Abstract
Aim
To assess the validity and potential clinical utility of evaluating MYC protein expression by immunohistochemistry (IHC) in mantle cell lymphoma (MCL).
Methods and results
MYC IHC was scored on a tissue microarray containing 62 MCL cases and 29 controls by two pathologists. Inter-observer correlation was high (intra-class correlation=0.98). MYC IHC scores correlated with MYC gene expression (Spearman’s 0.69, p<0.0001) and weakly with Ki-67 proliferation index (Spearman’s 0.30, p=0.03). Six cases of blastic MCL did not have higher mean MYC IHC scores or MYC mRNA expression than non-blastic MCL cases. None of 57 cases assessed, including all the blastic cases, showed MYC gene rearrangement by fluorescence in situ hybridization. Multivariate analysis using backward selection from potential predictors including age, lactate dehydrogenase, leukocyte count, MIPI score, ECOG performance status, blastic morphology, and Ki-67 index showed that MYC IHC score is an independent predictor of progression-free survival (hazard ratio=2.34, 95% CI 1.42 – 3.88, p=0.0009) and overall survival (hazard ratio=1.90, 95% CI 1.05 – 3.43, p=0.034).
Conclusions
We show that a new monoclonal anti-MYC antibody can enable accurate and reproducible visual assessment of MYC protein expression that is independently predictive of clinical outcomes in MCL.
Keywords: MYC, mantle cell lymphoma, immunohistochemistry, tissue microarray
Introduction
Mantle cell lymphoma (MCL) is an aggressive B-cell non-Hodgkin lymphoma whose molecular pathogenesis is partly attributed to the characteristic genetic feature of t(11;14)(q13;q32) resulting in over-expression of cyclinD1 and subsequent dysregulation of the cell cycle. While most cases of MCL are difficult to treat, with a median survival of 5 to 7 years, prognosis can vary considerably1–4. Gene expression studies performed in an effort to risk-stratify patients demonstrated heterogeneity in the expression of genes involved in cellular proliferation between MCL cases, with as much as a six year survival difference between cases having a proliferative signature versus those that did not5, 6. However, there has been little recent progress in translating these findings to the clinical laboratory. With the exception of the Ki-67 proliferative index7, there are no prognostic markers currently being assessed in the clinical laboratory to aid in risk-stratifying patients with MCL.
The MYC oncogene encodes a transcription factor that has the unique ability to globally activate transcription of over 15% of all cellular genes, many of which regulate cell growth and cell cycle progression8–10. Reciprocal translocations and amplifications of the MYC gene and over-expression of MYC RNA have been found in 5 to 40% of MCL11, 12. While MCL with MYC aberrations have been associated with blastic transformation and poor prognosis13–17, high MYC expression itself has also been shown to be one of the strongest predictors of poor outcome and only marginally associated with the Ki-67 proliferation index6, 18.
Assessment of MYC aberrations in the clinical laboratory has, up until now, required additional testing by fluorescence in-situ hybridization (FISH). Because FISH is relatively labor intensive to perform and interpret, assessment of MYC gene aberrations in B-cell lymphomas has largely been reserved for diagnosing cases of Burkitt lymphoma and the detection of an aggressive form of ‘double hit’ diffuse large B-cell lymphoma (DLBCL) which harbors a MYC translocation. Recently however, there have been several reports of prognostically valid MYC protein expression assessment by immunohistochemistry (IHC) in DLBCL with a new commercially available antibody19–21.
In the present study, our goal was to evaluate the validity and potential clinical utility of assessing MYC protein expression by IHC in MCL. We characterized the performance of a commercially available anti-MYC antibody on a MCL tissue microarray that included control cores of normal lymphoid tissue, low grade B-cell lymphoma, and high grade B-cell lymphoma with documented MYC-IgH translocation. MYC IHC scoring was compared to MYC gene expression and assessed for association with clinical outcomes by multivariate analysis.
Materials and Methods
Case selection
With approval from the appropriate institutional review boards (UW protocol M-2008-1011 and MCRF protocol SHA 10109), the pathologic archives of the University of Wisconsin Hospital and Clinics and Marshfield Clinic were searched for cases of MCL diagnosed between 1999 and 2010. 62 patients were identified to have archival formalin-fixed paraffin-embedded (FFPE) tissue from pre-treatment diagnostic biopsies available for tissue microarray (TMA) construction. All cases included expressed cyclinD1 and/or demonstrated t(11;14) by FISH with morphologic characteristics consistent with MCL. The median duration of follow-up was 5.47 years.
Tissue microarray construction
Representative areas of MCL were marked on hematoxylin and eosin (H&E) stained sections and triplicate 0.6mm cores from the corresponding paraffin block were punched and arranged 0.2mm apart vertically and horizontally using a Manual Tissue Arrayer (Beecher Instruments, Sun Prairie, WI). Included in the array were 6 small lymphocytic lymphoma (SLL), 6 grade 1 follicular lymphoma (FL), 5 Burkitt lymphoma (3 with MYC rearrangement and 2 without), 6 benign lymph nodes, and 6 benign tonsil cases.
Immunohistochemical staining and quantification
For MYC immunohistochemistry, antigen retrieval was performed in a Decloaking Chamber (Biocare Medical, Concorde, CA) for 4 minutes at 100°C. Endogenous peroxidases were blocked with Peroxidazed (Biocare Medical) and non-specific binding minimized with Biocare Terminator (Biocare Medical). Background protein staining was blocked with Background Punisher (Biocare Medical). Anti-MYC antibody (Y69, Epitomics, Burlingame, CA) was used at a 1:200 dilution (final concentration 0.56 μg/mL), followed by addition of Mach 2 Rabbit HRP-polymer secondary antibody (Biocare Medical). Protein expression was scored independently on an Olympus BX41 microscope at 20X objective magnification by two pathologists (MJO and DTY). MYC expression was identified as any clearly identifiable nuclear staining in lymphoid cells and quantified in 5% increments by visual estimation. MYC scores for triplicate cores were averaged for each case and each case then averaged between pathologists, after determining that overall inter-observer agreement was acceptable.
Ki-67 staining was performed using Ki-67 antibody (Clone SP6 Biocare Medical, Concord, USA). The Ki-67 index was defined as the percentage of Ki-67 positive tumor cells in representative areas of lymphoma evaluated by one observer (DTY) counting 100 cells in two representative high power fields at 400x magnification7 for each core and then averaged between the triplicate cores for each case.
Quantitative nuclease protection assay
MYC mRNA expression was determined by qNPA™ (High Throughput Genomics, Tucson, AZ) assay. 5μm thick tissue specimens from the corresponding paraffin blocks utilized to construct the TMA were dissolved in lysis buffer containing oligonucleotide probes for two housekeeping genes (TBP and B2M) and four 25-mer probes targeting position 333 (5′-GTCCGCAACCCTTGCCGCATCCACG-3′), 576 (5′-CCTCAACGRRAGCTTCACCAACAGG-3′), 1281 (5′-CCTGGTGCTCCATGAGGAGACACCG-3′), and 1534 (5′-CCAAGAGGGTCAAGTTGGACAGTGT-3′) of MYC mRNA. Unhybridized probe was digested by S1 nuclease and alkaline hydrolysis destroyed mRNA of mRNA-probe duplexes, leaving intact probe in stoichiometric concentrations proportional to expressed mRNA for chemiluminescent detection. MYC mRNA expression was normalized to the two housekeepers. Each case was run in triplicate and results averaged.
Fluorescence In Situ Hybridization
Interphase FISH was performed on two TMA sections according to described protocols22. 100 cells from a representative core of each case were assessed for % of split signals by a pathologist (DTY). The positive cut-off level was established as the mean number of split signals plus 3 times the standard deviation of 6 benign lymph node cores represented on the TMA.
Statistical Analysis
Agreement between mean MYC IHC scores generated by independent pathologists was assessed by intra-class correlation coefficient (ICC) with Bland Altman plot. Core to core variation was evaluated using a mixed effects model assessing variability in IHC score readings predicted by a fixed effect of inter-observer difference and a random effect of case differences, and was estimated as the residual variance (or within subject variance). Spearman’s rank correlation coefficient was used to assess the correlation between MYC IHC scores and MYC mRNA levels. Progression-free survival (PFS) was calculated from the date of diagnosis to progression of disease or death from any cause, whichever came first, and overall survival (OS) was calculated from the date of diagnosis to date of death. The survival functions of PFS and OS were estimated using the Kaplan-Meier (product-limit) method and compared between two groups using the log-rank test. Cox proportional hazards model was used to examine the association between the survival outcomes and prognostic factors. Results were quantified in terms of hazard ratios with 95% confidence intervals. Results were considered statistically significant if a two-tailed p-value was less than 0.05.
Results
Patient Characteristics
The median age of the 62 MCL patients included in this study was 61 years (range 42 to 91 years) and 77% were men (Table 1). At the time of diagnosis, 16% had ECOG performance status ≥ 2, 67% had Ann Arbor Stage 4 disease, 29% had a circulating leukocyte count ≥ 10 × 103/μL, 39% had elevated plasma lactate dehydrogenase (LDH) levels, 17% had an elevated Ki-67 proliferation index, and 10% had blastic histology. The MCL international prognostic index (MIPI) is a prognostic index developed specifically for MCL based on age, leukocyte count, ECOG performance status, and LDH23. 36% of the patients were in the MIPI high risk group. Patients included in this study were treated relatively similarly with 75% receiving frontline adriamycin-containing polychemotherapy.
Table 1.
Clinical and laboratory characteristics of 62 MCL patients
| Characteristics of patients at diagnosis | |
|---|---|
| Age, years | |
| Mean (SD) | 63 (11) |
| Median (Range) | 61 (42 – 91) |
|
| |
| Gender | |
| Male | 48/62 (77.4%) |
| Female | 14/62 (22.6%) |
|
| |
| ECOG Performance Status | |
| <2 | 9/58 (15.5%) |
| ≥2 | 49/58 (84.5%) |
|
| |
| Ann Arbor Stage | |
| I – III | 20/60 (33.3%) |
| IV | 40/60 (66.7%) |
|
| |
| WBC count | |
| <10 × 103/uL | 43/60 (71.7%) |
| ≥10 × 103/uL | 17/60 (28.3%) |
|
| |
| LDH | |
| Elevated | 22/57 (38.6%) |
| Normal | 35/57 (61.4%) |
|
| |
| Ki-67 index | |
| <30% | 48/58 (82.8%) |
| ≥30% | 10/58 (17.2%) |
|
| |
| MIPI risk | |
| High | 21/58 (36.2%) |
| Intermediate | 19/58 (32.8%) |
| Low | 18/58 (31.0%) |
|
| |
| Morphologic variant | |
| Non-Blastic | 56/62 (90.3%) |
| Blastic | 6/62 (9.7%) |
|
| |
| Treatment | |
| Frontline adriamycin-containing polychemotherapy | 45/60 (75.0%) |
Assessment of MYC Immunohistochemical Staining
In essentially all MCL cases and controls, MYC staining was clearly nuclear without any background cytoplasmic staining. Accordingly, visual semi-quantitative interpretation was relatively straightforward and agreement between two independent observers was excellent with intra-class correlation coefficient of 0.98 (95% CI = 0.96–0.99) (Figure 1A). Within the same case, there was little core to core variability in MYC staining with variance estimate of 30.3 between the replicate cores compared to 360.2 between patients.
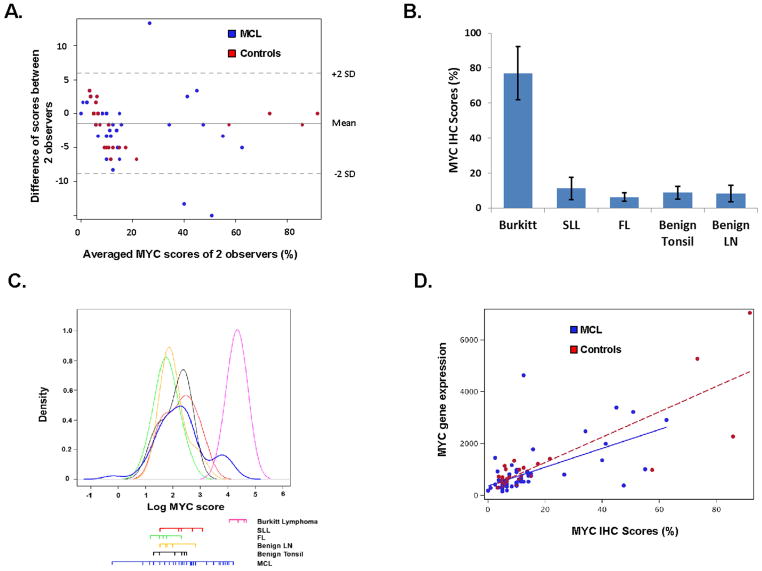
Figure 1. Validation of MYC immunohistochemical (IHC) staining through assessment of inter-observer correlation, staining pattern in control tissues, and correlation with MYC gene expression.
(A) Bland-Altman plot showing the degree of agreement between two independent pathologists scoring % MYC positive cells for 62 cases of MCL and 29 controls represented in a tissue microarray. Intra-class correlation coefficient = 0.98 (95% CI = 0.96–0.99). (B) MYC expression scores in control cases expressed as the average and standard deviation for 2 observers. Burkitt lymphoma (77.1 ± 15.1%), small lymphocytic lymphoma (11.3 ± 6.3%), follicular lymphoma (6.3 ± 2.5%), benign tonsil (8.8 ± 3.6%) and benign lymph node (8.3 ± 4.6%). (C) Frequency distribution of average MYC IHC scores for 62 MCL and 29 control cases that include 5 Burkitt lymphoma, 6 small lymphocytic lymphoma, 6 Grade 1 follicular lymphoma, 6 benign tonsil, and 6 benign lymph nodes. (D) Correlation of MYC IHC scores with MYC gene expression. Spearman’s correlation of Rho = 0.69 (p<0.0001) and 0.65 (p<0.0001) for MCL and control cases, respectively.
Mean MYC expression scores (± standard deviation) (Figure 1B) were highest in Burkitt lymphoma (77.1 ± 15.1%) and lower in the low grade B-cell lymphoma controls, small lymphocytic lymphoma (11.3 ± 6.3%) and follicular lymphoma (6.3 ± 2.5%). Expression scores were similarly low in the benign tonsil (8.8 ± 3.6%) and benign lymph node (8.3 ± 4.6%) controls. Positive staining cells were appropriately concentrated in the proliferation centers and germinal centers of the small lymphocytic lymphoma and benign lymph node/tonsil cases, respectively (Figure 2).
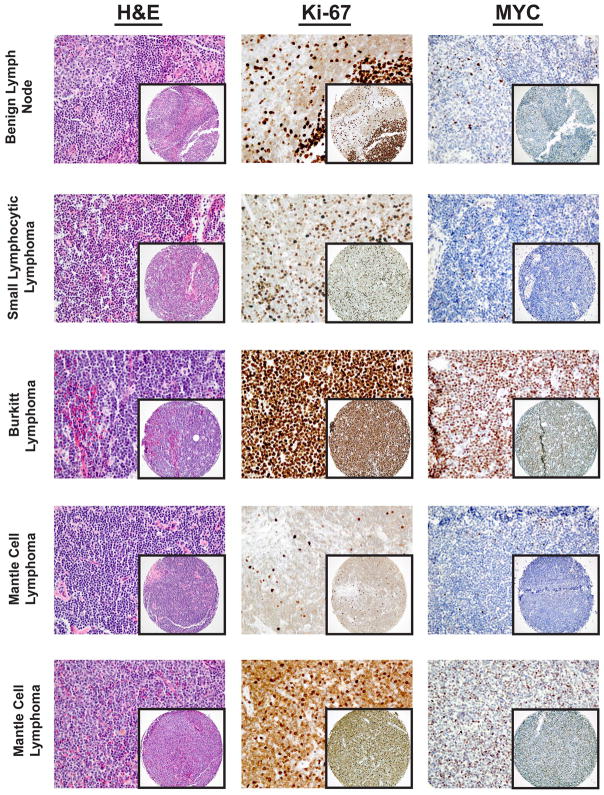
Figure 2. Examples of MYC immunohistochemical staining in control and MCL cases represented on a tissue microarray.
The benign lymph node and small lymphocytic lymphoma (SLL) shown are representative of controls with low MYC expression and were both scored as 5% MYC positive. Note that MYC expression is concentrated in the germinal centers of the benign lymph node and the proliferation centers of SLL, where Ki-67 expression is also the most brisk. Burkitt lymphoma with t(8;14) showed a diffuse distribution of numerous MYC positive cells (represented case was scored as 92% MYC positive). MCL cases varied greatly in MYC expression. The examples shown are of a low MYC expressing case scored as 5% MYC positive and a high MYC expressing case scored as 63% MYC positive.
The proportion of MYC positive tumor cells in MCL cases ranged from 0 to 63% with a median of 10% and mean of 14 ± 15%. The frequency distribution (Figure 1C) shows that the majority of MCL cases have MYC scores similar to that of the benign controls and low grade B-cell lymphomas. A subset of MCL cases have distinctly high MYC scores, in the range of Burkitt lymphoma cases.
Comparison of MYC Gene Expression with Immunohistochemical Staining
To further validate our assessment of MYC protein expression by IHC, MYC mRNA levels for each case was determined by quantitative nuclease protection assay performed on a macro-dissected 5 μm section of the same paraffin block from which the tissue cores were obtained. MYC mRNA levels correlated well with the IHC scores; Spearman’s rank correlation = 0.69 (p<0.0001) and 0.65 (p<0.0001) for MCL and control cases, respectively (Figure 1D).
MYC, Blastic Morphology, and Ki-67 Proliferation Index
Given that aberrations in the MYC gene have been associated with blastic transformation13–17, we evaluated the association between blastic histology and MYC expression (Figure 3A,B). Remarkably, the 6 blastic cases (Table 1) had a lower mean MYC IHC score and MYC gene expression than the remaining non-blastic MCL cases, with the gene expression comparison reaching statistical significance (Student t-test, p=0.009). MYC IHC scores showed a weak but statistically significant correlation with Ki-67 proliferation index (Spearman’s = 0.3, p = 0.03) (Figure 3C).
Figure 3. Association of MYC expression with blastic histology and Ki-67 proliferation index.
(A) Mean MYC IHC score of blastic (9.3 ± 8.5%) and non-blastic (15.1 ± 15.6%) MCL cases. (B) Mean MYC gene expression of blastic (469.4 ± 218.5) and non-blastic (885.1 ± 895.7) MCL cases. (C) MYC IHC scores and Ki-67 proliferation index show a weak Spearman’s correlation of 0.30 (p = 0.03).
To investigate whether MYC overexpression was associated with MYC gene rearrangement, FISH utilizing a dual color MYC break apart probe was performed on the TMA. 57 cases were successfully scored, including all the blastic cases. Remarkably, none of the MCL cases showed evidence of MYC rearrangement (Table 2).
Table 2.
MYC gene rearrangement analysis by fluorescent in-situ hybridization with dual color break apart probes
| Cases Scored | Number of cells with split signals (%)
|
Cases positive for MYC rearrangement * | ||
|---|---|---|---|---|
| Mean and SD | Range | |||
| Benign Lymph node | 6 | 2.5 ± 1.0 | 1 – 4 | |
| Burkitt Lymphoma | 2 | 94.0 ± 4.2 | 91 –97 | 2/2 |
| Mantle Cell Lymphoma | 57 | 1.3 ± 0.9 | 0 – 3 | 0/57 |
The positive cut-off level was 5.5%, established as the mean number of split signals plus 3 times the standard deviation of the 6 benign lymph node cores represented on the TMA.
MYC Immunohistochemistry and Clinical Outcomes
Of 62 MCL patients represented on the TMA, 60 had clinical outcomes available and had a median follow-up of 5.47 years (95% confidence interval (CI), 4.06 – 6.63) as of this analysis. By univariate analysis, MYC IHC score on a natural log scale was predictive of OS (hazard ratio = 2.25, p = 0.005) and PFS (hazard ratio = 2.35, p = 0.0007). Multivariate analysis using backward selection from potential predictors including age, LDH, WBC count, MIPI score, ECOG performance status, blastic morphology, and Ki-67 proliferation index in the parsimonious model showed MYC IHC score as an independent predictor of OS (hazard ratio = 1.90, p = 0.034) and PFS (hazard ratio = 2.34, p = 0.0009) (Table 3).
Table 3.
Multivariate analysis of MYC immunohistochemistry scores as a predictor of progression free survival and overall survival.*
| Parameter | Hazard Ratio
|
p-value
|
||
|---|---|---|---|---|
| PFS | OS | PFS | OS | |
| MYC score | 2.35 | 1.90 | 0.0009 | 0.034 |
| MIPI score | NS | 3.40 | NS | 0.0009 |
log(MYC score) was evaluated controlling for age, LDH, WBC count, MIPI score, ECOG performance status, blastic histology, and Ki-67 proliferation index. Predictors shown are valid in the final parsimonious model derived from backward selection. NS = not statistically significant
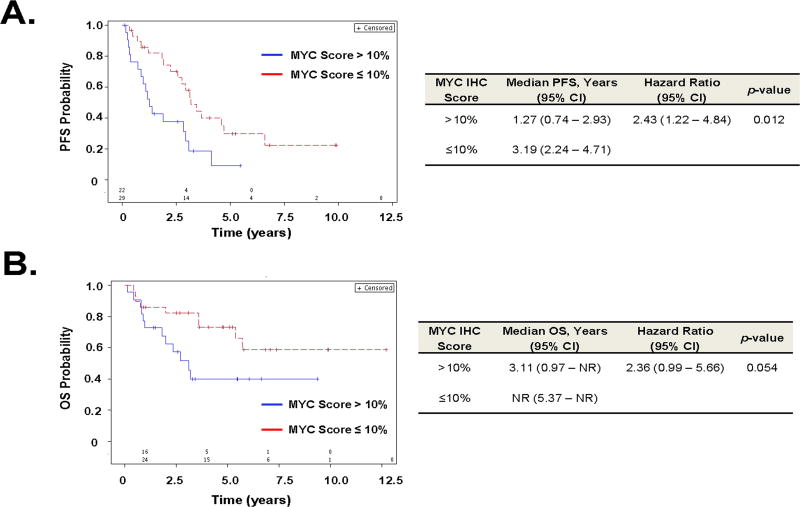
Unbiased dichotomization of the MCL cases at the median MYC IHC score of 10% shows patients with high MYC IHC scores >10% had shorter PFS than those with low MYC IHC scores ≤10% (p = 0.012) (Figure 4A). These two groups did not differ in terms of receiving frontline adriamycin-containing polychemotherapy (Chi-square test, p = 0.21). Statistical significance for a difference in OS at the cut-off score of 10% was not reached (p=0.054) (Figure 4B).
Figure 4. Association of MYC immunohistochemical staining scores with clinical outcomes in MCL.
(A) Kaplan–Meier plot of progression free survival (PFS) for MCL cases dichotomized at the median MYC IHC score of 10%. (B) Kaplan-Meier plot of overall survival (OS). Number of subjects at risk is listed above the X-axis. NR = not reached.
Discussion
Reciprocal translocations and amplifications of the MYC gene and over-expression of MYC RNA are found in up to 40% of MCL and have been associated with poor clinical outcome11, 12. We assessed the immunohistochemical performance of a new commercially available anti-MYC antibody and evaluated the ability of visually scoring MYC positivity to predict PFS and OS.
MYC immunohistochemical staining could be interpreted with excellent inter-observer reliability with an intra-class correlation coefficient of 0.98 (95% CI = 0.96 – 0.99) and showed appropriate expression levels in both high and low expression controls. In addition, average MYC IHC scores generated by 2 pathologists correlated well with MYC gene expression in both MCL cases as well as the controls with Spearman’s rank correlation = 0.69 and 0.65, respectively.
Ranking MCL cases by their MYC IHC scores showed a skewed distribution with a predominance of cases having scores similar to the low MYC expression controls. A subset of MCL cases had distinctly high MYC scores similar to the Burkitt lymphoma controls. Despite the fact that MCL with MYC aberrations have been associated with blastic transformation13–17, we did not find an association between high MYC scores and blastic histology. This discrepancy may be a consequence of selection bias in the previous studies that only included cases of MCL with MYC rearrangement. While many MCL cases with MYC rearrangement may be blastic, it may be that only a few blastic MCL cases have MYC rearrangement. Indeed, we show that not only do blastic cases have low MYC protein expression, none of the MCL cases in this cohort, including the 6 blastic cases, demonstrate MYC rearrangement by FISH analysis. MYC overexpression in MCL appears to be largely independent of MYC gene rearrangement.
Not only were MYC IHC scores not associated with blastic change, they showed only a weak correlation with Ki-67 proliferation index (Spearman’s = 0.3, p = 0.03), suggesting the oncogenic effect of MYC in MCL is likely not solely manifest as cellular proliferation, but may be imparting neoplastic cells a survival advantage through alternative mechanisms such as inhibition of TP53-mediated apoptosis23. Similar discordance between MYC gene and protein expression with both Ki-67 proliferation index and aggressive morphologic transformation have been previously described in MCL and DLBCL18,19, 25–26.
As a continuous variable, MYC score was an independent predictor of OS (p = 0.034) and PFS (p = 0.0009) after controlling for age, LDH, WBC count, MIPI score, ECOG performance status, blastic histology, and Ki-67 proliferation index.
Cognizant that the number of MCL cases in this series was insufficient to establish a reliable cut-off value, an unbiased dichotomization of the cases at the median MYC score (10%) was performed. At this arbitrary cut-off, MYC score was predictive of PFS (p = 0.012), but not OS (p = 0.054). Given the distribution of MYC scores, we speculate that a higher cut-off value would likely yield superior predictive results. However, analysis of a larger cohort of patients will be necessary to establish this.
While high MYC mRNA expression is one of the strongest predictors of poor outcome in MCL6, 18, technical factors have limited efforts to develop such an assay for the clinical laboratory. Likewise, FISH is useful for identifying MYC translocations, but is limited by its inability to identify altered MYC expression by mechanisms other than translocations and copy number amplification, and is not readily available in all clinical laboratories. We show that a new monoclonal anti-MYC antibody can enable simple, accurate, and reproducible visual assessment of MYC protein expression in MCL that is predictive of clinical outcomes. These findings are reflective of results from Johnson et al19 and Green et al20, who assessed MYC expression by immunohistochemistry in conjunction with BCL2 in diffuse large B-cell lymphoma. Both studies found immunohistochemical assessment of MYC and BCL2 expression capable of identifying a high-risk ‘double hit’ DLBCL subset that was originally characterized in the literature by FISH positive MYC and BCL2 translocations. A simple and robust method to risk-stratify patients based on MYC expression may be especially timely and pertinent given the recent reports of the anti-tumor effects of BET bromodomain inhibitors whose mechanism of action is through inhibition of MYC expression27, 28.
In conclusion, this study demonstrates that immunohistochemical assessment of MYC protein expression in MCL can be reliably performed and is predictive of clinical outcomes. A larger study is warranted to establish and validate potential cut-off values for risk-stratification.
Acknowledgments
This work was funded in part by Forward Lymphoma, the University of Wisconsin Carbone Cancer Center grant P30 CA014520 from the National Cancer Institute, and by the Clinical and Translational Science Award (CTSA) program, previously through the National Center for Research Resources (NCRR) grant 1UL1RR025011, and now by the National Center for Advancing Translational Sciences (NCATS), grant 9U54TR000021. The content is soley the responsibility of the authors and does not necessarily represent the official views of the NIH.
The authors thank the University of Wisconsin Carbone Cancer Center Translational Science BioCore and the Wisconsin State Laboratory of Hygiene for providing technical assistance and use of its facilities.
Footnotes
DISCLOSURE/CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Herrmann A, Hoster E, Zwingers T, et al. Improvement of overall survival in advanced stage mantle cell lymphoma. J Clin Oncol. 2009;27:511–518. doi: 10.1200/JCO.2008.16.8435. [DOI] [PubMed] [Google Scholar]
- 2.Orchard J, Garand R, Davis Z, et al. A subset of t(11;14) lymphoma with mantle cell features displays mutated IgVH genes and includes patients with good prognosis, nonnodal disease. Blood. 2003;101:4975–4981. doi: 10.1182/blood-2002-06-1864. [DOI] [PubMed] [Google Scholar]
- 3.Martin P, Chadburn A, Christos P, et al. Outcome of deferred initial therapy in mantle-cell lymphoma. J Clin Oncol. 2009;27:1209–1213. doi: 10.1200/JCO.2008.19.6121. [DOI] [PubMed] [Google Scholar]
- 4.Williams ME, Bernstein SH, Jares P, Kahl BS, Witzig TE, Gordon LI. Recent Advances in Mantle Cell Lymphoma: Report of the 2012 Mantle Cell Lymphoma Consortium Workshop. Leuk Lymphoma. 2013 doi: 10.3109/10428194.2013.771400. [DOI] [PubMed] [Google Scholar]
- 5.Rosenwald A, Wright G, Wiestner A, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 6.Hartmann E, Fernàndez V, Moreno V, et al. Five-gene model to predict survival in mantle-cell lymphoma using frozen or formalin-fixed, paraffin-embedded tissue. J Clin Oncol. 2008;26:4966–4972. doi: 10.1200/JCO.2007.12.0410. [DOI] [PubMed] [Google Scholar]
- 7.Klapper W, Hoster E, Determann O, et al. Ki-67 as a prognostic marker in mantle cell lymphoma-consensus guidelines of the pathology panel of the European MCL Network. J Hematop. 2009;2:103–111. doi: 10.1007/s12308-009-0036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin CY, Lovén J, Rahl PB, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nie Z, Hu G, Wei G, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klapproth K, Wirth T. Advances in the understanding of MYC-induced lymphomagenesis. Br J Haematol. 2010;149:484–497. doi: 10.1111/j.1365-2141.2010.08159.x. [DOI] [PubMed] [Google Scholar]
- 11.Aukema SM, Siebert R, Schuuring E, et al. Double-hit B-cell lymphomas. Blood. 2011;117:2319–2331. doi: 10.1182/blood-2010-09-297879. [DOI] [PubMed] [Google Scholar]
- 12.Hernández L, Hernández S, Beà S, et al. c-myc mRNA expression and genomic alterations in mantle cell lymphomas and other nodal non-Hodgkin’s lymphomas. Leukemia. 1999;13:2087–2093. doi: 10.1038/sj.leu.2401599. [DOI] [PubMed] [Google Scholar]
- 13.Au WY, Horsman DE, Viswanatha DS, Connors JM, Klasa RJ, Gascoyne RD. 8q24 translocations in blastic transformation of mantle cell lymphoma. Haematologica. 2000;85:1225–1227. [PubMed] [Google Scholar]
- 14.Hao S, Sanger W, Onciu M, Lai R, Schlette EJ, Medeiros LJ. Mantle cell lymphoma with 8q24 chromosomal abnormalities: a report of 5 cases with blastoid features. Mod Pathol. 2002;15:1266–1272. doi: 10.1097/01.MP.0000037310.82136.99. [DOI] [PubMed] [Google Scholar]
- 15.Au WY, Horsman DE, Gascoyne RD, Viswanatha DS, Klasa RJ, Connors JM. The spectrum of lymphoma with 8q24 aberrations: a clinical, pathological and cytogenetic study of 87 consecutive cases. Leuk Lymphoma. 2004;45:519–528. doi: 10.1080/10428190310001593120. [DOI] [PubMed] [Google Scholar]
- 16.Reddy K, Ansari-Lari M, Dipasquale B. Blastic mantle cell lymphoma with a Burkitt translocation. Leuk Lymphoma. 2008;49:740–750. doi: 10.1080/10428190701852024. [DOI] [PubMed] [Google Scholar]
- 17.Setoodeh R, Schwarz S, Papenhausen P, Zhang L, et al. Double-hit mantle cell lymphoma with MYC gene rearrangement or amplification: a report of four cases and review of the literature. Int J Clin Exp Pathol. 2013;6:155–167. [PMC free article] [PubMed] [Google Scholar]
- 18.Kienle D, Katzenberger T, Ott G, et al. Quantitative gene expression deregulation in mantle-cell lymphoma: correlation with clinical and biologic factors. J Clin Oncol. 2007;25:2770–2777. doi: 10.1200/JCO.2006.08.7999. [DOI] [PubMed] [Google Scholar]
- 19.Johnson NA, Slack GW, Savage KJ, et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30:3452–3459. doi: 10.1200/JCO.2011.41.0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green TM, Young KH, Visco C, et al. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30:3460–3467. doi: 10.1200/JCO.2011.41.4342. [DOI] [PubMed] [Google Scholar]
- 21.Tapia G, Lopez R, Muñoz-Mármol AM, et al. Immunohistochemical detection of MYC protein correlates with MYC gene status in aggressive B cell lymphomas. Histopathology. 2011;59:672–678. doi: 10.1111/j.1365-2559.2011.03978.x. [DOI] [PubMed] [Google Scholar]
- 22.Ventura RA, Martin-Subero JI, Jones M, et al. FISH analysis for the detection of lymphoma associated chromosomal abnormalities in routine parafin-embedded tissue. J Mol Diag. 2006;8:141–151. doi: 10.2353/jmoldx.2006.050083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoster E, Dreyling M, Klapper W, et al. A new prognostic index (MIPI) for patients with advanced-stage mantle cell lymphoma. Blood. 2008;111:558–565. doi: 10.1182/blood-2007-06-095331. [DOI] [PubMed] [Google Scholar]
- 24.Ceballos E, Delgado MD, Gutierrez P, et al. c-MYC antagonizes the effect of p53 on apoptosis and p21WAF1 transactivation in K562 leukemia cells. Oncogene. 19:2194–2204. doi: 10.1038/sj.onc.1203541. [DOI] [PubMed] [Google Scholar]
- 25.Snuderl M, Kolman OK, Chen YB, et al. B-cell lymphoma with concurrent IGH-BCL2 and MYC rearrangements are aggressive neoplasms with clinical and pathologic features distinct from Burkitt lymphoma and diffuse large B-cell lymphoma. Am J Surg Pathol. 2010;34:327–340. doi: 10.1097/PAS.0b013e3181cd3aeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kluk MJ, Chapuy B, Sinha P, et al. Immunohistochemical detection of MYC-driven diffuse large B-cell lymphomas. PLoS ONE. 2012;7:e33813. doi: 10.1371/journal.pone.0033813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mertz JA, Conery AR, Bryant BM, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]