Abstract
Background
Recently, bi-allelic mutations in the transcription factor RFX6 were described as the cause of a rare condition characterized by neonatal diabetes with pancreatic and biliary hypoplasia and duodenal/jejunal atresia.
Clinical Case
A male infant developed severe hyperglycemia (446 mg/dL) within 24 hours of birth. Acute abdominal concerns by day five necessitated exploratory surgery that revealed duodenal atresia, gallbladder agenesis, annular pancreas and intestinal malrotation. He also exhibited chronic diarrhea and feeding intolerance, cholestatic jaundice, and subsequent liver failure. He died of sepsis at four months old while awaiting liver transplantation. The phenotype of neonatal diabetes with intestinal atresia and biliary agenesis clearly pointed to RFX6 as the causative gene; indeed, whole exome sequencing revealed a novel homozygous RFX6 mutation c.779A>C; p.Lys260Thr (K260T). This missense mutation also changes the consensus 5’ splice donor site before intron 7 and is thus predicted to cause disruption in splicing. Both parents, who were not known to be related, were heterozygous carriers.
Conclusions
Targeted genetic testing based on consideration of phenotypic features may reveal a cause among the many genes now associated with heterogeneous forms of monogenic neonatal diabetes. Our study demonstrates the feasibility of using modern sequencing technology to identify one such rare cause. Continued research is needed to determine the possible cost-effectiveness of this approach, especially when clear phenotypic clues are absent. Further study of patients with RFX6 mutations should clarify its role in pancreatic, intestinal and enteroendocrine cellular development and explain features such as the diarrhea exhibited in our case.
Key terms: RFX6, Neonatal Diabetes, Mitchell-Riley Syndrome
BACKGROUND
Neonatal diabetes occurs rarely (1 per 1–200,000 births) but is very likely to have an underlying monogenic cause when diagnosed during the first six months of life (1). Monogenic neonatal diabetes is most commonly due to heterozygous mutations in KCNJ11, INS and ABCC8; however, mutations in nearly 20 other genes can cause rare and often syndromic forms usually involving a variety of extra-pancreatic manifestations (2).
The regulatory factor X (RFX) family consists of seven (RFX1-7) winged-helix transcription factors with a highly conserved RFX DNA binding domain (3). RFX proteins are expressed in many different tissues; however, RFX6 is the most restricted, with high expression only in mature pancreas and low levels in heart and liver. Human RFX6 is a 928 amino acid protein encoded by a 19 exon gene on chromosome 6q22. In mice, Rfx6 is expressed in the definitive endoderm early in development, but is later restricted to the gut and pancreatic bud, reactivated in endocrine progenitors, and ultimately restricted to the pancreatic islets in adults (4). Rfx6 deficient mice failed to generate any of the normal islet cell types except for pancreatic polypeptide producing cells (5).
Mutations in RFX6 were recently described as the cause of neonatal diabetes in seven cases who also had other digestive system defects including hypoplastic or annular pancreas, intestinal atresia or stenosis, intestinal malrotation, gallbladder hypoplasia or agenesis, cholestatic disease, and/or abnormal biliary tract (5,6). This phenotype has been termed the Mitchell-Riley syndrome (OMIM #601346). Here, we report a case with neonatal diabetes, gallbladder agenesis, duodenal atresia, and intestinal malrotation who we hypothesized would carry causal mutations in RFX6. We also provide a brief overview of the other 7 cases described previously.
CASE
Our patient was a male infant born at 34 weeks gestation to non-consanguineous Vietnamese parents, with no known family history of diabetes mellitus, familial intestinal disorders, or early death (UC proband 0250). Prenatally, he was known to have intrauterine growth restriction and duodenal atresia. He was transferred to CHOC Children’s hospital on day one of life due to need for a higher level of care.
He was small for gestational age with birth weight 1375 grams (<1%, −2.6 standard deviations), and length of 39.5 cm (<1%, −2.6 standard deviations). His physical exam was notable for minimal subcutaneous fat, but otherwise he had no dysmorphic features or apparent deformities.
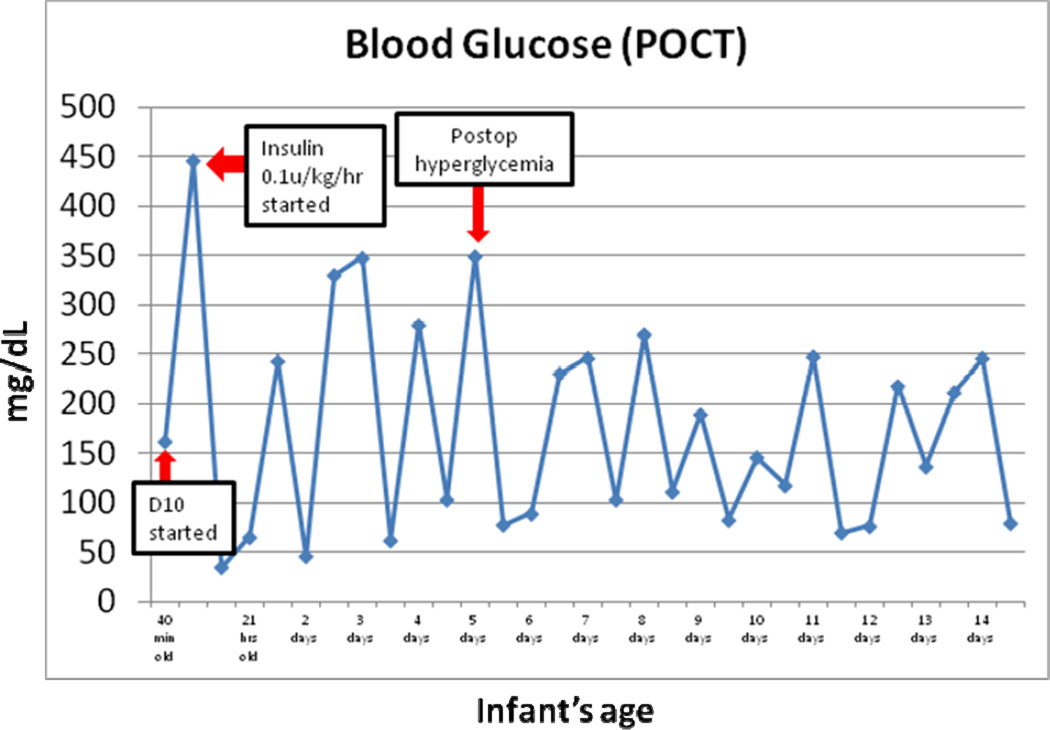
At 40 minutes of life, he was found to have an initial modestly elevated blood sugar of 163 mg/dL. He was placed on routine intravenous fluids of 10% dextrose (D10) while holding all feeds (NPO) and his blood glucose increased rapidly to 446 mg/dL. He was started on an insulin drip and required careful titration for wide fluctuations in glucose levels. He showed ready response to low insulin infusion rates (between 0.008 to 0.04 units/kg/hr), but blood glucoses ranged between 61–446 during the first week of life (Figure 1).
Figure 1.
Glucose fluctuations noted on days 1–14 of life. He responded readily to low insulin infusion rates with blood sugars between 60–250 by 2 weeks of age.
Radiologic imaging was performed to assess his gastrointestinal abnormalities including an abdominal x-ray on day of life one, which showed a double bubble consistent with duodenal atresia. A follow-up abdominal ultrasound showed malrotation and possible annular pancreas.
On day of life five, he developed an acute drop in hemoglobin and discoloration to his abdomen. He underwent urgent exploratory laparotomy, which confirmed duodenal atresia, intestinal malrotation, and jejunal perforation. He underwent malrotation correction and Roux-en-Y anastomosis. He remained on insulin drip post-operatively due to continued hyperglycemia.
Additional laboratory tests included a low C-peptide level (0.25 ng/mL at 10 days of life, with blood glucose of 246 mg/dL; normal 0.4–2.2 ng/mL), low amylase (8 units/L; normal 36–128 units/L), low lipase (13 units/L; normal 22–51 units/L), low stool pancreatic elastase (<100 mcg E1/g stool; normal >200 mcg E1/g stool), and high direct bilirubin (3.7 mg/dL at 2 months old; normal <0.5 mg/dL).
He had a prolonged neonatal course, complicated by cholestatic jaundice, recurrent infections, and feeding intolerance. He was ordered to have nothing by mouth (NPO) and placed on parenteral nutrition intermittently due to abdominal distension and profuse diarrhea. Attempts were made to retrial enteral feeds with different elemental formulas and he was also given pancreatic enzymes, but he exhibited little improvement in his continued severe diarrhea. He was also started on ursodiol and eventually phenobarbital for worsening cholestasis. At three months old, a repeat exploratory laparotomy for abdominal distension revealed gallbladder agenesis. A hepatobiliary scan was done at 3.5 months old, which showed non-visualization of the gallbladder and common bile duct consistent with gallbladder atresia. Liver biopsy results showed bile ductular proliferation with expansion of portal tracts. He was transferred to University of California Los Angeles for liver transplant evaluation due to liver failure, but unfortunately died at five months of age secondary to sepsis prior to transplant.
GENETIC ANALYSIS
Genomic DNA was isolated from the patient’s saliva, as well as both parents. Genomic DNA was extracted from salivary samples from the proband and parents using Oragene kits from DNA Genotek (Ottawa, Canada; http://www.dnagenotek.com). The NimbleGen SeqCap EZ Human Exome capture reagent and the NimbleGen CCDS defined exome library were used for targeted exome capture, and then sequenced using SOLiD sequencing. Variants were called using Atlas2 (7). Variant analysis (20x coverage; 10x in male X and Y chromosomes) was performed. To identify novel variants, ANNOVAR (8) was used to filter out variants in either dbSNP 129 or the July 2010 release of 1000 genomes. Novel variants were then annotated using ANNOVAR, with protein altering variants defined as those annotated to be nonsynonymous, nonsense, exonic indels, or splice site variants. dbNSFP1.3 was used to predict damaging effects of novel nonsynonymous and nonsense SNVs (9). dbNSFP contains conservation and functional predictions from phyloP (10); SIFT v4.0.3 (11); Polyphen2 v2.0.22 (12); MutationTaster, released March 2010 (13); and LRT, released November 20, 2009 (14).
Based on the phenotype in this case including duodenal atresia and malrotation, we prioritized variants in RFX6, which revealed a novel homozygous missense mutation in RFX6: c.779A>C, resulting in amino acid change in the codon just before intron 7: p.Lys260Thr (K260T). The novel K260T mutation was confirmed to be homozygous in the affected proband and heterozygous in both parents by bidirectional Sanger sequencing and analysis using Mutation Surveyor software (http://www.softgenetics.com).
The positively charged lysine is highly conserved at this position across multiple species (http://www.ncbi.nlm.nih.gov/homologene). Change to the non-charged threonine was predicted to be damaging using the PolyPhen-2 Humdiv prediction model (http://genetics.bwh.harvard.edu/pph2/), which tends to be more informative for rare variants. The mutation is downstream of the DNA-binding domain that characterizes all RFX proteins, but may interfere with the transcriptional activity of the B domain. The mutation also changes the consensus 5’ splice donor sequence for exon/intron 7: the normal sequence AAGgtatca becomes the mutant ACGgtatca (last three nucleotides of exon in caps, and 6 intronic nucleotides in lowercase). Two analysis tools (www.fruitfly.org/seq_tools/splice.html and www.cbs.dtu.dk/services/NetGene2/) predicted that the mutant sequence is no longer likely to be used as a 5’ donor splice site (15,16). This would cause exon skipping or the use of alternative splice sites, with all outcomes resulting in a modified transcript that might result in nonsense-mediated decay or a greatly altered and/or truncated protein sequence and resultant loss of function of the RFX6 transcription factor.
Runs of homozygosity were detected using SNP and Variation Suite v7.6.9 (Golden Helix, Inc.; http://www.goldenhelix.com). There were a total of 20 autosomal runs of homozygosity (minimum of 25 variant sites and 500 kb), with length of average 5.5 (SD 8.1) Mb. We recognize that because this is exome data, this is an overestimate; however, it suggests that there may be a low level of relatedness between the parents.
DISCUSSION
We report the eighth case with neonatal diabetes, hepatobiliary anomalies and intestinal malformations due to mutations in RFX6. Our case was not known to be consanguineous but carried a novel homozygous missense mutation K260T that also changes the consensus 5’ donor splice site sequence just before intron 7. Although tissue was not available to confirm absence of a normally spliced transcript, the highly similar and unusual phenotypic features shared with other cases with damaging RFX6 mutations makes this mutation very likely to be the cause of these problems in our patient (Table 1). The fact that this RFX6 variant is not present in publically available repositories (1000 genomes, dbSNP) suggests that it is rare and that the parents of the proband may be distantly related. However, this does not exclude the possibility of a founder effect whereby the variant may be present at some uncertain frequency within the Vietnamese population. Addressing this question most directly would require testing of a large number of individuals within that population.
Table 1.
RFX6 mutations in eight patients (seven probands) with neonatal diabetes and multiple congenital digestive tract anomalies.
| Original report (all reports) |
Mitchell, et al (5,17) | Chappell, et al (5,6,21) |
Martinovici, et al (5,22) |
Smith, et al (5) |
Spiegel et al (6) |
Current | ||
|---|---|---|---|---|---|---|---|---|
| Designation* | Proband 1 | 1B (sister of 1) | Proband 3 | Proband 2 | Proband 4 | Proband 5 | Proband 6 | Proband 7 |
| Gestation age (wks) | 36 | 34 | 39 | 35 | 38 | 35 | 38 | 34 |
| Gender | M | F | F | F | M | M | M | M |
| Origin | Pakistani | Pakistani | Unknown | Pakistani | Moroccan | French | Arab Israeli | Vietnamese |
| Consanguinity | Yes | Yes | No | Yes | Yes | No | No | No |
| Birth weight (g) | 1540 | 1310 | 2295 | 1700 | 1340 | NR | 1490 | 1375 |
| Age at death | 5 months | 6 months | Alive at 9 yrs old | Alive at 6 years | 3 months | 2.5 months | Alive at 21 months | 5 months |
| Neonatal diabetes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Age when insulin started | 1 day | 2 days | Not mentioned | 8 days | 1 day | 2 days | 10 days | 1 day |
| Abnormal pancreas | Hypoplastic | Annular | Hypoplastic | No | Hypoplastic | NR | Annular | Annular |
| Intestinal atresia/stenosis | Duodenal & Jejunal | Duodenal & Jejunal | Duodenal | Duodenal & Anal | Duodenal & Jejunal | Duodenal | Duodenal & Jejunal | Duodenal |
| Intestinal malrotation | No | No | Duodenal | Duodenal | Duodenal | Duodenal | No | Yes |
| Gallbladder | Hypoplastic | Hypoplastic | Hypoplastic | Agenesis | Agenesis | NR | Agenesis | Agenesis |
| Cholestatic disease | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Abnormal biliary tract | Intrahepatic atresia | Intrahepatic atresia | No | No | No | No | No | Yes |
| Neonatal hemochromatosis | No | No | No | No | No | No | Yes | No |
| Chronic diarrhea | Yes | Yes | No | Yes | No | NR | Yes | Yes |
| Parenteral nutrition dependent | Yes | Yes | No | No | No | NR | Yes | Yes |
| Liver failure | Yes | Yes | No | No | No | Yes | No | Yes |
| RFX mutation | Homozygous IVS2+2T>C | Homozygous IVS2+2T>C | IVS1-12A>G/IVS6+2T>G | Homozygous p.S217P | Homozygous out-of-frame exon 7 deletion | Homozygous p.R181Q | Homozygous c.781-2_787 del insG | Homozygous p.K260T (also disrupts intron 7 splice donor site) |
Multiple publications from 2004–2010 reported cases with neonatal diabetes along with intrauterine growth restriction, intestinal atresia, intestinal malrotation, biliary atresia, gallbladder hypoplasia, and hypoplastic or annular pancreas, a constellation of features now termed the Mitchell-Riley syndrome (17–22). In 2010, Smith et al. reported a total 6 cases from five families (from previous reports or new) with similar features who all carried damaging mutations in RFX6 (5). An additional isolated seventh case (Proband 6) was subsequently found also to have an RFX6 mutation (6).
The longest surviving case (Proband 3, alive at 9 years), was the only compound heterozygote, with two different splice site mutations, while the next oldest (Proband 2, alive at 6 years) had a homozygous missense mutation that was shown to reduce but not abrogate DNA binding (5). This genotype-phenotype correlation is also reflected in the birth weights of these two cases being the highest of any reported. However, our case had a phenotype similar in severity to the other cases with homozygous mutations either disrupting a splice site (Proband 1 and sister 1B), introducing a frameshift (Proband 4) or a substitution of a highly conserved residue within the DNA-binding domain shown to completely abrogate DNA binding (Proband 5).
A third surviving case still alive at 21 months (Proband 6) had a homozygous nine-nucleotide deletion spanning the intron 7/exon 8 border and exhibited intractable malabsorptive diarrhea causing parenteral nutrition dependence with frequent life-threatening infections (6). Importantly, this case did not respond to numerous dietary interventions, bile acid sequestrants or exocrine pancreatic supplementation. This clinical picture is reminiscent of cases with neonatal diabetes and intractable diarrhea due to mutations in NEUROG3, a transcription factor upstream of RFX6 important in endocrine pancreatic differentiation and development of the enteroendocrine cells of the gut (23,24). A role for RFX6 in the development of these cells remains unclear. Although chronic diarrhea has been reported in the majority of cases with RFX6 mutations (Table 1), it has not been emphasized as a consistent feature of the syndrome. Careful reporting of such details will be important in establishing genotype/phenotype correlations reflecting underlying molecular genetic mechanisms.
Our case did exhibit feeding intolerance and chronic diarrhea, and was frequently on parenteral feedings, consistent with malabsorption. Although enteroendocrine cellular dysfunction is clearly a possible explanation, the intestinal atresias and subsequent repairs in our case and others may make it clinically difficult to ascertain the cause of diarrhea and/or malabsorption. Another possibility in our case is exocrine pancreatic insufficiency related to pancreatic hypoplasia that is suggested by his low but detectable serum amylase and lipase with undetectable stool pancreatic elastase. However, he did not exhibit significant improvement with pancreatic enzyme replacement. Furthermore, autopsy of the pancreas in one previous case suggested preservation of exocrine pancreatic tissue despite the absence of staining for insulin, glucagon or somatostatin (17), it is unclear if RFX6-related disruption of pancreatic development leads to clinically significant loss of exocrine pancreatic function. Further study of human patients and animal models should clarify the role of RFX6 in pancreatic development and function, as well as in intestinal and enteroendocrine cellular development.
Acknowledgements
These studies were supported in part by a grant from the American Diabetes Association (1–11-CT-41), grants from National Institute of Diabetes and Digestive and Kidney Diseases (DK020595 and K23 DK094866) and National Human Genome Research Institute (HG003273 and HG006542) of the National Institutes of Health, and gifts from the Kovler Family Foundation and Lewis-Sebring Foundation.
REFERENCES
- 1.Slingerland AS, Shields BM, Flanagan SE, Bruining GJ, Noordam K, Gach A, et al. Referral rates for diagnostic testing support an incidence of permanent neonatal diabetes in three European countries of at least 1 in 260,000 live births. Diabetologia. 2009 Aug;52(8):1683–1685. doi: 10.1007/s00125-009-1416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greeley SAW, Naylor RN, Philipson LH, Bell GI. Neonatal diabetes: an expanding list of genes allows for improved diagnosis and treatment. Curr Diab Rep. 2011 Oct 13;11(6):519–532. doi: 10.1007/s11892-011-0234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aftab S, Semenec L, Chu J, Chen N. Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol Biol. 2008;8(1):226. doi: 10.1186/1471-2148-8-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scharfmann R, Polak M. Transcribing neonatal diabetes mellitus. N Engl J Med. 2010 Apr 22;362(16):1538–1539. doi: 10.1056/NEJMcibr1001845. [DOI] [PubMed] [Google Scholar]
- 5.Smith SB, Qu H-Q, Taleb N, Kishimoto NY, Scheel DW, Lu Y, et al. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010 Feb 11;463(7282):775–780. doi: 10.1038/nature08748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spiegel R, Dobbie A, Hartman C, de Vries L, Ellard S, Shalev SA. Clinical characterization of a newly described neonatal diabetes syndrome caused by RFX6 mutations. Am J Med Genet. 2011 Sep 30;155A(11):2821–2825. doi: 10.1002/ajmg.a.34251. [DOI] [PubMed] [Google Scholar]
- 7.Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010 Sep;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Jian X, Boerwinkle E. dbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011 Jul 26;32(8):894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siepel A, Pollard K, Haussler D. New methods for detecting lineage-specific selection. Proceedings of the 10th international conference on research in computational molecular biology (RECOMB 2006); 2006. pp. 190–205. [Google Scholar]
- 11.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 12.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010 Apr;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010 Aug;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 14.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009 Sep;19(9):1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4(3):311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 16.Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 1991 Jul 5;220(1):49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell J, Punthakee Z, Lo B, Bernard C, Chong K, Newman C, et al. Neonatal diabetes, with hypoplastic pancreas, intestinal atresia and gall bladder hypoplasia: search for the aetiology of a new autosomal recessive syndrome. Diabetologia. 2004 Dec 8;47(12):2160–2167. doi: 10.1007/s00125-004-1576-3. [DOI] [PubMed] [Google Scholar]
- 18.Verwest AM, Poelman M, Dinjens WN, Batstra MR, Oostra BA, Lequin MH, et al. Absence of a PDX-1 mutation and normal gastroduodenal immunohistology in a child with pancreatic agenesis. Virchows Arch. 2000 Dec 1;437(6):680–684. doi: 10.1007/s004280000305. [DOI] [PubMed] [Google Scholar]
- 19.Ashraf A, Abdullatif H, Hardin W, Moates JM. Unusual case of neonatal diabetes mellitus due to congenital pancreas agenesis. Pediatr Diabetes. 2005 Dec 1;6(4):239–243. doi: 10.1111/j.1399-543X.2005.00114.x. [DOI] [PubMed] [Google Scholar]
- 20.Galán-Gómez E, Sánchez EB, Arias-Castro S, Cardesa-García JJ. Intrauterine growth retardation, duodenal and extrahepatic biliary atresia, hypoplastic pancreas and other intestinal anomalies: Further evidence of the Martínez-Frías syndrome. Eur J Med Genet. 2007 Mar;50(2):144–148. doi: 10.1016/j.ejmg.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Chappell L, Gorman S, Campbell F, Ellard S, Rice G, Dobbie A, et al. A further example of a distinctive autosomal recessive syndrome comprising neonatal diabetes mellitus, intestinal atresias and gall bladder agenesis. Am J Med Genet. 2008 Jul 1;146A(13):1713–1717. doi: 10.1002/ajmg.a.32304. [DOI] [PubMed] [Google Scholar]
- 22.Martinovici D, Ransy V, Vanden Eijnden S, Ridremont C, Pardou A, Cassart M, et al. Neonatal hemochromatosis and Martinez-Frias syndrome of intestinal atresia and diabetes mellitus in a consanguineous newborn. Eur J Med Genet. 2010 Jan;53(1):25–28. doi: 10.1016/j.ejmg.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Pinney SE, Oliver-Krasinski J, Ernst L, Hughes N, Patel P, Stoffers DA, et al. Neonatal diabetes and congenital malabsorptive diarrhea attributable to a novel mutation in the human neurogenin-3 gene coding sequence. J Clin Endocrinol Metab. 2011 Jul;96(7):1960–1965. doi: 10.1210/jc.2011-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubio-Cabezas O, Jensen JN, Hodgson MI, Codner E, Ellard S, Serup P, et al. Permanent Neonatal Diabetes and Enteric Anendocrinosis Associated With Biallelic Mutations in NEUROG3. Diabetes. 2011 Mar 29;60(4):1349–1353. doi: 10.2337/db10-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]