Abstract
Tobacco smoke has been shown to produce both DNA damage and epigenetic alterations. However, the potential role of DNA damage in generating epigenetic changes is largely underinvestigated in human studies. We examined the effects of smoking on the levels of DNA methylation in genes for tumor protein p53, cyclin-dependent kinase inhibitor2A, hypermethylated-in-cancer-1 (HIC1), interleukin-6, Long Interspersed Nuclear Element type1, and Alu retrotransposons in blood of 177 residents in Thailand using bisulfite-PCR andpyrosequencing. Then, we analyzed the relationship of this methylation with the oxidative DNA adduct, M1dG (a malondialdehyde adduct), measured by 32P-postlabeling. Multivariate statistical analyses showed that HIC1 methylation levels were significantly increased in smokers compared with nonsmokers (p ≤ .05). A dose response was observed, with the highest HIC1 methylation levels in smokers of ≥ 10 cigarettes/day relative to nonsmokers and intermediate values in smokers of 1–9 cigarettes/day (p for trend ≤ .001). No additional relationships were observed. We also evaluated correlations between M1dG and the methylation changes at each HIC1 CpG site individually. The levels of this adduct in smokers showed a significant linear correlation with methylation at one of the 3 CpGs evaluated in HIC1: hypermethylation at position 1904864340 was significantly correlated with the adduct M1dG (covariate-adjusted regression coefficient (β) = .224 ± .101 [SE], p ≤ .05). No other correlations were detected. Our study extends prior work by others associating hypermethylation of HIC1 with smoking; shows that a very specific hypermethylation event can arise from smoking; and encourages future studies that explore a possible role for M1dG in connecting smoking to this latter hypermethylation.
Key Words: tobacco smoking, overall and site specific methylation, HIC1, oxidative DNA damage, M1dG.
Tobacco cigarette smoking is causally associated with cancer at different sites (IARC, 2012). Although tobacco control remains the most appropriate strategy to decrease the incidence of smoking-related cancer cases, epigenetic studies may improve the understanding of the underlying mechanisms and the development of appropriate cancer prevention strategies.
DNA methylation occurs mainly in the context of CpG sites, which are present in the regulatory regions of many genes (Esteller, 2008). Aberrant DNA methylation, especially in genes involved in growth control and the maintenance of genomic integrity, is an important event in carcinogenic process, which may ultimately lead to tumor formation (Esteller, 2008). DNA hypomethylation is linked to the reactivation of mobile retroelements, genomic instability, and proto-oncogene activation (Esteller, 2008). Conversely, hypermethylation, both in gene-specific promoter regions and at whole-genome level, has been linked to many cancers, even when it is measured in peripheral blood (Marsit and Christensen, 2013). For instance, hypermethylation of the promoter regions of tumor suppressor genes (TSGs) concomitant with their transcriptional silencing has been observed in lung cancer (Rauch et al., 2008). However, only a few reports have associated smoking with changes in DNA methylation patterns of gene-specific promoter regions in blood and bronchi (Lokk et al., 2012; Terry et al., 2011).
Recently, we observed dose-response patterns of DNA methylation at 5 of 6 interrogated loci, including genes for tumor protein p53 (TP53), cyclin-dependent kinase inhibitor2A (CDKN2A), hypermethylated-in-cancer-1 (HIC1), and interleukin-6 (IL-6), as well as Long Interspersed Nuclear Element type1 (LINE-1) and Alu retrotransposons, in participants representing high, moderate, and low levels of exposure to undefined mixtures of airborne emissions of a steel-petrochemical complex in Rayong, Thailand (Peluso et al., 2012a). Intriguingly, biomarkers of DNA damage and oxidative stress were associated with methylation at several loci. Herein, we aimed to identify epigenetic changes due to smoking habit in the same 6 loci. TSGs and repeated elements were selected for their biological functions and associations with lung cancer (Leng et al., 2012; Li et al., 2012; Tekpli et al., 2013; Zheng et al., 2012).
Additionally, our previous analysis showed correlations of hypomethylation of LINE-1 and IL-6 methylation with the levels of exocyclic DNA adducts, indicated from 3-(2-deoxy-β-D-erythro-pentafuranosyl)pyrimido[1,2-α]purin-10(3H)-one deoxyguanosine (M1dG) adducts, a marker of oxidative stress and peroxidation of lipids (Marnett, 1999), suggesting that environmentally induced DNA methylation loss may be mediated by DNA damage (Peluso et al., 2012a). Interestingly, the levels of M1dG tended to be associated with tobacco smoking in the study participants (Peluso et al., 2010b, 2012b). M1dG is considered a biomarker of environmental (Peluso et al., 2010b, 2012b, 2013a) and dietary exposures (Leuratti et al., 2002; Peluso et al., 2012b; van Helden et al., 2009; Vanhees et al., 2013), predictive of development of larynx, breast, and lung cancers and tumor progression (Munnia et al., 2004, 2006; Peluso et al., 2011; Wang et al., 1996). In healthy people, M1dG adducts occur at levels comparable to the most abundant form of base oxidation, eg, 7,8-dihydro-8-oxo-2′-deoxyguanosine (8-oxodG) (Marnett, 1999). Our studies have also indicated a relationship between inflammation, M1dG, and myeloperoxidase-catalyzed production of hypochlorous acid, a main reactive oxygen species, in lungs of C57Bl6 mice (Güngör et al., 2010a,b). Recently, it has been suggested that oxidatively damaged DNA may play a role in the regulation of gene expression (Khobta et al., 2010). Therefore, our next objective was to examine the correlation between the levels of a specific kind of exocyclic DNA adducts, eg, M1dG, and DNA methylation changes in smokers. Our aim was to provide insights on the potential mechanisms of carcinogens contained in tobacco smoke correlated with epigenomic changes.
MATERIALS AND METHODS
Study population.
The study was conducted by testing peripheral blood leukocytes of 62 nonsmokers (58% of males), 7 former smokers (100% of males), and 108 current smokers (92.5% of males), all resident in Rayong province, Thailand. The mean age of nonsmokers, ex-smokers, and current smokers was 33.5 years ± 7.4, 39.1±9.4, and 33.8±7.6, respectively. Smokers reported smoking 11±4.9 cigarettes per day for at least 4 years. A questionnaire that collected standard demographic and lifestyle data, including age, gender, tobacco smoking habit, number of cigarettes smoked per day, residence, and occupation, was administered to the participants before blood sampling collection. Blood samples were collected at the recruitment. The Map Ta Phut study was approved by the ethics committee of National Cancer Institute of Bangkok, Thailand. Participation rate was about 95%. Written informed consent was obtained before blood sampling. M1dG data using the 32P-postlabeling assay were provided from our previous studies in Rayong region (Peluso et al., 2010b, 2012b). Details on the Map Ta Phut study were reported elsewhere (Peluso et al., 2008, 2013b,c).
Bisulfite-PCR and pyrosequencing.
DNA was purified from buffy-coat samples using phenol extraction (Peluso et al., 1990). DNA methylation was quantified using bisulfite-PCR and pyrosequencing techniques, as previously described (Peluso et al., 2012a). DNA samples from smokers and nonsmokers were interspersed across plates through a single-blind sampling procedure to avoid any possible bias from plate effects. In brief, the samples were bisulfite treated by EZ-96 DNA Methylation-Gold KitTM (Zymo Research, Orange, California) and PCR amplified. A PCR was conducted in 50 µl of GoTaq Green Master mix (Promega, Madison, Wisconsin), 1 pmol of the forward primer, 1 pmol of the reverse primer, 50ng of bisulfite-treated genomic DNA, and water. Primer sequences and PCR conditions are reported in Table 1. One of the forward or reverse primers was biotin labeled, depending on the proximity to CpG sites to be quantified, and used to purify the final PCR product using Sepharose beads. The PCR product was bound to Streptavidin Sepharose HP (Amersham Biosciences, Uppsala, Sweden), and the Sepharose beads containing the trapped PCR product were purified, washed, denatured using a 0.2M NaOH solution, and washed again using the Pyrosequencing VacuumPrep Tool (Pyrosequencing, Westborough, Massachusetts), as recommended by the manufacturer. A 0.3µM pyrosequencing primer was then annealed to the purified single-stranded PCR product, and pyrosequencing was performed using the Pyromark Pyrosequencing System (Pyrosequencing). The size of bisulfite PCR products was 168bp for LINE-1, 148bp for Alu (Genbank Accession (GA) no.X58075), 220bp for TP53 (GA no.AF307851), 169bp for CDKN2A (GA no.L27211), 268 pb for HIC1 (GA no. L41919), and 111 pb for IL-6 (GA no.M18403). The levels of methylation were measured in 5 CpG sites of CDKN2A, 3 CpG positions in HIC1 and TP53, and 2 in IL-6. Methylation quantification was performed using the provided software. The degree of methylation was expressed as the mean for each gene of the percent 5-methylcytosine (% 5-mC = 5-methylated cytosines/(unmethylated 5-cytosines + methylated 5-cytosines). The within-sample coefficients of variation, defined as the ratio of the standard deviation to the mean of triplicates, were 0.009 for LINE-1, 0.03 for Alu, 0.12 for CDKN2A, 0.32 for TP53, 0.08 for IL-6, and 0.35 for HIC1. We used built-in controls to verify bisulfite conversion efficiency. After PCR amplification, every DNA product was tested twice for each marker to confirm the reproducibility of the results (technical replicates).
Table 1.
Adjusted Marginal Meansa of the Levels of Methylation in Line-1 and Alu Repeated Elements as Well as in the Promoter Regions of Tp53, Cdk2a, Hic1, and Il-6 According to Smoking Status and Number of Cigarettes Smoked Per Day
| N | LINE-1 | Alu | TP53 | CDKN2A | HIC1 | IL-6 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SE | p | Mean ± SE | p | Mean ± SE | p | Mean ± SE | p | Mean ± SE | p | Mean ± SE | P | ||
| Tobacco smoking status | |||||||||||||
| Nonsmokerb | 62 | 76.0±0.4 | 24.9±0.2 | 9.8±0.6 | 5.9±0.4 | 16.6±0.9 | 42.6±1.8 | ||||||
| Ex-smoker | 7 | 77.0±1.2 | 0.388 | 25.7±0.6 | 0.174 | 13.0±2.4 | 0.151 | 6.7±1.2 | 0.536 | 25.3±3.8 | 0.012 | 44.2±5.6 | 0.786 |
| Smoker | 108 | 75.9±0.3 | 0.903 | 25.0±0.1 | 0.862 | 10.6±0.5 | 0.310 | 6.1±0.3 | 0.723 | 19.9±0.8 | 0.013 | 39.8±1.2 | 0.226 |
| Daily cigarette smoking | |||||||||||||
| 1–9 cigarettes/day | 35 | 76.0±0.5 | 0.981 | 25.0±0.2 | 0.719 | 10.0±0.8 | 0.850 | 6.0±0.5 | 0.953 | 17.6±1.2 | 0.503 | 36.4±1.9 | 0.023 |
| ≥ 10 cig/day | 73 | 75.9±0.4 | 0.904 | 24.9±0.2 | 0.974 | 11.1±0.7 | 0.170 | 6.2±0.4 | 0.656 | 21.0±1.0 | 0.002 | 41.7±1.6 | 0.719 |
| 0.900c | 0.985c | 0.149c | 0.637c | 0.001c | 0.961c | ||||||||
| Comparison: 1–9 vs ≥ 10 cigarettes/day | 0.930 | 0.713 | 0.279 | 0.718 | 0.029 | 0.029 | |||||||
Note. aEstimated from multiple regression models including as independent variables age, gender, smoking status, tobacco consumption, residence and employment, and percent neutrophils in differential blood counts, as appropriated. The adjusted marginal means of the levels of methyaltion were computed at the average values the other covariates.
bReference category.
c p Value for trend between nonsmokers, 1–9 cigarettes/day, and ≥ 10 cigarettes/day.
Reference standard.
A reference adduct standard was prepared: calf thymus DNA was treated with 10mM malondialdehyde (MDA) (ICN Biomedicals, Irvine, California), as previously reported (Bono et al., 2010). MDA-treated DNA was diluted with untreated DNA to obtain decreasing levels of the reference adduct standard to generate a calibration curve.
Mass spectrometry.
DNA adducts in MDA-treated calf thymus DNA sample were analyzed by mass spectrometry (Voyager DE STR from Applied Biosystems, Framingham, Massachusetts), as reported elsewhere (Wang et al., 2012a,b), through the following sequence of steps: (1) Reaction of DNA with NaBH4 followed by precipitation with isopropanol (Goda and Marnett, 1991); (2) digestion with snake venom phosphodiesterase and nuclease P1; (3) extraction of DNA adducts that are less polar than normal nucleotides on an OASIS cartridge (Waters Corp.); (4) tagging with an isotopologue pair of benzoylhistamines (do and d4) in a phosphate-specific labeling reaction in the presence of carbodiimide; (5) removal of residual reagents by ion exchange solid-phase extraction; (6) resolution of tagged adducts by capillary reversed-phase high-pressure liquid chromatography (HPLC) with a collection of drops onto a MALDI plate; (7) addition of matrix (α-cyano-4-hydroxycinnamic acid); and (8) analysis by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS).
32P-DNA postlabeling assay.
M1dG adducts were measured using 32P-DNA postlabeling assay (Peluso et al., 2010b, 2012b). Briefly, DNA (2 μg) was hydrolyzed by incubation with micrococcal nuclease (21.45 mU/μl) and spleen phosphodiesterase (6.0 mU/μl) at 37°C for 4.5h. Hydrolyzed DNA was treated with nuclease P1 (0.1U/μl) at 37°C for 30min (Munnia et al., 2007). After enzymatic treatment, samples were incubated with 12 μCi of carrier-free [γ-32P]ATP (3000 Ci/mM) and polynucleotide kinase T4 (0.75U/μl) to generate 32P-labeled adducts at 37°C for 30min. 32P-labeled adducts were applied on polyethyleneimine cellulose thin-layer chromatography plates (Macherey-Nagel, Germany). The chromatography analysis was performed using 0.35M MgCl2 for the preparatory chromatography; and 2.1M lithium formate, 3.75M urea pH 3.75 and 0.24M sodium phosphate, and 2.4M urea pH 6.4 for the two-dimensional chromatography (van Helden et al., 2009). Detection and quantification of M1dG adducts and total nucleotides were performed using storage phosphor imaging techniques and intensifying screens from Molecular Dynamics. The intensifying screens were scanned using Typhoon 9210. ImageQuant from Molecular Dynamics was used. M1dG adducts were expressed as relative adduct labeling = pixels in adducted nucleotides/pixels in total nucleotides. The levels of M1dG adducts were corrected across experiments based on the recovery of reference standard (Munnia et al., 2007).
Statistical analysis.
Standard descriptive analysis was used to evaluate the mean levels of DNA methylation in the regulatory regions of CDKN2A, TP53, HIC1, and IL-6, as well as in LINE-1 and Alu repetitive elements by smoking status. Multivariate analyses were performed by fitting linear regression models adjusted by age, gender, residence, and employment to evaluate the changes of DNA methylation due to smoking cigarettes. Also, we adjusted for percent neutrophils in differential blood counts to account for possible differences in the proportion of leukocyte subtypes. The regression parameters estimated from multiple regression models were presented as adjusted marginal means. The adjusted means of DNA methylation were computed at the average values of all the other variables in the models. Some studies have shown a relationship between folic acid and certain B vitamins with DNA methylation; thus, we conducted sensitivity analyses by adding as a covariate in the multiple regression models the weekly intake of fruit and vegetables (Peluso et al., 2012a).
Smokers were stratified into 2 groups considering the number of cigarette consumed per day: (1) 1–9 cigarettes/day and (2) ≥ 10 cigarette/day. A log-normal multiple regression model corrected for age, gender, residence, and employment and percent neutrophils in differential blood counts were used to analyze the adjusted mean changes of DNA methylation in the 2 groups.
Our previous study showed that the mean methylation level at 2 of 6 interrogated loci, eg, LINE-1 and IL-6, was inversely associated with the generation of oxidative DNA damage (Peluso et al., 2012a); herein, we evaluated the correlation between the M1dG adduct and methylation changes at each CpG site of TSGs individually to increase the sensitivity of statistical analysis. A multiple regression model adjusted for age, gender, number of cigarettes/day, and percent neutrophils in differential blood counts was used to evaluate the correlation between the levels of the M1dG adduct with the changes of DNA methylation. Multiple regression analysis was performed on logarithmic transformed adduct and methylation data to normalize the distribution of experimental data (Siegmund et al., 2006). A value of p ≤ .05 (two tailed) was considered significant. Data were analyzed using SPSS 13.0 (IBM SPSS Statistics, New York, New York).
RESULTS
DNA Methylation and Tobacco Smoking
Multivariate analyses using linear regression models showed that the adjusted marginal means of the levels of methylation in the promoter region of HIC1 of smokers was significantly increased compared with nonsmokers (Table 1). Conversely, no significant relationships were observed with the adjusted mean methylation levels of CDKN2A, TP53, and IL-6, as well as in LINE-1 and Alu repeated elements. Specifically, the adjusted marginal means of the levels of methylation at the regulatory region of HIC1 were 16.6% in nonsmokers, 25.3% in ex-smokers (p ≤ 0.05 vs nonsmokers), and 19.9% in smokers (p ≤ 0.05 vs nonsmokers). All the associations reported did not show major differences after correction for the average of weekly intake of fruit and vegetables (data not shown).
The association between specific DNA methylation changes and smoking status was strengthened by the presence of a statistical significant difference between the adjusted mean levels of methylation in the promoter region of HIC1 of smokers of ≥ 10 cigarettes/day in respect to nonsmokers (Table 1). No relationships with cigarette smoking were found with the other 6 methylation markers.
In particular, the adjusted marginal means of the levels of methylation in the regulatory region of HIC1 was 17.6% in smokers of 1–9 cigarettes/day (p = .503 vs nonsmokers) and 21.0% in the individuals who smoked ≥ 10 cigarettes/day (p ≤ .01 vs nonsmokers). A significant trend was also observed (p for trend ≤ .001), with the highest mean levels of HIC1 methylation in smokers of ≥ 10 cigarettes/day in respect to nonsmokers and with intermediate mean level of HIC1 methylation in smokers of 1–9 cigarettes/day.
Reference Standard by 32P-Postlabeling and MALDI-TOF-MS
There were 5.0 M1dG adducts ± 0.6 per 106 total nucleotides in MDA-treated calf thymus DNA based on 32P-postlabeling. The presence of the M1dG adduct in this sample was confirmed by mass spectrometry as reported before (Wang et al., 2012a,b), and we are using the nomenclature reported by Goda and Marnett (1991) for this adduct. A calibration curve (not shown) then was set up by diluting this sample with DNA and measuring the decreasing levels of M1dG, r 2 = .99.
HIC1 Methylation and M1dG
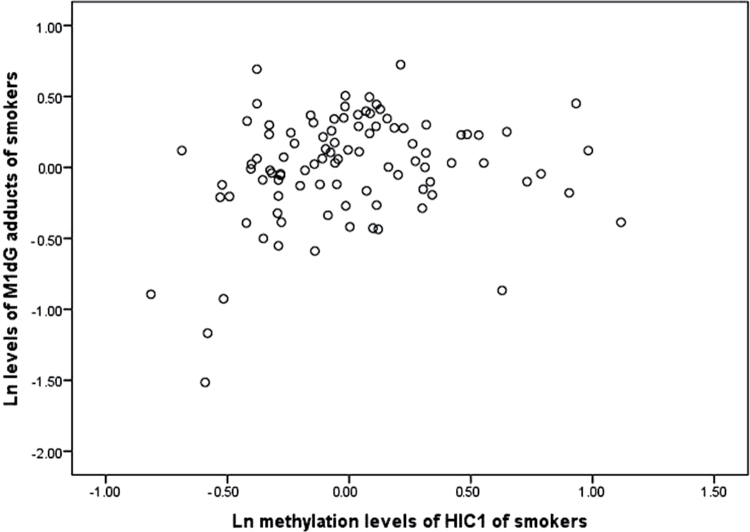
When we have previously examined the association between the mean methylation changes of 3 CpG sites in the promoter region of HIC1 and the levels of M1dG adducts, no statistically significant association was found in overall methylation (Peluso et al., 2012a). Encouraged by a recent work that showed that the analysis of single CpG methylation changes may be more effective in determining methylation changes in smokers (Wangsri et al., 2012), we decided to consider individually the methylation levels in each of the 3 CpG sites studied. Interestingly, the statistical analysis showed that the levels of methylation in one of the sites were linearly associated with those of M1dG adducts (Fig. 1). Specifically, the multivariate regression analysis showed that the level of methylation at the CpG site, position 1904864340, was significantly correlated with the generation of M1dG adduct, regression coefficient (β) = .224 ± .101 (SE), p ≤ .05, after correction for age, gender, number of cigarettes/day, and percent neutrophils in differential blood counts. A scatter plot of this data is reported in Figure 1. No significant correlations were found at the other 2 CpG sites under study. The levels of M1dG adducts were not correlated with HIC1 methylation changes in nonsmokers.
Fig. 1.
Relationship between the ln levels of M1dG adducts per 108 total nucleotides, expressed such as relative adduct labeling, and the ln levels of methylation, expressed such as percent 5-methylcytosine, at a specific CpG site, pos.1904864340, within the regulatory region of HIC1 in smokers after correction for age, gender, number of cigarettes/day, and percent neutrophils in differential blood counts. Abbreviations: HIC1, hypermethylated-in-cancer-1; M1dG, malondialdehyde-deoxyguanosine
DISCUSSION
In this study, we examined the association between tobacco smoking and the changes of methylation in selected TSGs and repeated elements. Our findings show that the promoter region of HIC1 was significantly hypermethylated in the blood of smokers relative to nonsmokers, whereas the levels of methylation in the promoter regions of TP53, CDKN2A, IL-6, as well as in LINE-1 and Alu repeated elements, were not. The levels of methylation in the regulatory regions of HIC1 were significantly higher in subjects who smoked more than ≥ 10 cigarettes/day in respect to nonsmokers. A significant trend was observed, with the highest levels of methylation of HIC1 in the smokers who smoked ≥ 10 cigarettes/day compared with nonsmokers, with intermediate levels of methylation in those who reported to smoke 1–9 cigarettes/day. Furthermore, the levels of methylation of a specific CpG site in HIC1 were positively and significantly associated with those of a specific type of exocyclic DNA adducts, eg, M1dG, a malondialdehyde DNA adduct that is a marker of oxidative stress and lipid peroxidation.
Our results indicate that the lifestyle habit of smoking tobacco is significantly associated with hypermethylation of the promoter region of HIC1. Higher levels of methylation were detected in the smokers who reported to smoke ≥ 10 cigarettes/day, with intermediate levels in smokers of 1–9 cigarettes/day. An association between cigarette smoke and HIC1 methylation changes has been also shown from a recent in vitro experimental study (Brait et al., 2013). In that study, cigarette smoke extracts induced a significant hypermethylation of the promoter region of HIC1 in experimental cells compared with untreated cells. Interestingly, the exposure to smoke extracts also induces hypermethylation of melanoma cell adhesion molecule and “deleted in colon cancer,” 2 genes abnormally expressed in a variety of tumors. HIC1 is a key gene with a main transcriptional regulator role in the control of cell proliferation and in the regulation of apoptosis in response to DNA damage. Alteration of the HIC1 signaling pathway may induce abnormal cell growth and oxidative stress responses contributing to a cancerous phenotype. HIC1 is epigenetically silenced in a variety of tumors, including non–small cell lung cancer (Fleuriel et al., 2009). HIC1 silencing is considered to be a smoking-related precancerous event because the hypermethylation of HIC1 occurs with a greater incidence in lung cancer smokers than in those who have never smoked (Brait et al., 2013).
The previous research focusing on changes of DNA methylation in gene-specific promoter regions in the peripheral blood of current smokers relative to nonsmokers has shown different effects of tobacco smoking on epigenetic changes. Two specific sites within factor II receptor-like 3, a gene encoding thrombin protease–activated receptor-4, and G protein–coupled receptor 15, a gene which encodes a G protein–coupled receptor that acts as a chemokine receptor for human immunodeficiency virus type 1 and 2, showed significant differences in the levels of methylation between smokers and nonsmokers (Shenker et al., 2013; Terry et al., 2011; Wan et al., 2012). One intragenic region of the aryl hydrocarbon receptor repressor gene and 2 intergenic regions on 2q37.1 and 6p21.33 were also found to be hypomethylated in smokers compared with nonsmokers (Shenker et al., 2013). On the other hand, studies looking at methylation at whole-genome level have not found a relationship between smoking habit and the levels of DNA methylation in repetitive elements (Terry et al., 2011). Nevertheless, methylation changes associated to smoking habit and cancer risk were found when single CpG sites were considered (Wangsri et al., 2012), indicating that the study of specific methylation pattern may be more effective in determining smoking effects and cancer risk in respect to the analysis of overall methylation changes (Patchsung et al., 2012; Wangsri et al., 2012).
Exposure to tobacco smoke constituents, which include more than 60 chemical carcinogens (IARC, 2012), is a key example of environmental exposure that causes both mutations in cancer-related genes and epigenetic alterations. However, the mechanisms by which chemical carcinogens disrupt the epigenetic code are largely unknown. Accumulating evidence from our group and others suggests that DNA damage can bring on DNA methylation changes (Ha et al., 2011; Khobta et al., 2010; Peluso et al., 2012a). For an example, our previous study in the Amphur-Muang district, Rayong, Thailand, has shown that the genetic damage induced by polycyclic aromatic hydrocarbons and reactive oxygen species, as reflected in aromatic DNA adducts and M1dG, was associated with significant methylation changes at several loci in all study participants (Peluso et al., 2012a). In particular, the levels of aromatic DNA adducts, a biomarker of environmental carcinogen exposures (Peluso et al., 1997, 1998, 2013c; Phillips and Venitt, 2012), genetic susceptibility, and cancer risk (Agudo et al., 2012; Airoldi et al., 2005; Matullo et al., 2001; Peluso et al., 2004, 2010a; Veglia et al., 2008), showed negative correlation with p53 methylation. Conversely, the levels of M1dG adducts, a biomarker of oxidative stress and peroxidation of lipids (Marnett, 1999), also derived from the generation of base-propenal intermediates in DNA sequence (Zhou et al., 2005), showed significant negative correlations with LINE-1 and IL-6 methylation in the analysis of all participants.
Herein, prompted by a recent study, which showed that tobacco smoking may alter methylation of specific CpG sites (Wangsri et al., 2012), we have analyzed individually the association of a specific type of exocyclic DNA adducts with the methylation levels in each of the 3 CpG sites studied in the regulatory region of HIC1. We found that the generation of M1dG adduct of smokers was linearly and significantly correlated with methylation changes at a specific CpG site in the promoter region of HIC1. Even though the methylation correlation with oxidative DNA damage was with only at 1 CpG site, the alteration may play a role in carcinogenesis by, eg, impairing transcriptional factor binding. This finding fits well with emerging data showing transcriptional repression after oxidative DNA damage at the promoter region of CpG island-containing genes (O’Hagan et al., 2011). Indeed, the recruitment of enzymes catalyzing DNA methylation, such as DNA methyltransferase1 (DNMT1) and DNMT3B (O’Hagan et al., 2011), to sites of oxidative damage along with transcriptional factors, which generally mediate transcriptional repression, such as SIRT1, has been implicated in gene silencing (O’Hagan et al., 2011). Tekpli et al. (2013) have recently associated aberrant DNA methylation of cytochrome P4501A1 enhancer with DNA damage and mRNA levels in histologically normal lung tissue of lung cancer smokers (Tekpli et al., 2013). The correct restoration of the original methylation pattern of DNA damaged region is essential to ensure the maintenance and fidelity of the epigenetic component of the genome. A gain of methylation in the promoter regions with high CpG content may be associated to the generation of oxidative damage, an event that induces the recruitment of large silencing complexes that may explain cancer-specific aberrant DNA methylation and transcriptional silencing (O’Hagan et al., 2011). Although this scenario may be true, another mechanism should be considered: hypomethylation changes at the site of DNA repair may also reflect an inappropriate de novo methylation of cytosines during the process of restoration of epigenetic information (Cuozzo et al., 2007; Jin and Robertson, 2013).
Traditionally, epigenetic and DNA sequence alterations have been considered of as 2 parallel mechanisms involved in carcinogenesis. In this study, we show that exocyclic DNA adducts have the potential to influence DNA methylation patterns and consequently gene expression of cigarette smokers. Although the mechanisms by which DNA damage increase or decrease methylation changes may be different (O’Hagan et al., 2011), the observed cross talk between the genome and epigenome offers new insights into the mechanisms of action of carcinogens contained in tobacco smoke and subsequent disease susceptibility, including carcinogenesis.
FUNDING
National Cancer Institute, Bangkok, Thailand; Tuscan Tumor Institute, Florence, Italy; the “Associazione Italiana per la Ricerca sul Cancro”, Milan, Italy; the U.S. National Institute of Environmental Health Sciences (R01ES021733, R21ES021895, R21ES020010, P30ES000002, P42ES017198).
REFERENCES
- Agudo A., Peluso M., Munnia A., Luján-Barroso L., Sánchez M. J., Molina-Montes E., Sánchez-Cantalejo E., Navarro C., Tormo M. J., Chirlaque M. D., et al. (2012). Aromatic DNA adducts and risk of gastrointestinal cancers: A case-cohort study within the EPIC-Spain. Cancer Epidemiol. Biomarkers Prev. 21, 685–692 [DOI] [PubMed] [Google Scholar]
- Airoldi L., Vineis P., Colombi A., Olgiati L., Dell’Osta C., Fanelli R., Manzi L., Veglia F., Autrup H., Dunning A., et al. (2005). 4-Aminobiphenyl-hemoglobin adducts and risk of smoking-related disease in never smokers and former smokers in the European Prospective Investigation into Cancer and Nutrition prospective study. Cancer Epidemiol. Biomarkers Prev. 14, 2118–2124 [DOI] [PubMed] [Google Scholar]
- Bono R., Romanazzi V., Munnia A., Piro S., Allione A., Ricceri F., Guarrera S., Pignata C., Matullo G., Wang P., et al. (2010). Malondialdehyde-deoxyguanosine adduct formation in workers of pathology wards: The role of air formaldehyde exposure. Chem. Res. Toxicol. 23, 1342–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brait M., Munari E., LeBron C., Noordhuis M. G., Begum S., Michailidi C., Gonzalez-Roibon N., Maldonado L., Sen T., Guerrero-Preston R., et al. (2013). Genome-wide methylation profiling and the PI3K-AKT pathway analysis associated with smoking in urothelial cell carcinoma. Cell Cycle 12, 1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuozzo C., Porcellini A., Angrisano T., Morano A., Lee B., Di Pardo A., Messina S., Iuliano R., Fusco A., Santillo M. R., et al. (2007). DNA damage, homology-directed repair, and DNA methylation. PLoS Genet. 3, e110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Esteller M. (2008). Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159 [DOI] [PubMed] [Google Scholar]
- Fleuriel C., Touka M., Boulay G., Guérardel C., Rood B. R., Leprince D. (2009). HIC1 (Hypermethylated in Cancer 1) epigenetic silencing in tumors. Int. J. Biochem. Cell Biol. 41, 26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda Y., Marnett L. J. (1991). High-performance liquid chromatography with electrochemical detection for determination of the major malondialdehyde-guanine adduct. Chem. Res. Toxicol. 4, 520–524 [DOI] [PubMed] [Google Scholar]
- Güngör N., Knaapen A. M., Munnia A., Peluso M., Haenen G. R., Chiu R. K., Godschalk R. W., van Schooten F. J. (2010a). Genotoxic effects of neutrophils and hypochlorous acid. Mutagenesis 25, 149–154 [DOI] [PubMed] [Google Scholar]
- Güngör N., Pennings J. L., Knaapen A. M., Chiu R. K., Peluso M., Godschalk R. W., Van Schooten F. J. (2010b). Transcriptional profiling of the acute pulmonary inflammatory response induced by LPS: role of neutrophils. Respir. Res. 11, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha K., Lee G. E., Palii S. S., Brown K. D., Takeda Y., Liu K., Bhalla K. N., Robertson K. D. (2011). Rapid and transient recruitment of DNMT1 to DNA double-strand breaks is mediated by its interaction with multiple components of the DNA damage response machinery. Hum. Mol. Genet. 20, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC (2012). Personal habits and indoor combustions. Volume 100 E. A review of human carcinogens. IARC Monogr. Eval. Carcinog. Risks Hum. 100, 1–538 [PMC free article] [PubMed] [Google Scholar]
- Jin B., Robertson K. D. (2013). DNA methyltransferases, DNA damage repair, and cancer. Adv. Exp. Med. Biol. 754, 3–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khobta A., Anderhub S., Kitsera N., Epe B. (2010). Gene silencing induced by oxidative DNA base damage: Association with local decrease of histone H4 acetylation in the promoter region. Nucleic Acids Res. 38, 4285–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng S., Stidley C. A., Liu Y., Edlund C. K., Willink R. P., Han Y., Landi M. T., Thun M., Picchi M. A., Bruse S. E., et al. (2012). Genetic determinants for promoter hypermethylation in the lungs of smokers: A candidate gene-based study. Cancer Res. 72, 707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuratti C., Watson M. A., Deag E. J., Welch A., Singh R., Gottschalg E., Marnett L. J., Atkin W., Day N. E., Shuker D. E., et al. (2002). Detection of malondialdehyde DNA adducts in human colorectal mucosa: Relationship with diet and the presence of adenomas. Cancer Epidemiol. Biomarkers Prev. 11, 267–273 [PubMed] [Google Scholar]
- Li L., Choi J. Y., Lee K. M., Sung H., Park S. K., Oze I., Pan K. F., You W. C., Chen Y. X., Fang J. Y., et al. (2012). DNA methylation in peripheral blood: A potential biomarker for cancer molecular epidemiology. J. Epidemiol. 22, 384–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokk K., Vooder T., Kolde R., Välk K., Võsa U., Roosipuu R., Milani L., Fischer K., Koltsina M., Urgard E., et al. (2012). Methylation markers of early-stage non-small cell lung cancer. PLoS One 7, e39813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnett L. J. (1999). Lipid peroxidation-DNA damage by malondialdehyde. Mutat. Res. 424, 83–95 [DOI] [PubMed] [Google Scholar]
- Marsit C., Christensen B. (2013). Blood-derived DNA methylation markers of cancer risk. Adv. Exp. Med. Biol. 754, 233–252 [DOI] [PubMed] [Google Scholar]
- Matullo G., Guarrera S., Carturan S., Peluso M., Malaveille C., Davico L., Piazza A., Vineis P. (2001). DNA repair gene polymorphisms, bulky DNA adducts in white blood cells and bladder cancer in a case-control study. Int. J. Cancer 92, 562–567 [DOI] [PubMed] [Google Scholar]
- Munnia A., Amasio M. E., Peluso M. (2004). Exocyclic malondialdehyde and aromatic DNA adducts in larynx tissues. Free Radic. Biol. Med. 37, 850–858 [DOI] [PubMed] [Google Scholar]
- Munnia A., Bonassi S., Verna A., Quaglia R., Pelucco D., Ceppi M., Neri M., Buratti M., Taioli E., Garte S., et al. (2006). Bronchial malondialdehyde DNA adducts, tobacco smoking, and lung cancer. Free Radic. Biol. Med. 41, 1499–1505 [DOI] [PubMed] [Google Scholar]
- Munnia A., Saletta F., Allione A., Piro S., Confortini M., Matullo G., Peluso M. (2007). 32P-Post-labelling method improvements for aromatic compound-related molecular epidemiology studies. Mutagenesis 22, 381–385 [DOI] [PubMed] [Google Scholar]
- O’Hagan H. M., Wang W., Sen S., Destefano Shields C., Lee S. S., Zhang Y. W., Clements E. G., Cai Y., Van Neste L., Easwaran H., et al. (2011). Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 20, 606–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patchsung M., Boonla C., Amnattrakul P., Dissayabutra T., Mutirangura A., Tosukhowong P. (2012). Long interspersed nuclear element-1 hypomethylation and oxidative stress: Correlation and bladder cancer diagnostic potential. PLoS One 7, e37009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso M., Amasio E., Bonassi S., Munnia A., Altrupa F., Parodi S. (1997). Detection of DNA adducts in human nasal mucosa tissue by 32P-postlabeling analysis. Carcinogenesis 18, 339–344 [DOI] [PubMed] [Google Scholar]
- Peluso M., Bollati V., Munnia A., Srivatanakul P., Jedpiyawongse A., Sangrajrang S., Piro S., Ceppi M., Bertazzi P. A., Boffetta P., et al. (2012a). DNA methylation differences in exposed workers and nearby residents of the Ma Ta Phut industrial estate, Rayong, Thailand. Int. J. Epidemiol. 41, 1753–60; discussion 1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso M., Castegnaro M., Malaveille C., Talaska G., Vineis P., Kadlubar F., Bartsch H. (1990). 32P-postlabelling analysis of DNA adducted with urinary mutagens from smokers of black tobacco. Carcinogenesis 11, 1307–1311 [DOI] [PubMed] [Google Scholar]
- Peluso M., Merlo F., Munnia A., Valerio F., Perrotta A., Puntoni R., Parodi S. (1998). 32P-postlabeling detection of aromatic adducts in the white blood cell DNA of nonsmoking police officers. Cancer Epidemiol. Biomarkers Prev. 7, 3–11 [PubMed] [Google Scholar]
- Peluso M., Munnia A., Ceppi M., Giese R. W., Catelan D., Rusconi F., Godschalk R. W., Biggeri A. (2013a). Malondialdehyde-deoxyguanosine and bulky DNA adducts in schoolchildren resident in the proximity of the Sarroch industrial estate on Sardinia Island, Italy. Mutagenesis 28, 315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso M., Munnia A., Piro S., Armillis A., Ceppi M., Matullo G., Puntoni R. (2010a). Smoking, DNA adducts and number of risk DNA repair alleles in lung cancer cases, in subjects with benign lung diseases and in controls. J. Nucleic Acids 2010, 386798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso M., Munnia A., Piro S., Jedpiyawongse A., Sangrajrang S., Giese R. W., Ceppi M., Boffetta P., Srivatanakul P. (2012b). Fruit and vegetable and fried food consumption and 3-(2-deoxy-β-D-erythro-pentafuranosyl)pyrimido[1,2-α] purin-10(3H)-one deoxyguanosine adduct formation. Free Radic. Res. 46, 85–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso M., Munnia A., Risso G. G., Catarzi S., Piro S., Ceppi M., Giese R. W., Brancato B. (2011). Breast fine-needle aspiration malondialdehyde deoxyguanosine adduct in breast cancer. Free Radic. Res. 45, 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso M., Neri M., Margarino G., Mereu C., Munnia A., Ceppi M., Buratti M., Felletti R., Stea F., Quaglia R., et al. (2004). Comparison of DNA adduct levels in nasal mucosa, lymphocytes and bronchial mucosa of cigarette smokers and interaction with metabolic gene polymorphisms. Carcinogenesis 25, 2459–2465 [DOI] [PubMed] [Google Scholar]
- Peluso M., Srivatanakul P., Jedpiyawongse A., Sangrajrang S., Munnia A., Piro S., Ceppi M., Boffetta P., Godschalk R. W., van Schooten F. J. (2013b). Aromatic DNA adducts and number of lung cancer risk alleles in Map-Ta-Phut Industrial Estate workers and nearby residents. Mutagenesis 28, 57–63 [DOI] [PubMed] [Google Scholar]
- Peluso M., Srivatanakul P., Munnia A., Jedpiyawongse A., Ceppi M., Sangrajrang S., Piro S., Boffetta P. (2010b). Malondialdehyde-deoxyguanosine adducts among workers of a Thai industrial estate and nearby residents. Environ. Health Perspect. 118, 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso M., Srivatanakul P., Munnia A., Jedpiyawongse A., Meunier A., Sangrajrang S., Piro S., Ceppi M., Boffetta P. (2008). DNA adduct formation among workers in a Thai industrial estate and nearby residents. Sci. Total Environ. 389, 283–288 [DOI] [PubMed] [Google Scholar]
- Peluso M. E., Munnia A., Srivatanakul P., Jedpiyawongse A., Sangrajrang S., Ceppi M., Godschalk R. W., van Schooten F. J., Boffetta P. (2013c). DNA adducts and combinations of multiple lung cancer at-risk alleles in environmentally exposed and smoking subjects. Environ. Mol. Mutagen. 54, 375–383 [DOI] [PubMed] [Google Scholar]
- Phillips D. H., Venitt S. (2012). DNA and protein adducts in human tissues resulting from exposure to tobacco smoke. Int. J. Cancer 131, 2733–2753 [DOI] [PubMed] [Google Scholar]
- Rauch T. A., Zhong X., Wu X., Wang M., Kernstine K. H., Wang Z., Riggs A. D., Pfeifer G. P. (2008). High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 105, 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenker N. S., Polidoro S., van Veldhoven K., Sacerdote C., Ricceri F., Birrell M. A., Belvisi M. G., Brown R., Vineis P., Flanagan J. M. (2013). Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum. Mol. Genet. 22, 843–851 [DOI] [PubMed] [Google Scholar]
- Siegmund K. D., Levine A. J., Chang J., Laird P. W. (2006). Modeling exposures for DNA methylation profiles. Cancer Epidemiol. Biomarkers Prev. 15, 567–572 [DOI] [PubMed] [Google Scholar]
- Tekpli X., Landvik N. E., Anmarkud K. H., Skaug V., Haugen A., Zienolddiny S. (2013). DNA methylation at promoter regions of interleukin 1B, interleukin 6, and interleukin 8 in non-small cell lung cancer. Cancer Immunol. Immunother. 62, 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry M. B., Delgado-Cruzata L., Vin-Raviv N., Wu H. C., Santella R. M. (2011). DNA methylation in white blood cells: Association with risk factors in epidemiologic studies. Epigenetics 6, 828–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden Y. G., Keijer J., Heil S. G., Picó C., Palou A., Oliver P., Munnia A., Briedé J. J., Peluso M., Franssen-van Hal N. L., et al. (2009). Beta-carotene affects oxidative stress-related DNA damage in lung epithelial cells and in ferret lung. Carcinogenesis 30, 2070–2076 [DOI] [PubMed] [Google Scholar]
- Vanhees K., van Schooten F. J., van Waalwijk van Doorn-Khosrovani S. B., van Helden S., Munnia A., Peluso M., Briedé J. J., Haenen G. R., Godschalk R. W. (2013). Intrauterine exposure to flavonoids modifies antioxidant status at adulthood and decreases oxidative stress-induced DNA damage. Free Radic. Biol. Med. 57, 154–161 [DOI] [PubMed] [Google Scholar]
- Veglia F., Loft S., Matullo G., Peluso M., Munnia A., Perera F., Phillips D. H., Tang D., Autrup H., Raaschou-Nielsen O., et al. ; Genair-EPIC Investigators (2008). DNA adducts and cancer risk in prospective studies: A pooled analysis and a meta-analysis. Carcinogenesis 29, 932–936 [DOI] [PubMed] [Google Scholar]
- Wan E. S., Qiu W., Baccarelli A., Carey V. J., Bacherman H., Rennard S. I., Agusti A., Anderson W., Lomas D. A., Demeo D. L. (2012). Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum. Mol. Genet. 21, 3073–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Dhingra K., Hittelman W. N., Liehr J. G., de Andrade M., Li D. (1996). Lipid peroxidation-induced putative malondialdehyde-DNA adducts in human breast tissues. Cancer Epidemiol. Biomarkers Prev. 5, 705–710 [PubMed] [Google Scholar]
- Wang P., Fisher D., Rao A., Giese R. W. (2012a). Nontargeted nucleotide analysis based on benzoylhistamine labeling-MALDI-TOF/TOF-MS: Discovery of putative 6-oxo-thymine in DNA. Anal. Chem. 84, 3811–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Gao J., Li G., Shimelis O., Giese R. W. (2012b). Nontargeted analysis of DNA adducts by mass-tag MS: Reaction of p-benzoquinone with DNA. Chem. Res. Toxicol. 25, 2737–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangsri S., Subbalekha K., Kitkumthorn N., Mutirangura A. (2012). Patterns and possible roles of LINE-1 methylation changes in smoke-exposed epithelia. PLoS One 7, e45292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Xiong D., Sun X., Wang J., Hao M., Ding T., Xiao G., Wang X., Mao Y., Fu Y., et al. (2012). Signification of Hypermethylated in Cancer 1 (HIC1) as Tumor Suppressor Gene in Tumor Progression. Cancer Microenviron. 5, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Taghizadeh K., Dedon P. C. (2005). Chemical and biological evidence for base propenals as the major source of the endogenous M1dG adduct in cellular DNA. J. Biol. Chem. 280, 25377–25382 [DOI] [PubMed] [Google Scholar]