Abstract
Mutations in several sarcomeric proteins have been linked to various human myopathies. Therefore, having an in vivo developmental model available that develops quickly and efficiently is key for investigators to elucidate the critical steps, components and signaling pathways involved in building a myofibril; this is the pivotal foundation for deciphering disease mechanisms as well as the development of myopathy-related therapeutics. Although striated muscle cell culture studies have been extremely informative in providing clues to both the distribution and functions of sarcomeric proteins, myocytes in vivo develop in an irreproducible 3D environment. Xenopus laevis (frog) embryos are cost effective, compliant to protein level manipulations and develop relatively quickly (≤ a week) in a petri dish, thus providing a powerful system for de novo myofibrillogenesis studies. Although fluorophore-conjugated phalloidin labeling is the gold standard approach for investigating actin-thin filament architecture, it is well documented that phalloidin-labeling can be challenging and inconsistent within Xenopus embryos. Therefore we highlight several techniques that can be utilized to preserve both antibody and fluorophore-conjugated phalloidin labeling within Xenopus embryos for high-resolution fluorescence microscopy.
Keywords: Immunofluorescence microscopy, Xenopus laevis, myofibrillogenesis, phalloidin, sarcomere
1. Introduction
The sarcomere is an ordered, semi-crystalline, macro-molecular complex consisting of thousands of contractile and regulatory proteins that interact with one another to create the smallest contractile unit of striated muscle. Sarcomeric and sarcomeric-associated proteins are involved in numerous cellular processes (e.g., calcium signaling, mechano-sensing and structural maintenance) to facilitate and maintain optimal acto-myosin interactions for muscle contraction. How the dynamic sarcomeric structure is assembled and maintained is currently an active area of investigation. There are a plethora of skeletal and cardiac cell culture studies that have demonstrated the temporal and spatial localization of contractile proteins. Although they have been extremely informative and have provided important insights to deciphering the events occurring during the stages of myofibrillogenesis, they fall short from providing an accurate de novo perspective of muscle development that a growing embryo can provide. Therefore, identifying models that allow for gene and/or protein manipulations within a developing embryo that are amenable to high resolution microscopy is crucial for obtaining direct insights into the functional significance of individual sarcomeric components during myofibril assembly.
For more than 40 years, the Xenopus laevis (frog) embryo has been an excellent model for studying and imaging the dynamic steps of embryonic development in vivo [e.g., 1–5]. Due to the Xenopus embryos’ large size (~1mm), visualization of both gross morphology and subcellular structure during development is relatively straightforward. Since the embryo can develop into a tadpole quickly (within four days at room temperature) and externally (not internally like its mammalian counterparts), investigators can view any stage of embryonic development in less than a week. These features of Xenopus development allow investigators to observe potential effects of a protein knockdown or overexpression at every developmental time point from a single cell to a swimming tadpole. In fact, several groups have used Xenopus embryos or Xenopus myotomal cell cultures to study events occurring during myofibrillogenesis [6–9]. In all, Xenopus laevis is not only an emerging but also an excellent model for assessing protein localization and function during de novo myofibrillogenesis in a developing embryo.
Determining the timing and the localization of where a protein is expressed is an important first step to deciphering the physiological function of a protein. Whole mount immunofluorescence microscopy is an approach that most often uses combinations of unconjugated primary antibodies and fluorophore-conjugated secondary antibodies as molecule-specific probes to localize and image proteins of interest within different regions of the embryo and/or at the subcellular level. Fluorophore-conjugated phalloidin is also frequently used to localize filamentous actin (F-actin) in combination with antibodies generated against actin-binding proteins to provide important clues to how actin-binding proteins may functionally associate with F-actin. Staining for the Z-line component alpha-actinin is often used for identifying the specific stages of myofibrillogenesis [e.g., 10–12]. Informative immunofluorescence studies stem from acquiring clear and highly detailed images from transparent tissue samples. Unfortunately, tissues from Xenopus embryos are greatly pigmented and opaque due to the epidermis and yolk granules, respectively. In fact, younger embryos (stage 32 and younger) tend to have more yolk granules and thus are potentially more challenging with respect to unwanted autofluorescence signals. These features often pose challenges when acquiring images but can be overcome with several techniques that have been developed. Some of these approaches include the addition of a bleaching and/or a clearing step. Bleaching removes the embryo’s pigmentation, while clearing makes the embryo (including the yolk granules) transparent. Transparency is achieved when the embryo’s refractive index is matched by the mounting medium (i.e., Murray’s clear) [13]. Although both approaches are effective in improving image quality, they compromise phalloidin labeling.
Here we discuss several different approaches for preserving both antibody and phalloidin staining in Xenopus embryo skeletal muscle. We highlight a variation of whole mount immunofluorescence staining which entails manually removing the epidermis of the embryo; this bypasses the bleaching step. Another approach is the preparation of cryosections. This method not only preserves the structural integrity of the myofibrils to be analyzed but also greatly increases the penetrance of both antibody and phalloidin into the samples. Finally, we describe a “clearing” technique that markedly improves microscopic analysis and preserves both antibody and phalloidin stains. Fortunately, as outlined above, there are many options that can enhance acquisition of high-resolution images for the study of sarcomeric protein localization in developing Xenopus embryos.
2. Fixation of Embryos
Fixation of embryos is a crucial step in capturing and preserving the fine structural detail of a tissue sample. In the process of holding molecules and structures in place, fixatives can also alter or mask the target antigen, making it poorly or completely unrecognizable by the antibody. Therefore, it is important to select a fixative according to the antibody being used.
After the Xenopus eggs are fertilized and de-jellied [i.e., removal of the gelatinous matrix coat of proteins and carbohydrates that surround the embryo via incubation in a L-cysteine solution in 0.2× Marc’s modified buffer (MMR; pH 7.95) for 4–8 minutes], they are placed into a petri dish filled with 0.2× MMR for further development (see Section 2.1.1 for formula). Embryos develop between 15–25°C (the rate of development is temperature dependent). The development of the embryo is monitored via the use of a stereomicroscope. Once they reach the desired stage, they are fixed in the solution of choice.
There are two general types of fixatives used for immunofluorescence microscopy: alcohol- and aldehyde-based fixatives. In short, alcohol-based fixatives are “denaturing” fixatives due to their ability to reduce the solubility and frequently perturb the tertiary structure of proteins. Aldehyde fixatives function by creating covalent bonds between proteins, often preserving the tertiary structure. The chemistry behind each fixative type is well understood relative to their antigen-alteration and fixing mechanisms of action and has been described previously [e.g., 13–15].
2.1. Fixation Types and Components
There are several aldehyde- and alcohol-based fixative solutions that can be used to fix Xenopus embryos. Some of the aldehyde-based fixatives solutions are MEMPFA (MOPS/EGTA/magnesium sulfate/paraformaldehyde), 4% paraformaldehyde (PFA), 0.37% formaldehyde and Millonig’s fix [16–18]. Alcohol-based fixative solutions include 100% methanol, 1 methanol: 1 acetone, and Dent’s fix [19, 20].
Based on our experience and from others, immunofluorescence staining is often more effective on aldehyde-based fixed embryos than on alcohol-based fixed embryos. It is important to note that the ability of an antibody to recognize its antigen can vary between fixatives. As an example, in our hands, anti-α-actinin antibodies and not anti-tropomodulin antibodies are effective at recognizing their respective proteins in Dent’s-fixed embryos (data not shown). Thus, it is clear that in Dent’s fix, some epitopes are preserved while others are not recognizable.
With respect to the fixatives we tested, MEMPFA was the most effective for antibody and phalloidin use. 0.37% formaldehyde fixed embryos exhibited similar results to MEMPFA; hence it is a viable alternative. Surprisingly, we observed that not all aldehyde-based fixatives that we tested were optimal for both antibody and phalloidin use. For example, Xenopus embryos fixed in Millonig’s Fix, a fix specifically designed for preservation of actin filaments in sea urchins [18], exhibit strong phalloidin labeling but poor antibody staining. Phalloidin staining of alcohol-fixed embryos is well known to be poor since it loses its F-actin binding ability once exposed to methanol [17]. In certain instances, the variability in the contractile state of each sarcomere (i.e., the distance between Z lines) can prove to be challenging for accurate visualization and quantification of specific sarcomeric structures (for example, for high resolution of thin filament pointed ends) hence the use of a “relaxing” buffer (EGTA, K+ and ATP containing buffer) can be utilized before (~30 min incubation) and during the fixation step.
2.1.1. Formulas for Fixatives
Unless mentioned otherwise, fixatives should be prepared fresh before use. All fixatives should be prepared in a chemical fume hood while wearing gloves, goggles and a mask. The institution’s chemical waste disposal policies should be followed.
i. MEMPFA (MOPS/EGTA/magnesium sulfate/paraformaldehyde).
4% paraformaldehyde (PFA)
2mM EGTA
100mM MOPS, pH 7.4 (needs to be protected from light; once yellow, should be discarded).
1mM MgSO4
ii. 4% Paraformaldehyde
4% PFA from an 8% stock (made from PFA prill: Electron Microscopy Sciences Cat#19202)
dH2O at ~50°C
Stir and bring PFA prill into solution with 10N NaOH.
or
8% PFA from a 16% aqueous solution (from Electron Microscopy Sciences Cat#15710)
8% PFA can be stored in an airtight glass bottle at 4°C for ≤ two weeks.
iii. Dent’s Fix [20]
4 methanol (100%):1 dimethyl sulfoxide (DMSO)
iv. 0.37% Formaldehyde Adapted from [6]
0.37% (w/v) formaldehyde (diluted from formalin containing 37% formaldehyde)
80mM NaCl
10mM EGTA
10mM MgCl2
10mM HEPES, pH 7.4
v. Methanol
100% ice-cold methanol
vi. 1 Methanol: 1 acetone
1 methanol (100%): 1 acetone (100%)
vii. Millonig’s Fix [18]
3% formaldehyde (diluted from formalin containing 37% formaldehyde)
136mM NaCl
200mM NaH2PO4, pH 7.4
2.1.2. Relaxing buffer formula (adapted from [16, 21])
100mM MOPS, pH 7.4
1mM MgSO4
5mM EGTA
5mM ATP
2.1.3. Fixation Procedure
Materials
Petri dishes (60×15mm)
Glass storage vials (15×44mm)
2ml plastic transfer pipettes
Stereomicroscope
Protocol
Using a transfer pipet move embryos from 0.2× MMR in a petri dish into labeled glass vials and remove excess liquid.
With a transfer pipette, add fixative solution to each glass vial (substitute relaxing/fixative procedure if desired).
Cap and allow embryos to fix at room temperature for 2–4 hours.
Transfer vials to 4°C overnight.
Store fixed embryos the next day (see below). Avoid over-fixing the embryos (i.e., fixing embryos > 48 hours at room temperature).
2.1.4. Storage of Fixed Embryos
Traditionally, embryo storage entails transferring the fixed embryos to vials filled with 100% methanol at −20°C. This approach is effective at preserving embryos for approximately two months. Although this storage procedure is conducive for many antibody stains, the opportunity to stain with fluorescently-conjugated phalloidin is lost. In order to preserve phalloidin staining, fixed embryos can be stored in a lower percentage fixative (1% MEMPFA) for up to a month at 4°C.
3. Embryonic Tissue Preparation: Whole Mount and Cryosection Approaches
3.1. Tissue Whole Mount Preparation
Whole mount immunohistochemistry (IHC) typically involves staining the entire embryo and this approach takes at least 3 days. We discovered, however, that dissecting the somitic region of the embryo and removing the skin before staining not only increases antibody and phalloidin penetration into the skeletal muscle area (somites), but also reduces the protocol to 1.5 days without compromising the quality of the stain.
3.1.1. Somitic tissue dissecting procedure
Somites develop in the dorso-lateral region of the Xenopus embryo. Under a stereomicroscope, use two pairs of forceps (one in each hand) to dissect somitic tissue from fixed embryos. The No. 5 forceps provide a tip that is helpful in pinning down the tissue. The No. 55 forceps are much sharper with a curved point and are useful for grasping the somite tissue and epidermis during the dissection process.
Materials
Positively charged (poly-lysine coated) glass slides
Nail polish to make slide wells
Petri dishes (60×15mm)
Stereomicroscope
Dissection tools: Forceps No. 5 and No. 55
Plastic 1ml pipette tip box (i.e., humidifying chamber)
200µl pipette and tips
10µl pipette and tips
Reagents
i. Dissection Solution
1× Marc’s Modified Ringers stock (MMR)
0.1M NaCl
2mM KCl
1mM MgSO4
2mM CaCl2
5mM HEPES, pH 7.8
0.1mM EDTA
(Note this is typically diluted to 0.2× for use).
ii. Permeabilization Solution
0.2% Triton-X100 in 1× PBS
Protocol
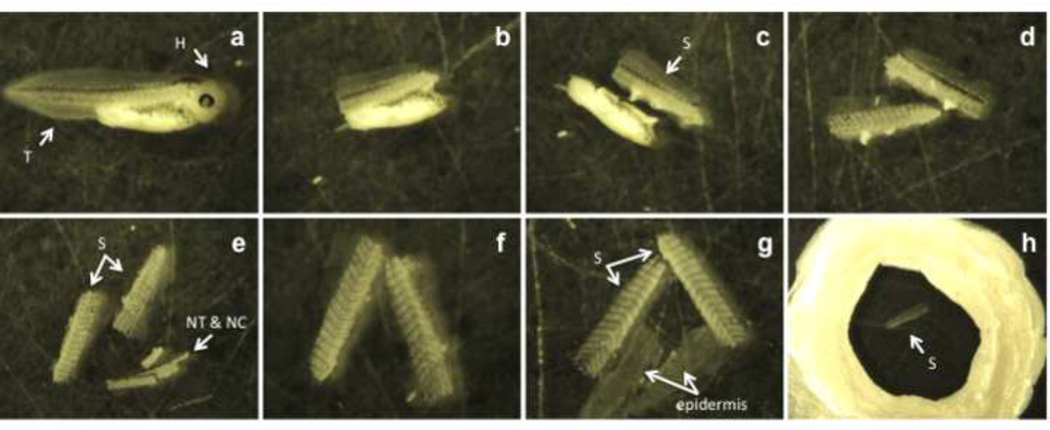
Fill a petri dish with 0.2× MMR and place a fixed embryo into the dish with a transfer pipette (Fig. 1a).
Using both forceps, remove the head and tail region from the trunk of the embryo (Fig. 1b).
Remove the yolky endodermal tissue by cutting just below the somites (Fig. 1c).
Separate the somitic regions on each side of the embryo. To do this, place the remaining trunk of the embryo dorsal side down against the petri dish, so that the somites are “up”. With forceps, split the somites apart, cutting through the center of the dorsal region (Fig. 1d).
Pin the tissue down with the No. 5 forceps, and peel off the notochord (NC) and neural tube (NT) with the No. 55 forceps (Fig. 1e,f). Typically, one somitic side will have the notochord and neural tube attached to it.
Peel the epidermis away from somites with forceps. While pinning down the somites with the No. 5 forceps, the epidermis is peeled away with the No. 55 forceps (Fig.1g); this should be done for both somite pieces. This process may take several attempts. Complete removal of the epidermis is essential since its’ high pigmentation obstructs acquisition of high-quality images (Fig. 4a & 4b).
Fill the circular “slide well” with 50µl of permeabilization solution and place the dissected pieces of somitic tissue into the well (Fig. 1h). Slide wells are created earlier on glass slides by layering several coats of nail polish (minimum of 3) to build the walls of the well (diameter approximately 1.5mm). Each layer is applied after the initial coat dries.
Section 4.1. describes the subsequent immunostaining procedure.
Figure 1. Somitic tissue dissecting procedure.
The fixed stage 40 embryo is placed into a petri dish (a). With forceps, both the head (H) and tail (T) are removed (b). The yolk-filled belly region is separated from the somitic tissue (S) (c). The somites are split into two pieces (d). One somitic side will have the neural tube (NT) and notochord (NC) still attached and both structures are removed with forceps (e, f). The epidermis is peeled away from the somitic regions (g). The somitic tissue (S) is placed into a well formed from nail polish for immunostaining (h).
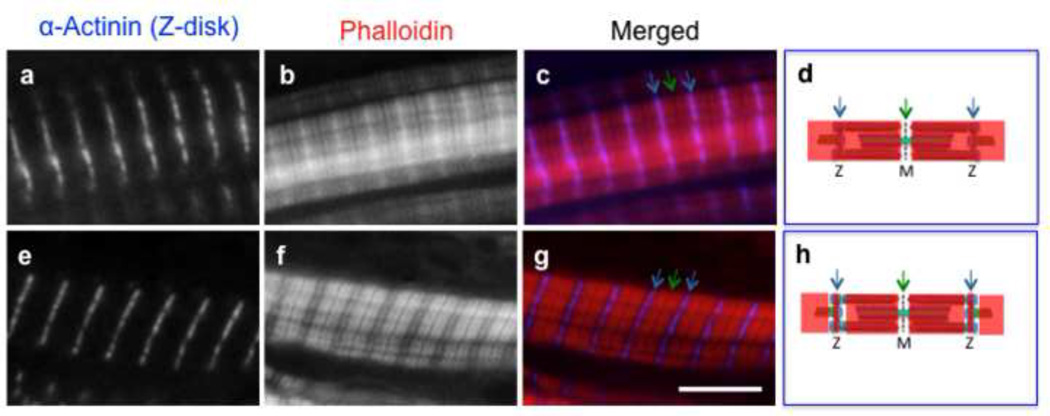
Figure 4. Two different phalloidin staining patterns are observed in somitic tissue.
Fluorescently-labeled phalloidin can exhibit two distinct staining patterns in the Xenopus muscle cells. Green arrows point to the Z-disk (Z) and the blue arrows point to the M-line (M) (c, d, g, h). For whole-mounts, phalloidin exhibits a staining pattern that spans the length of the actin-thin filament (b, d). Phalloidin will also exhibit a staining pattern that spans the length of the actin-thin filament except for a region within the I-band in close proximity to the Z-disc (f, h). With cryosections, fluorophore-conjugated phalloidins consistently stain the entire actin-thin filaments (Fig. 5, j–l). Scale bar is 5 µm.
3.2. Tissue Cryosection Preparation
As an alternative to whole mount staining, we have found that utilizing frozen sections of developing Xenopus embryos preserves outstanding structural details of developing muscle. Although individual sections only provide a snapshot (single slice) within an embryo (in comparison to whole mounts), it does enhance antibody and phalloidin access. This approach also allows for the use of higher resolution (high numerical aperture (NA)) objectives since the working distance between the sample and the objective can be reduced (compared with whole mounts). Here, we describe a cryosection preparation optimized in our laboratory based on protocols described previously [22, 23].
Embryos are fixed as described (Section 2.1.2). Although embryos that have been fixed for days to weeks can be used for this procedure, the staining quality of stored embryos is often compromised (e.g., lower signal to noise ratio) in comparison with samples that are processed immediately after fixation. Gelatin-coated coverslips need to be prepared before sectioning [24]. Once samples are frozen, trained personnel on a cryostat can section them. For further details on cryosection preparation and sectioning, see reviews by [25, 26].
Materials
2ml plastic transfer pipettes
Positively charged (poly-lysine coated) glass slides
Stereomicroscope
Embryo dissection tools: Forceps No. 5 and No. 55
Bench top liquid nitrogen container
Metal Ladle (~4” diameter)
Aluminum foil
Tissue-Tek Cryomolds (25×20×5mm) (Sakura® Finetek)
Delicate task wipers (e.g., most commonly used are Kimwipes from Kimberly-Clark)
Gelatin-coated (18×18mm) coverslips (See [24] for preparation, or commercially available from NeuVitro, Cat# GG-18-gelatin)
0.5 ml micro-centrifuge tubes
Reagents
Isopentane
Liquid nitrogen
30% sucrose in 1× PBS
Optimal cutting temperature (OCT) freezing medium (Sakura® Finetek)
Protocol
Rinse fixed embryos with 1× PBS. Embryos can be stored in 1× PBS at 4°C up to a week before moving on to step 2.
Incubate embryos overnight in 30% sucrose/PBS at 4°C. Sucrose acts as a cryoprotectant. Ensure embryos sink to the bottom or assist with a quick shake; this indicates that the embryos are saturated with sucrose. If the embryos remain afloat, more time is needed in the sucrose solution. Typically, embryos stage 37 or younger, require two days of saturation.
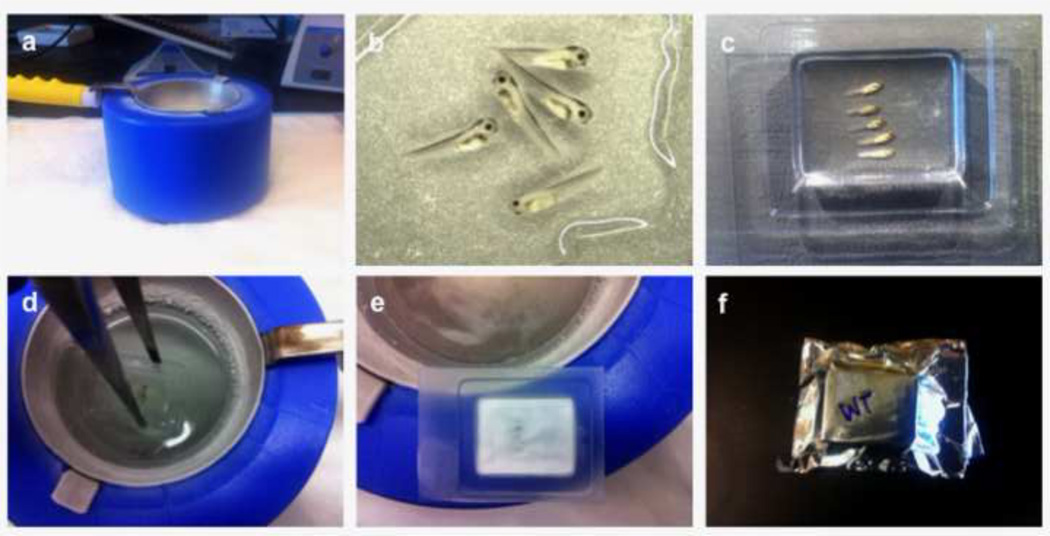
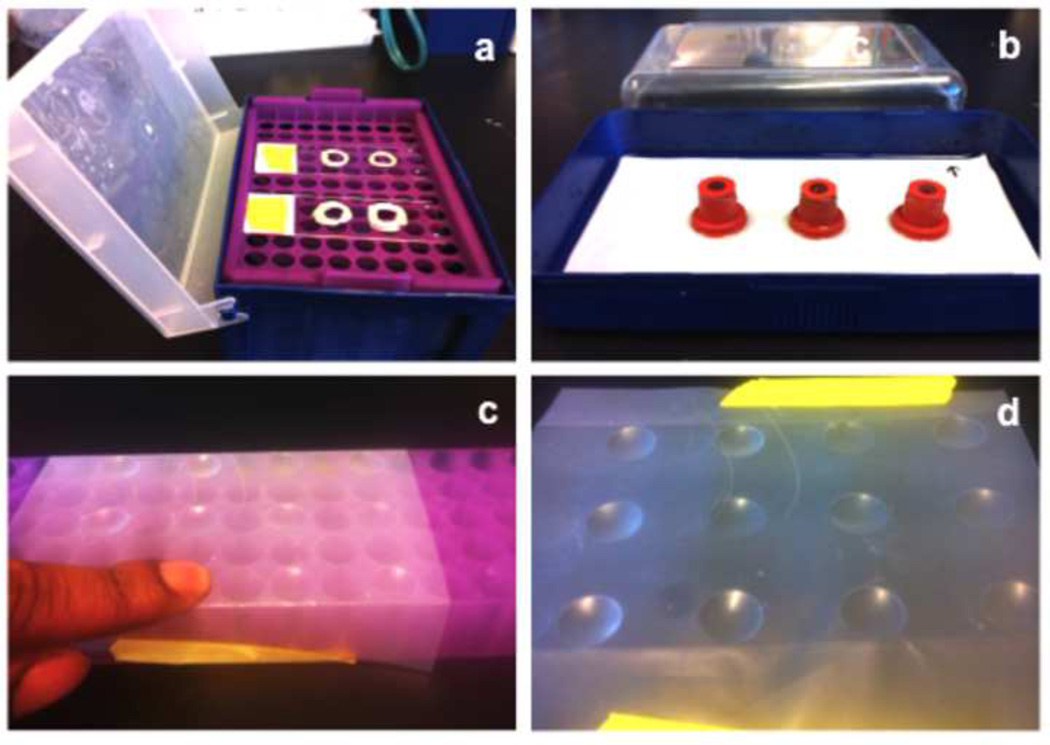
Set up the isopentane/liquid nitrogen ice bath. Fill container with liquid nitrogen. Place the ladle into the nitrogen-filled container and fill ladle with isopentane. Let the liquid nitrogen cool the isopentane. Ice sludge will form at the bottom of the ladle indicating that the isopentane has reached the optimal temperature for freezing samples (Fig. 2a). Alternatively, isopentane can be stored at −80°C until use.
Transfer 3–4 embryos onto a glass slide with a plastic transfer pipette. Absorb as much of the sucrose as possible with a kimwipe. Cover the embryos with OCT freezing medium (Fig. 2b).
Place a drop of OCT medium into a labeled cryomold. Do not fill mold up.
Transfer embryos from the glass slide into the cryomold with the OCT medium. Using forceps, gently lift the embryos one at a time and place them in the cryomold. Position the embryos on their side for sagittal sections. Align the embryos, ensuring that they are flat against the bottom of the mold. Fill mold with OCT (just enough to completely cover the embryos) (Fig. 2c). Re-align the embryos with forceps if necessary. Ensure that the cryomold is bubble free (i.e., bubbles can damage tissue during cryosections).
Freeze the samples. Immerse the cryomold with embryos into the cooled isopentane and hold until the OCT turns opaque (white). Hold for another thirty seconds to ensure the sample is completely frozen (Fig. 2d, e). Freeze-thaws will cause tissue damage.
Store samples in aluminum foil. Allow for a quick air dry of frozen samples before placing them onto a piece of foil. Wrap up samples tightly (Fig. 2e).
Store the aluminum foil-wrapped frozen samples. Place wrapped samples in a plastic bag. Remove as much air as possible from the storage bag. Store at −80°C (Fig. 2f).
Section samples in cryostat. Have a trained person generate ~5 µm thick sections and “melt” them onto gelatin-coated coverslips.
Store sections in 1× PBS at 4° C, until ready for staining. For longer storage, keep sections well wrapped in foil in an airtight container at −80°C until use. For immunostaining of cryosections, see Section 4.2.
Figure 2. Cryosection preparation.
A liquid nitrogen bath setup with an isopentane filled ladle (a). 4–5 embryos are cleared of all residual sucrose solution and covered with OCT freezing medium (b). The embryos are transferred to, and aligned in a cryomold (c) and then immersed into the isopentane bath (d) until they turn opaque (e). Samples are quickly allowed to dry, wrapped in aluminum foil and stored (e, f).
4. Immunostaining
Antibody stains not only provide important information about the localization of endogenous proteins but also can be used to monitor the localization of exogenous proteins produced in the embryo by mRNA injection. Most importantly, deliberate alterations in protein levels may result in perturbations of sarcomere structure, leading to important clues about a protein’s function that can be assessed by immunofluorescence staining. Fortunately, several groups engaged in deciphering the roles of sarcomeric proteins during striated muscle development have demonstrated that many antibodies raised against human, chicken or mouse sarcomeric proteins robustly recognize Xenopus orthologues due to the high conservation of contractile proteins across species [6–9, 27–36]. See Table 1 for a list of commercial anti-sarcomeric protein antibodies that recognize Xenopus skeletal muscle proteins. It is worth noting that antibodies raised against “cardiac-specific” isoforms (i.e., α-cardiac actin) may recognize a similar antigen in skeletal muscle. For example, anti-α-cardiac actin antibodies (clone Ac1-20.4.2 from Sigma-Adrich) effectively recognize the actin-thin filaments in Xenopus skeletal myotomal cells.
Table 1. Some primary antibodies against sarcomeric proteins that cross-react with Xenopus orthologues.
In most cases the concentrations of the antibodies are not given, thus only the working dilutions for whole mount IHC are provided.
| Antibody | Species | Supplier & Catalog No# |
Final Dilution |
Reference |
|---|---|---|---|---|
| Anti-α-actinin EA-53 | Mouse monoclonal | Sigma A7811 | 1:200 | [33] |
| Anti-α-cardiac actin Ac1-20.4.2 | Mouse monoclonal | American Research Products 03-61075 | 1:400 | [34] |
| Anti-pan α-actin | Rabbit polyclonal | Sigma A 2103 | 1:200 | [35] |
| Anti-actin 5C5 | Mouse monoclonal | Sigma A2172 | 1:500 | [27] |
| Anti-α-skeletal actin JLA20 | Mouse monoclonal | Developmental Studies Hybridoma Bank | unknown | [28] |
| Anti-α-tropomyosin TM311 | Mouse monoclonal | Sigma T 2780 | 1:200 | [36] |
| Anti-tropomyosin CH1 | Mouse monoclonal | Developmental Studies Hybridoma Bank | 1:50 | [9] |
| Anti-leiomodin | Rabbit polyclonal | Proteintech 14948-1-AP | 1:400 | |
| Anti-titin 9D10 | Mouse monoclonal | Developmental Studies Hybridoma Bank | unknown | [6] |
| Anti-myosin F59 | Mouse monoclonal | Developmental Studies Hybridoma Bank | 1:50 | [6] |
| Anti-myosin MF20 | Mouse monoclonal | Developmental Studies Hybridoma Bank | 1:1 | [29] |
| Anti-myosin IIa 56-396-5 | Mouse monoclonal | Developmental Studies Hybridoma Bank | 1:300 | [30] |
| Anti-cofilin2 | Rabbit polyclonal | Millipore 07-300 | 1:300 | [31] |
| Anti-phospho cofilin1 | Rabbit polyclonal | Santa Cruz SC-12912-R | 1:3000 | [31] |
| Anti-phospho cofilin2 | Rabbit polyclonal | Abcam AB14134 | 1:300 | [31] |
| Anti-desmin DB-E-5 | Mouse monoclonal | Millipore MAB3430 | 1:200 | [32] |
| Anti-cardiac troponin T CT3 | Mouse monoclonal | Developmental Studies Hybridoma Bank | 1:10 | [9] |
| Anti-capZ 1E5.25 | Rabbit polyclonal | Developmental Studies Hybridoma Bank | unknown | [6] |
Immunofluorescence staining approaches allow investigators the ability to detect and gather information on multiple target antigens within a cell and/or tissues. Fluorescently conjugated-secondary antibodies are one of many ways to visualize a target protein. Described below is a straightforward “single” stain. However, most often double or triple localization of different molecules is required; these approaches take advantage of the wide range of fluorophores available as well as the different species that antibodies are generated in. The protocols provided below can be adapted seamlessly for multiple stain approaches. Specifically, multiple labeling can be done sequentially or in combination (e.g., mixing the primary antibodies).
Note that primary and secondary antibodies have to be carefully titrated since each lot can differ with respect to its concentration and fluorophore coupling efficiency. Additionally, “secondary alone” control samples (i.e., samples where the primary antibodies are not used) should be performed to determine optimal working concentrations; that is, dilutions that do not result in nonspecific “background” signals.
Fluorescently-conjugated phalloidin can also be added to any of the staining protocols listed below. Note, most vendors distribute fluorophore-conjugated phalloidin in a lyophilized form with instructions to re-suspend and store the lyophilized product in 100% methanol. Therefore to achieve optimal actin-filament staining, it is important to evaporate off the methanol before diluting the phalloidin. Dilute stock in wash buffer to a final concentration of 0.001 unit/µl for whole mounts and 0.0005 unit/µl for sections.
From our experience, the staining patterns demonstrated by fluorophore-conjugated phalloidins on Xenopus somitic tissue prepared in whole mount is often dependent upon how the sample was prepared. In dissected somitic tissue, phalloidin exhibit either a staining pattern that spans the entire actin-thin filament or a staining pattern that is devoid of detectable staining within the I band (on each side of the Z-disc) (Fig. 4). Interestingly with sections, fluorophore-conjugated phalloidins consistently stain the length of the actin-thin filaments, likely due to better accessibility (Fig. 5j–l).
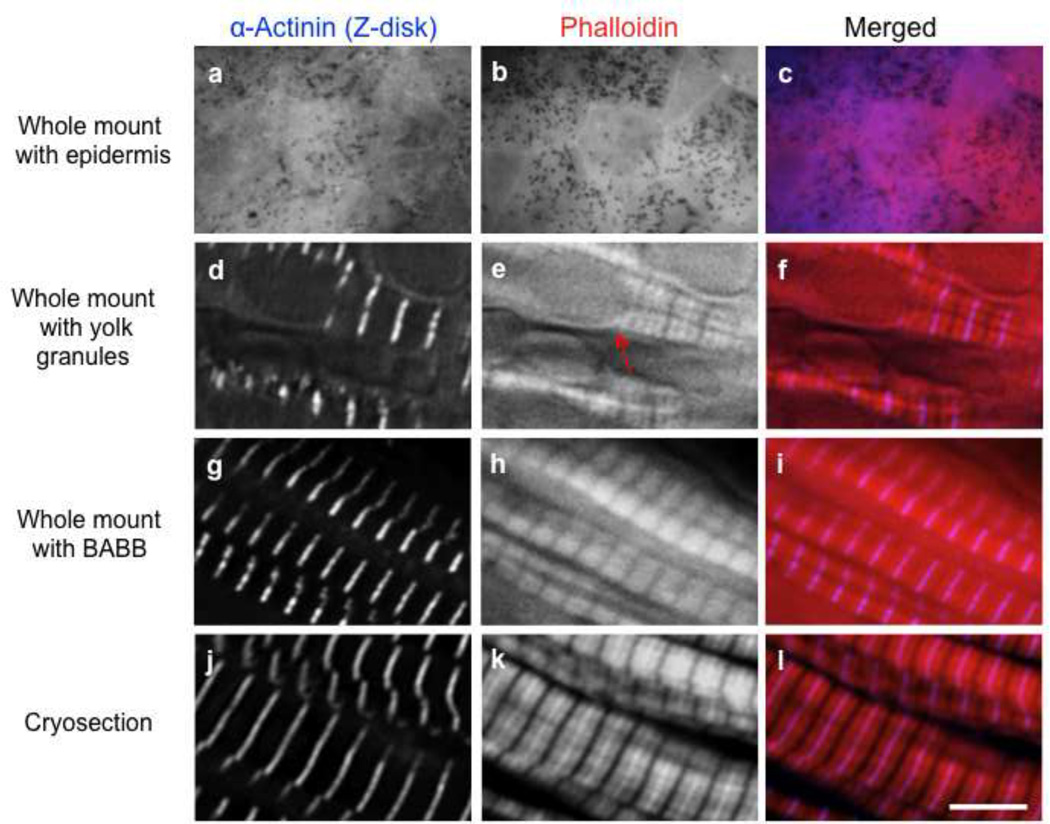
Figure 5. Comparison of stains from whole-mount, cryosection and BABB approaches.
When the epidermis is left on the whole-mount tissue, it obstructs the observation of myofibrils/sarcomeric proteins (a–c). Whole-mounts free of the epidermis exhibit staining for sarcomeric proteins (d–f) but yolk granules remain visible and obstruct subcellular structures (e, arrow). After staining, clearing somitic tissue with BABB masks yolk platelets; phalloidin labeling is preserved (g–i). Cryosections are superior for staining of actin filaments and sarcomeric proteins (j–l). Scale bar is 5 µm.
4.1. Whole Mount Immunostaining Protocol
The approach described here is a cost effective way to do whole mount immunostaining on Xenopus embryo tissues. The entire immunostaining procedure is carried out in a slide well (Fig. 1h). To prevent the tissue from drying out during overnight incubations, slide wells are placed into a humidifying chamber constructed from a 1ml pipette tip box with a cover that is filled (half-way) with water. The slide wells are placed on top of the tip rack with tissue samples inside the wells (Fig. 3a).
Figure 3. Setup for whole-mount immunohistochemistry (IHC), cryosection IHC and BABB preparation.
For whole-mount IHC, the entire procedure is executed in a nail polish slide well (a). For cryosection IHC, sections on coverslips are incubated on rubber stoppers in a moist covered chamber (b). For BABB preparation of the somitic tissue, the isopropanol washes are carried out in parafilm wells (c, d).
As mentioned above, fluorophore-conjugated phalloidin can be added to this protocol, following any wash step. If the phalloidin stain does not appear to be optimal, transfer tissue samples into 0.5ml micro-centrifuge tubes filled with 100µl of the diluted phalloidin solution. Wrap tubes in aluminum foil to protect from the light and rotate on a neutator for two hours. Afterwards, remove tissue samples from each tube and place back into the slide wells for washes.
Materials
Charged slides with nail polish wells (described above)
1ml pipette tip box (humidifying chamber)
200µl pipette and tips
Forceps
Coverslips (18x18mm)
Stereoscope
Solutions
i. Permeabilization solution
0.2% Triton-X100 in 1× PBS
ii. Blocking solution
2% BSA plus 1% normal donkey serum in 1× PBS
iii. Wash solution
0.02% Triton-X 100 in 1× PBS
iv. Antibody solutions
Dilute primary antibodies (see Table 1) or secondary fluorescently-conjugated antibodies in blocking solution.
v. Mounting medium specialized for immunofluorescence microscopy (e.g., Aqua-Poly/Mount from Polysciences, Inc.)
Protocol
Permeabilization. This is a continuation from step 8 of 3.1.1 (somitic tissue dissecting procedure). Incubate in permeabilization solution for at least one hour at room temperature.
Blocking. Remove permeabilization solution and add blocking solution (50µl) to each well. Block for at least 2 hours at room temperature. If nonspecific background is a problem, the samples can be blocked initially at 37°C for 30min, prior to the 2 hour room temperature incubation. This step is used to prevent non-specific binding of antibodies by binding to any “sticky” parts of the cells/tissue that are being stained.
Primary Antibody. Remove as much blocking solution from the wells as possible to prevent dilution of the subsequent primary antibodies. Add 50µl of the diluted primary antibodies to each well. Close humidifying chamber to prevent samples from drying out and incubate overnight at 4°C. If time is limited, immunofluorescence staining can be successful with a one-hour incubation in primary antibody; this is a viable option for antibodies with high affinity and/or when the antigen is abundant.
First Wash. Rinse tissue of excess primary antibodies with five 50µl washes, each for 5 minutes. Be careful when adding and removing liquid from the wells. If not, tissue will be lost.
Secondary Antibody. Remove as much washing solution as possible and add 50µl of the secondary antibody solution to each well. Close humidifier and cover samples to prevent exposure to light. Incubate tissue for at least 45 minutes at room temperature.
Second wash. Wash as described for First Wash.
Continue protocol or go to 4.1.1 for Clearing Procedure.
Preparation for Mounting of Stained Tissue. Place a glass slide on the platform of the stereomicroscope. Adjust the light source to best view the tissue. Using a 200µl pipette, transfer the tissue to the center of the slide. At this stage, remove any remaining epidermis with forceps. Gently shear apart the somitic tissue with the forceps. This flattens the tissue to be viewed and thus, enhances imaging of myofibrils. Remove as much wash buffer as possible as to not dilute the mounting medium (i.e., this will cause premature fading of the fluorescent signal). Rinse tissue with dH2O to clear of excessive salts from wash buffer (i.e., this will reduce background when imaging).
Final Mounting Steps. Place ~4 drops of mounting media in close proximity, surrounding the stained tissue. Gently place the coverslip on top of the tissue. Once the coverslip has been laid down, with forceps tap the four corners of the coverslip. This approach removes air bubbles and keeps the tissue centered under the coverslip. Dry the mounted slides away from light. For long-term storage, place slides at 4°C in the dark.
4.1.1. Clearing Procedure
As mentioned above, it is difficult to acquire clear images from Xenopus embryos due to the abundance of yolk granules that auto-fluoresce. Implementing a clearing step before imaging can alleviate this problem. The clearing protocol includes dehydrating the tissue after staining, with a series of alcohol washes. Methanol is often the method of choice (over other alcohols) since there is little or no shrinkage of the tissue; however, the ability to stain with phalloidin is compromised. Isopropanol can replace methanol, and if done quickly, phalloidin labeling is preserved [17]. The unfortunate tradeoff of using isopropanol is that there is significant tissue shrinkage.
Materials
2ml Plastic transfer pipette
Positive charged (poly-lysine coated) glass slides
Stereomicroscope
Coverslips (18×18mm)
10µl and 200µl pipettes and tips
1.5ml microcentrifuge tube rack
Aluminum foil
Self-sealing film (e.g., Parafilm® M)
Kimwipes
Permanent Marker
Solutions
Alcohol Dilution Series (these can be stored and reused)
100% 95% 85% 70%, 50%, and 25% methanol (or isopropanol)
Clearing solution
1 benzyl alcohol: 2 benzyl benzoate (BABB): protect from light and make fresh each time. BABB is highly corrosive and toxic.
Protocol
Create wells for alcohol dehydration. Place a piece of parafilm on top of a 1.5 ml plastic tube rack and tape it in place (Fig. 3c,d). Use fingers to press the parafilm into the holes of the tube rack (do not break the parafilm). This will create “wells”. If necessary, leave empty wells between the alcohol solutions to prevent inadvertent mixing.
Dehydration. Each incubation is 15 seconds long. Transfer between wells using a 200µl pipette. The series is as follows: 25%, 50%, 70%, 85%, 95%, 100% and 100%. Ensure all water is extracted from the tissue. If there is residual water, it will mix with the subsequent clearing solution (see below) and create a precipitate that renders the tissue opaque.
Transfer dehydrated tissue. With a 200µl pipette, transfer tissue to a marked area on a prepared charged slide under a stereomicroscope. Remove excess 100% isopropanol from the tissue. To prepare the slide, prepare draw a circle to designate the location for the tissue to be placed.
Clear the tissue. Add 2–5µl of BABB to the tissue and incubate for 5 min. Repeat three times with fresh BABB solution. Watch the tissue “disappear”. The circle helps with identifying the location of the tissue once it becomes transparent. Finally, add 5–10µl of BABB to the tissue for mounting.
Place a coverslip onto the tissue. Remove excess BABB surrounding coverslip with a kimwipe. Seal the coverslip with nail polish. Let mounted slides air dry away from light.
4.2. Cryosection Immunostaining Protocol
The immunostaining approach for cryosections follows a similar protocol as for whole mount staining: the solutions are the same, while some different materials are used. Unlike the whole mount approach, cryosections have increased tissue and antigen accessibility; therefore antibodies can be used more dilute and for shorter incubation times. For example, the anti-sarcomeric α-actinin antibody (Sigma-Aldrich) is used at a dilution factor of 1:3000 with an overnight incubation period when conducting whole mount immunofluorescence staining. Conversely with staining sections, the anti-α-actinin antibody works well at 1:6000 with an incubation time of one hour.
As with whole mount staining, fluorescently-conjugated phalloidin can be added or substituted into the protocol. Specifically, add 50µl of diluted phalloidin solution onto the coverslip and incubate for two hours at room temperature. Cover the humidifying chamber to protect coverslips from light.
Materials
Standard 6-well cell culture plate, non-treated, flat-bottom with lid
200µl pipette and 1ml pipette with tips
Forceps
Humidifying chamber (container with lid)
Rubber stoppers (flanged rubber stoppers for vaccine bottles) (Plasticoid Corp: Cat#20190)
beaker
Filter paper (0.34mm)
Parafilm
Kimwipes
Slides
Mounting Medium for Fluorescence Microscopy
Light Box
Solutions
Same as described above for Whole Mount staining without Clearing
Permeabilization. Place one coverslip (section facing up) into each well of a 6 well culture dish with forceps. Gently fill wells with 2 ml of permeabilization solution. Incubate for 15 minutes at room temperature on a light box (i.e., this step photo-bleaches the tissue, often reducing autofluorescence from the tissue, particularly the yolk granules). Check and make sure coverslips do not float to the top (sections will dry out and compromise image quality if not submerged). If they float up, poke down with forceps.
Blocking. Remove permeabilization buffer and add 2ml of blocking solution to each well. Incubate for 45 minutes at room temperature on a light box.
Primary antibody. Set up the humid chamber (Fig. 3b). Place filter paper into a plastic container (humid chamber) and wet it completely with water (pour off excess water). Place the rubber stoppers on top of the wet filter paper. The wet filter paper holds the rubber stoppers in place. With a pair of forceps, remove the coverslip from the well, remove excess blocking buffer by touching a corner of the coverslip on a kimwipe and place the coverslip face-up on top of a rubber stopper. Note, care must be taken to assure that sections remain on coverslips during the staining procedure. Add 50µl of primary antibody solution onto the coverslip. Incubate for one hour at room temperature (or overnight at 4°C). For overnight incubations, parafilm the humidifier to prevent sections from drying out.
First wash. Remove primary antibody by touching edge of coverslip to a Kimwipe. Place the coverslips back into 6-well cell culture dish for washing steps. Wash with 2ml of wash buffer, 3 times for 5 minutes each.
Secondary Antibody. Transfer the coverslip from the six well plate to the rubber stoppers in the humid chamber. Remove excess wash buffer and place the coverslip on top of the rubber stopper. Add 50µl of the secondary antibody solution onto each coverslip. Cover humidifying chamber to protect coverslips from light. Incubate 45 minutes at room temperature.
Second wash. Remove secondary antibody by touching edge of coverslip to a Kimwipe. Wash as described for First Wash.
Rinse coverslip several times in beaker filled with dH2O. Remove excess dH2O from the coverslip with a Kimwipe.
Place a glass slide onto the platform of the stereomicroscope. Add a single drop of mounting medium onto the slide. Work quickly to prevent mounting medium from drying. Drying reduces the medium’s spreading ability and will prevent complete immersion of the tissue.
Place coverslip, section down, onto mounting medium drop. Gently press the coverslip against the glass slide with the back-end of the forceps to ensure equal spreading of the mounting medium around the sample and to remove trapped air. Let mounted slides air dry away from light.
5. Results
Traditionally, bleaching and clearing approaches have been useful in optimizing immunofluorescence imaging of Xenopus embryos. The methods we describe here illustrate different approaches investigators can utilize for optimal imaging of proteins and filamentous actin structures within Xenopus somitic tissue. If removing the epidermis is done correctly (Fig. 1h), the whole mount dissection yields intact tissue and clear visualization of sarcomeric components [e.g., α-actinin and phalloidin (Fig. 5d,e)]. If the epidermis is not removed properly, it will obstruct viewing of the myofibrils within the somitic tissue (Fig. 5a,b). The presence of abundant yolk granules can also be detrimental for imaging of the myofibrils. This is especially true for stage 32 and younger embryos, where large numbers of yolk granules can block the visualization of subcellular structures and add to the autofluorescent background (Fig. 5e). Clearing tissue with an isopropanol dehydration/BABB protocol after immunofluorescence staining effectively masks yolk granules, and preserves phalloidin labeling (Fig. 5g,h). Unfortunately, somitic tissue undergoes shrinkage after the isopropanol/BABB process therefore compromising quantitative analysis of any subcellular structure. Of the plethora of protocols tested by our laboratory, we found that generating Xenopus embryo cryosections was the best approach for optimal imaging of developing muscle tissue. Because the cryosection technique yields a ~5µm section of the embryo, the amount of autofluorescence from other structures is greatly reduced. This advantage along with the enhancement of tissue penetrance results in robust and clear imaging of sarcomeric components within the somitic tissue (e.g., Fig. 5j,k).
Although this article focuses primarily on studying myofibril assembly in developing skeletal muscle tissue, these techniques can affectively be applied to studying myofibril assembly during heart development. However, while many cardiac studies in Xenopus have primarily focused on elucidating the molecular network leading to cardiac induction or capturing the morphological transformation of the myocardial tube into a threechambered heart [e.g., 9, 37–38], there is great potential for assessing the temporal and spatial localization of sarcomeric proteins at the subcellular level [9, 39].
6. Concluding Remarks
One of the most fascinating examples of cytoskeletal assembly is the sarcomere; perhaps the most highly ordered macromolecular structure in eukaryotic cells. When striated muscle cells differentiate, thousands of structural and regulatory molecules assemble to the precision of single molecules. In the last several decades, significant progress has been made in understanding the complex process involved in discerning the steps and specific molecules required for myofibrillogenesis. Initial investigations stemmed from remarkable, in-depth electron microscopy studies that allowed investigators to visually identify the stages involved in myofibrillogenesis, without the identification of most molecules involved [e.g., 40–42]. These findings were followed by a plethora of immunofluorescence and biochemical studies in isolated myocytes (from rat, mouse, chick and frog) and various model organisms including Drosophila melanogaster, zebra fish, C. elegans as well as the gold standard of many laboratories, genetically altered mice. With these studies providing an important framework for understanding the molecular players involved in myofibrillogenesis and the Xenopus laevis embryo in place as an in vivo developmental model of de novo myofibril assembly, the field is now poised to investigate the mechanisms by which perturbations in specific sarcomeric components lead to the development of human myopathies. The optimized protocols for preparing embryonic Xenopus somitic tissue, phalloidin labeling and immunofluorescence staining for high resolution imaging presented in this publication lay a solid foundation for these mechanistic, disease-related investigations in the Xenopus embryo.
Highlights.
Staining protocols were optimized to study myofibrillogenesis in Xenopus embryos.
A dissection approach was developed for ideal staining of Xenopus somitic regions.
Optimized phalloidin labeling and immunostaining for Xenopus embryo cryosections.
Demonstrated methods to preserve phalloidin labeling after Murray’s clear.
Acknowledgements
We thank members of the Gregorio and Krieg laboratories for thoughtful discussions on the techniques presented here, especially Daniel Schnurr and Candace Myers. CCG would also like to thank Elizabeth Repasky (Roswell Park Cancer Center, Buffalo, NY) for developing the original frozen section immunofluorescence staining protocol that we adapted for staining Xenopus embryos. PAK is supported by the Sarver Heart Center and by the NIH grant HL093694. CCG is supported by NIH grants HL108625 and HL083146. CUN is supported by More Graduate Education at Mountain States Alliance (MGEMSA) and Western Alliance to Expand Student Opportunities (WAESO).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Muntz L. Myogenesis in the trunk and leg during development of the tadpole of Xenopus laevis (Daudin 1802) J. Embryol. Exp. Morph. 1975;33:757–774. [PubMed] [Google Scholar]
- 2.Boucaut J, Thierry D. Fibronectin in early amphibian embryos. Cell Tissue Res. 1983;234:135–145. doi: 10.1007/BF00217407. [DOI] [PubMed] [Google Scholar]
- 3.Fagotto F, Gumbiner BM. Beta-catenin localization during Xenopus embryogenesis: accumulation at tissue and somite boundaries. Development. 1994;120:3667–3679. doi: 10.1242/dev.120.12.3667. [DOI] [PubMed] [Google Scholar]
- 4.Schohl A, Fagotto F. Beta-catenin, MAPK and Smad signaling during early Xenopus development. Development. 2000;129:37–52. doi: 10.1242/dev.129.1.37. [DOI] [PubMed] [Google Scholar]
- 5.Kieserman EK, Lee C, Gray RS, Park TJ, Wallingford JB. High-magnification in vivo imaging of Xenopus embryos for cell and developmental biology. Cold Spring Harbor Protocols. 2010;5 doi: 10.1101/pdb.prot5427. pdb-prot5426. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Cook JD, Terry M, Spitzer NC, Ferrari MB. Calcium transients regulate patterned actin assembly during myofibrillogenesis. Dev. Dyn. 2004;229:231–242. doi: 10.1002/dvdy.10428. [DOI] [PubMed] [Google Scholar]
- 7.Campbell NR, Podugu SP, Ferrari MB. Spatiotemporal characterization of short versus long duration calcium transients in embryonic muscle and their role in myofibrillogenesis. Dev. Biol. 2006;292:253–264. doi: 10.1016/j.ydbio.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 8.Geach TJ, Zimmerman LB. Paralysis and delayed Z-disc formation in the Xenopus tropicalis unc45b mutant dicky ticker. BMC Dev. Biol. 2010;10:75. doi: 10.1186/1471-213X-10-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolker SJ, Tajchman U, Weeks DL. Confocal imaging of early heart development in Xenopus laevis. Dev. Biol. 2000;218:64–73. doi: 10.1006/dbio.1999.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang SM, Greaser ML, Schultz E, Bulinski JC, Lin JJ, Lessard JL. Studies on cardiac myofibrillogenesis with antibodies to titin, actin, tropomyosin, and myosin. J. Cell Biol. 1988;107:1075–1083. doi: 10.1083/jcb.107.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dukhee R, Sanger JM, Sanger JW. The premyofibril: evidence for its role in myofibrillogenesis. Cell Motil. Cytoskel. 2005;28:1–24. doi: 10.1002/cm.970280102. [DOI] [PubMed] [Google Scholar]
- 12.Schultheiss T, Lin ZX, Lu MH, Murray J, Fischman DA, Weber K, Masaki T, Imamura M, Holtzer H. Differential distribution of subsets of myofibrillar proteins in cardiac nonstriated and striated myofibrils. J. Cell Biol. 1990;110:1159–1172. doi: 10.1083/jcb.110.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klymkowsky MW, Hanken J. Whole mount staining in Xenopus and other vertebrates. In: Kay BK, Peng HB, editors. Methods in Cell Biology. Vol. 36. San Diego: Academic Press Inc.; 1991. pp. 419–441. [DOI] [PubMed] [Google Scholar]
- 14.French D, Edsall JT. The reactions of formaldehyde with amino acids and proteins. In: Edsall ML, Anson JT, editors. Advances in Protein Chemistry. Vol. 2. New York: Academic Press Inc.; 1945. pp. 277–335. [Google Scholar]
- 15.Pearse AGE. Histochemistry: theoretical and applied. Am. J. Med. Sci. 1961;241:136. [Google Scholar]
- 16.Sive HL, Grainger RM, Harland RM. Early development of Xenopus laevis: a laboratory manual. Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 17.Strickland L, Dassow G, Ellenberg J, Foe V, Lenart P, Burgess D, Charles A, Ettensohn GAW, Gary MW. Light Microscopy of Echinoderm Embryos. In: Ettensohn CA, Wray GA, Wessel GM, editors. Methods in Cell Biology. Vol. 74. San Diego: Elsevier Academic Press Inc.; 2004. pp. 371–409. [DOI] [PubMed] [Google Scholar]
- 18.Millonig G. A study on the formation and structure of the sea urchin spicule. J. Submicrosc. Cytol. 1970;2:157–165. [Google Scholar]
- 19.Klymkowsky MW, Maynell LA, Polson AG. Polar asymmetry in the organization of the cortical cytokeratin system of Xenopus laevis oocytes and embryos. Development. 1987;100:543–557. doi: 10.1242/dev.100.3.543. [DOI] [PubMed] [Google Scholar]
- 20.Dent JA, Polson AG, Klymkowsky MW. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development. 1989;105:61–74. doi: 10.1242/dev.105.1.61. [DOI] [PubMed] [Google Scholar]
- 21.Gokhin D, Kim N, Lewis S, Hoenecke HR, D’Lima DD, Fowler VM. Thin-filament length correlates with fiber type in human skeletal muscle. Am. J. Physiol. Cell. 2012;302:555–565. doi: 10.1152/ajpcell.00299.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown D, Martz SN, Binder O, Goetz SC, Price BMJ, Smith JC, Conlon FL. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development. 2005;132:553–563. doi: 10.1242/dev.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer AH, Jacobson KA, Rose J, Zeller R. Cryosectioning Tissues. Vol. 8. Cold Spring Harbor Protocols; 2008. pdb-prot4991. [DOI] [PubMed] [Google Scholar]
- 24.Fischer AH, Jacobson KA, Rose J, Zeller R. Preparation of slides and coverslips for microscopy. Vol. 5. Cold Spring Harbor Protocols; 2008. pdbprot4988. [DOI] [PubMed] [Google Scholar]
- 25.Webster P. The production of cryosections through fixed and cryoprotected biological material and their use in immunocytochemistry. In: Hajibagheri N, editor. Methods in Molecular Biology. Vol. 117. Totowa, NJ: Humana Press Inc.; 1999. pp. 49–76. [DOI] [PubMed] [Google Scholar]
- 26.Tokuyasu KT. A technique for ultracryotomy of cell suspensions and tissues. J. Cell Biol. 1973;57:551–565. doi: 10.1083/jcb.57.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vernon AE, Philpott A. A single cdk inhibitor, p27Xic1, functions beyond cell cycle regulation to promote muscle differentiation in Xenopus. Development. 2003;130:71–83. doi: 10.1242/dev.00180. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz LM, Kay BK. Differential expression of the Ca2+ -binding protein parvalbumin during myogenesis in Xenopus laevis. Dev. Biol. 1988;128:441–452. doi: 10.1016/0012-1606(88)90306-5. [DOI] [PubMed] [Google Scholar]
- 29.Lee Y, Saint-Jeannet JP. Cardiac neural crest is dispensable for outflow tract septation in Xenopus. Development. 2011;138:2025–2034. doi: 10.1242/dev.061614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugita S, Adachi T, Ueki Y, Sato M. A novel method for measuring tension generated in stress fibers by applying external forces. Biophys. J. 2011;101:53–60. doi: 10.1016/j.bpj.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hocking JC, Hehr CL, Bertolesi G, Funakoshi H, Nakamura T, McFarlane S. LIMK1 acts downstream of BMP signaling in developing retinal ganglion cell axons but not dendrites. Dev. Biol. 2009;330:273–285. doi: 10.1016/j.ydbio.2009.03.027. [DOI] [PubMed] [Google Scholar]
- 32.Cary RB, Klymkowsky MW. Desmin organization during the differentiation of the dorsal myotome in Xenopus laevis. Differentiation. 1994;56:31–38. doi: 10.1046/j.1432-0436.1994.56120031.x. [DOI] [PubMed] [Google Scholar]
- 33.Sadikot T, Hammond C, Ferrari MB. Distinct roles for telethonin N- versus C- terminus in sarcomere assembly and maintenance. Dev. Dyn. 2010;239:1124–1135. doi: 10.1002/dvdy.22263. [DOI] [PubMed] [Google Scholar]
- 34.Mudry RE, Perry CN, Richards M, Fowler VM, Gregorio CC. The interaction of tropomodulin with tropomyosin stabilizes thin filaments in cardiac myocytes. J. Cell. Biol. 2003;162:1057–1068. doi: 10.1083/jcb.200305031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pappas CT, Bhattacharya N, Cooper JA, Gregorio CC. Nebulin interacts with Cap Z and regulates thin filament architecture within the Z-disc. Mol. Biol. Cell. 2008;19:1837–1847. doi: 10.1091/mbc.E07-07-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishikawa A, Hayashi H. Spatial, temporal and hormonal regulation of programmed muscle cell death during metamorphosis of the frog Xenopus laevis. Differentiation. 1995;59:207–214. doi: 10.1046/j.1432-0436.1995.5940207.x. [DOI] [PubMed] [Google Scholar]
- 37.Mohun TJ, Leong LM, Weninger WJ, Sparrow DB. The Morphology of Heart Development in Xenopus laevis. Dev. Biol. 2000;218:74–88. doi: 10.1006/dbio.1999.9559. [DOI] [PubMed] [Google Scholar]
- 38.Kaltenbrun E, Tandon P, Amin NM, Waldron L, Showell C, Conlon FL. Xenopus: an emerging model for studying congenital heart disease. Birth Defects Res. A Clin. Mol. Teratol. 2011;91:495–510. doi: 10.1002/bdra.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith SJ, Ataliotis P, Kotecha S, Towers N, Sparrow DB, Mohun TJ. The MLC1v gene provides a transgenic marker of myocardium formation within developing chambers of the Xenopus heart. Dev. Dyn. 2005;232:1003–1012. doi: 10.1002/dvdy.20274. [DOI] [PubMed] [Google Scholar]
- 40.Raworth AE, Pepe FA. Ultrastructure of developing muscle cells in the chick embryo. Am. J. Anat. 1965;116:115–147. doi: 10.1002/aja.1001160107. [DOI] [PubMed] [Google Scholar]
- 41.Peng BH, Wolosewick JJ, Cheng P. The development of myofibrils in cultured muscle cells: a whole-mount and thin-section electron microscopic study. Dev. Biol. 1981;88:121–136. doi: 10.1016/0012-1606(81)90224-4. [DOI] [PubMed] [Google Scholar]
- 42.Fischman DA. An electron microscope study of myofibril formation in embryonic chick skeletal muscle. J. Cell Biol. 1967;32:557–575. doi: 10.1083/jcb.32.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]