002. [Google Scholar]
- Digman MA, Brown CM, Sengupta P, Wiseman PW, Horwitz AR, Gratton E. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophysical Journal. 2005;89:1317–1327. doi: 10.1529/biophysj.105.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankena J, Vanverseveld HW, Stouthamer AH. Substrate and energy costs of the production of exocellular enzymes by Bacillus licheniformis. Biotechnology and Bioengineering. 1988;32:803–812. doi: 10.1002/bit.260320612. [DOI] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V. Plant cell responses to arbuscular mycorrhizal fungi: getting to the roots of the symbiosis. Plant Cell. 1996;8:1871–1883. doi: 10.1105/tpc.8.10.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooday GW. The ecology of chitin degradation. Advances in Microbial Ecology. 1990;11:387–430. [Google Scholar]
- Gooday GW. Cell membrane. In: Gow NAR, Gadd GM, editors. Growing Fungus. Chapman & Hall; London: 1994a. pp. 43–62. [Google Scholar]
- Gooday GW. Physiology of microbial degradation of chitin and chitosan. In: Ratledge C, editor. Biochemistry of Microbial Degradation. Kluwer Academic; Netherlands: 1994b. pp. 279–312. [Google Scholar]
- Hobbie JE, Hobbie EA. N-15 in symbiotic fungi and plants estimates nitrogen and carbon flux rates in Arctic tundra. Ecology. 2006;87:816–822. doi: 10.1890/0012-9658(2006)87[816:nisfap]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Hodge A, Campbell CD, Fitter AH. An arbuscular mycorrhizal fungus accelerates decomposition and acquires nitrogen directly from organic material. Nature. 2001;413:297–299. doi: 10.1038/35095041. [DOI] [PubMed] [Google Scholar]
- Hodge A, Fitter AH. Substantial nitrogen acquisition by arbuscular mycorrhizal fungi from organic material has implications for N cycling. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13754–13759. doi: 10.1073/pnas.1005874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Gao X. Plasmonic fluorescent quantum dots. Nature Nanotechnology. 2009;4:571–576. doi: 10.1038/nnano.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerley SJ, Read DJ. The biology of mycorrhiza in the Ericaceae. XVIII. Chitin degradation by Hymenoscyphus ericae and transfer of chitin-nitrogen to the host plant. New Phytologist. 1995;131:369–375. doi: 10.1046/j.1469-8137.1997.00778.x. [DOI] [PubMed] [Google Scholar]
- Koske RE, Gemma JN. A modified procedure for staining roots to detect VA mycorrhizas. Mycological Research. 1989;92:486–505. [Google Scholar]
- Leake JR, Read DJ. Chitin as a nitrogen source for mycorrhizal fungi. Mycological Research. 1990;94:993–995. [Google Scholar]
- Lin Z, Su Z, Wan Y, Zhang H, Mu Y, Yang B, Jin Q. Labeled avidin bound to water-soluble nanocrystals by electrostatic interactions. Russian Chemical Bulletin. 2004;53:2690–2694. [Google Scholar]
- Lundeberg G. Utilization of various nitrogen sources, in particular bound soil nitrogen, by mycorrhizal fungi. Studia Forestalia Suecica. 1970;79:1–95. [Google Scholar]
- Mack MC, Treseder KK, Manies KL, Harden J, Schuur EAG, Vogel J, Randerson J, Chapin FS. Recovery of aboveground plant biomass and productivity after fire in mesic and dry black spruce forests on inland Alaska. Ecosystems. 2008;11:209–225. [Google Scholar]
- McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA. A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytologist. 1990;115:495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x. [DOI] [PubMed] [Google Scholar]
- Muir DCG, Howard PH. Are there other persistent organic pollutants? A challenge for environmental chemists. Environmental Science & Technology. 2006;40:7157–7166. doi: 10.1021/es061677a. [DOI] [PubMed] [Google Scholar]
- Näsholm T, Ekblad A, Nordin A, Giesler R, Hogberg M, Hogberg P. Boreal forest plants take up organic nitrogen. Nature. 1998;392:914–916. [Google Scholar]
- Näsholm T, Kielland K, Ganeteg U. Uptake of organic nitrogen by plants. New Phytologist. 2009;182:31–48. doi: 10.1111/j.1469-8137.2008.02751.x. [DOI] [PubMed] [Google Scholar]
- Newman EI, Reddell P. The distribution of mycorrhizas among families of vascular plants. New Phytologist. 1987;106:745–751. doi: 10.1111/j.1469-8137.1987.tb00175.x. [DOI] [PubMed] [Google Scholar]
- Paul EA, Clark FE. Soil Microbiology and Biochemistry. 2. Academic Press; San Diego: 1996. [Google Scholar]
- Peng ZA, Peng X. Formation of high-quality CdTe, CdSe, and CdS nanocrystals using CdO as precursor. Journal of American Chemical Society. 2000;123:183–184. doi: 10.1021/ja003633m. [DOI] [PubMed] [Google Scholar]
- Read DJ, Perez-Moreno J. Mycorrhizas and nutrient cycling in ecosystems – a journey towards relevance? New Phytologist. 2003;157:475–492. doi: 10.1046/j.1469-8137.2003.00704.x. [DOI] [PubMed] [Google Scholar]
- Regent_Instruments. WinRHIZO Tron MF. Regent Instruments Inc; Canada: 2003. [Google Scholar]
- Reich PB, Grigal DF, Aber JD, Gower ST. Nitrogen mineralization and productivity in 50 hardwood and conifer stands on diverse soils. Ecology. 1997;78:335–347. [Google Scholar]
- Rossow MJ, Sasaki JM, Digman MA, Gratton E. Raster image correlation spectroscopy in live cells. Nature Protocols. 2010;5:1761–1774. doi: 10.1038/nprot.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell E. Soil Conditions and Plant Growth. Longmans, Green and Company; London, UK: 1912. [Google Scholar]
- Schimel JP, Bennett J. Nitrogen mineralization: challenges of a changing paradigm. Ecology. 2004;85:591–602. [Google Scholar]
- Smith SE, Read DJ. Mycorrhizal Symbiosis. 3. Academic Press; San Diego: 2008. [Google Scholar]
- Staddon PL, Ramsey CB, Ostle N, Ineson P, Fitter AH. Rapid turnover of hyphae of mycorrhizal fungi determined by AMS microanalysis of 14C. Science. 2003;300:1138–1140. doi: 10.1126/science.1084269. [DOI] [PubMed] [Google Scholar]
- Talbot JM, Allison SD, Treseder KK. Decomposers in disguise: mycorrhizal fungi as regulators of soil C dynamics in ecosystems under global change. Functional Ecology. 2008;22:955–963. [Google Scholar]
- Toth R, Miller RM, Jarstfer AG, Alexander T, Bennett EL. The calculation of intraradical fungal biomass from percent colonization in vesicular-arbuscular mycorrhizae. Mycologia. 1991;83:553–558. [Google Scholar]
- Treseder KK. A meta-analysis of mycorrhizal responses to nitrogen, phosphorus, and atmospheric CO2 in field studies. New Phytologist. 2004;164:347–355. doi: 10.1111/j.1469-8137.2004.01159.x. [DOI] [PubMed] [Google Scholar]
- Treseder KK, Cross A. Global distributions of arbuscular mycorrhizal fungi. Ecosystems. 2006;9:305–316. [Google Scholar]
- Treseder KK, Mack MC, Cross A. Relationships among fires, fungi, and soil dynamics in Alaskan boreal forests. Ecological Applications. 2004;14:1826–1838. [Google Scholar]
- Treseder KK, Turner KM, Mack MC. Mycorrhizal responses to nitrogen fertilization in boreal ecosystems: potential consequences for soil carbon storage. Global Change Biology. 2007;13:78–88. [Google Scholar]
- Uhlenbeck GE, Ornstein LS. On the theory of the Brownian motion. Physical Review. 1930;36:823. [Google Scholar]
- Whiteside MD, Treseder KK, Atsatt PR. The brighter side of soils: quantum dots track organic nitrogen through fungi and plants. Ecology. 2009;90:100–108. doi: 10.1890/07-2115.1. [DOI] [PubMed] [Google Scholar]
- Xiao-Wei W, Ming G, Chun-Hua D, Jin-Ming C, Yong Y, Fang-Wen S, Guang-Can G, Zheng-Fu H. Anti-bunching and luminescence blinking suppression from plasmon-interacted single CdSe/ZnS quantum dot. Optics Express. 2010;18:6340–6346. doi: 10.1364/OE.18.006340. [DOI] [PubMed] [Google Scholar]
- Zimmermann T, Rietdorf J, Pepperkok R. Spectral imaging and its applications in live cell microscopy. FEBS Letters. 2003;546:87–92. doi: 10.1016/s0014-5793(03)00521-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.