Summary
Microsatellites - simple tandem repeats present at millions of sites in the human genome - can shorten or lengthen due to a defect in DNA mismatch repair. We present here the first comprehensive genome-wide analysis of the prevalence, mutational spectrum and functional consequences of microsatellite instability (MSI) in cancer genomes. We analyzed MSI in 277 colorectal and endometrial cancer genomes (including 57 microsatellite-unstable ones) using exome and whole-genome sequencing data. Recurrent MSI events in coding sequences showed tumor type-specificity, elevated frameshift-to-inframe ratios, and lower transcript levels than wildtype alleles. Moreover, genome-wide analysis revealed differences in the distribution of MSI versus point mutations, including overrepresentation of MSI in euchromatic and intronic regions compared to heterochromatic and intergenic regions, respectively, and depletion of MSI at nucleosome-occupied sequences. Our results provide a panoramic view of MSI in cancer genomes, highlighting their tumor type-specificity, impact on gene expression, and the role of chromatin organization.
Introduction
About 15% of sporadic colorectal cancers (CRC) harbor widespread alterations in the length of microsatellite (MS) sequences, known as microsatellite instability (MSI) (Boland and Goel, 2010). Sporadic MSI CRC tumors display unique clinicopathological features including near-diploid karyotype, higher frequency in older populations and in females, and a better prognosis (de la Chapelle and Hampel, 2010; Popat et al., 2005). MSI is known to occur due to a defective DNA mismatch repair (MMR) system with key MMR genes inactivated by various mechanisms such as germline mutation in MSH2 or MLH1 in most Lynch syndrome cases (Bronner et al., 1994; Leach et al., 1993) and epigenetic silencing of MLH1 in most sporadic cases (Herman et al., 1998; Veigl et al., 1998). The DNA slippage within coding sequences can induce frameshifting mutations that result in the production of truncated, functionally inactive proteins. For CRC genomes, cancer-related genes frequently targeted by MSI (e.g., TGFBR2, ACVR2A and BAX) have been studied (Jung et al., 2004; Markowitz et al., 1995; Rampino et al., 1997). MSI is also present in other tumors, such as in endometrial cancer (EC) of the uterus, the most common gynecological malignancy (Duggan et al., 1994). The same reference Bethesda panel originally developed to screen an inherited genetic disorder (Lynch syndrome) (Umar et al., 2004) is currently applied to test MSI for CRCs and ECs. However, the genes frequently targeted by MSI in CRC genomes rarely harbor DNA slippage events in EC genomes (Gurin et al., 1999) and it is largely unknown whether MS-unstable EC genomes have similar molecular origins or functional consequences as CRC genomes.
In this study, we utilize the exome and whole-genome sequencing data for CRC and EC genomes from The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas Network, 2012; Cancer Genome Atlas Network, 2013) to profile the genomic landscape of MSI in these two tumor types, including the patterns of single nucleotide variations (SNV) in MMR pathways, a comprehensive catalog of genomic loci with frequent MSI, the genomic distribution and sequence properties of the affected microsatellites, and correlations with other genomic and epigenetic features.
Results
The mutational spectrum of exome-wide MSI in cancer genomes
To examine the impact of MSI on protein-coding sequences, we analyzed the exome sequencing data for 147 CRC and 130 EC patients (Table S1). The initial cohorts included 27 CRC/30 EC MSI-H (MSI-high), 23/11 MSI-L (MSI-low) and 97/89 MSS (MS-stable) genomes, as evaluated by the revised Bethesda guidelines (Umar et al., 2004). We used computational methods to identify MS contained within sequencing reads and to detect significant differences in their lengths between tumor and matched normal genomes (see Experimental Procedures). Whereas the current Bethesda panel categorization simply classifies cancer genomes into MSI-H, MSI-L and MSS based on the number of markers altered, our analysis shows that MSI-H genomes show a dramatically higher number of MSI events (median of 290 and 126 MSI events per MSI-H CRC and EC genome, respectively) compared to MSI-L (median of 5 and 2) and MSS (median of 4 and 1) in both cancer types (Figures 1A, 1B and S1). The difference in the number of MSI events is not significant between MSI-L and MSS (P = 0.22 and 0.42 for CRC and EC; Figure S1). Details on the identified MSI events are in Tables S2 and S3. When corrected for the background distribution of different repeat types in the exome reference set of MS, we observe a depletion of MSI events in coding sequences, likely reflecting purifying selection of mutations involving coding sequences (Figure 1C).
Figure 1. The mutational spectrum of MSI events and MMR genes in CRC and EC genomes.
(A) The number of MS loci with significant tumor genome-specific DNA slippage events is shown for each of CRC genomes (141 cases with data on MLH1 promoter hypermethylation are displayed out of 147; also see Figure S1), along with the SNV mutation rate. The samples are sorted in decreasing order of MSI events. The MSI status based on the Bethesda criteria (25 MSI-H, 23 MSI-L and 93 MSS cases) are noted. The functional status of selected MMR genes and POLE are classified into MSI events (frameshift and in-frame), nonsilent point mutations (missense or nonsense) and transcriptional silencing of MLH1 by hypermethylation. See main Text for the sample with an arrow. (B) Similar to (A) for EC genomes (115 cases with MLH1 promoter hypermethylation data are displayed out of 130). (C) (Left) For the 27 MSI-H CRC genomes, the numbers of MSI events in the four different categories of genomic regions (coding, noncoding and 5′/3′ UTR) are shown in the upper panel. In the lower panel, the number of MSI events was normalized by the total number of MS in the exome reference set for each category. (Right) Same analysis for MSI-H EC genomes (three samples with <10 MSI were removed). See also Figure S1 and Table S1, S2 and S3.
We next examined the relationship between MSI events and SNV mutation rates as well as the mutation status of key MMR genes (Figures 1A and 1B). Our combined mutational profiles highlight three main features. First, we observe the vulnerability of specific MMR genes to different types of somatic mutations as their inactivating mechanism. Although most of the MSI-H CRC and EC genomes harbor transcriptional silencing of MLH1 by promoter hypermethylation, frameshifting DNA slippage events are the primary inactivating mechanism for MSH3 and, to a lesser extent, for MSH6 in MS-unstable CRC and EC genomes. Other MMR genes such as MSH2, PMS1 and PMS2 only harbor nonsilent (missense or nonsense) SNVs, mostly in the hypermutated samples. Second, complementary mechanisms of inactivation are observed for some genes. For example, nonsilent SNVs and DNA slippage events are mutually exclusive for both MSH3 and MSH6 in MS-unstable genomes, suggesting that these two may be alternative mechanisms for inactivation of those genes (Ciriello et al., 2012). Third, a number of samples show highly elevated SNV mutation rates, most of them harboring missense mutations of POLE (Cancer Genome Atlas Network, 2012; Cancer Genome Atlas Network, 2013), but there is no relationship between SNV mutation rates and MSI. In addition, POLE-mutated genomes can be largely classified into two classes depending on the MLH1 status: MS-unstable genomes (inactivation of MLH1) and MS-stable ones (functional MLH1). The highly elevated mutation rates are observed for the latter. It is possible that POLE mutations in MS-unstable genomes are late events. Alternatively, MSI is sufficient to achieve the phenotypes required by cancer cells in MS-unstable genomes and/or these genomes do not tolerate the additional mutation burden from SNVs. Our observations also highlight the primary role of MLH1 inactivation in the establishment of an MSI phenotype since POLE-mutated genomes with functional MLH1 maintain the MS stability in the presence of frequent nonsilent SNVs in MMR genes. We observe two POLE-mutated MSI-H genomes (1 CRC and 1 EC; arrows in Figure 1) with nonsilent MLH1 mutations but not transcriptional silencing of MLH1, in which the genomic instability associated with POLE mutation might have triggered inactivation of MLH1 leading to the MSI phenotype.
Loci frequently targeted by MSI show a higher rate of frameshift events
For each MSI event, we examined the distribution of changes in the length of the mutant MS allele compared to its germline counterpart. After clustering the MSI events, the heatmap, which mimics the electrophoretic autoradiogram in a conventional MSI study, illustrates the extent of allelic shift for each MSI event (Figures 2A and 2D). Most allelic shifts are deletions and a higher allelic shift in the length of the mutant allele is more frequent in 3′ UTR than in coding regions. Figure 2A is for MSI events at mononucleotide repeats; a similar pattern is also observed for dinucleotide repeats (Figure S2). We further classified MSI events into low- and high-allelic shift (LAS and HAS, respectively) depending on whether the mode (most frequent value) of the MS allele lengths is equal to its germline length or not. The ratio of LAS/HAS events is higher in coding regions than in 5′/3 UTRs or noncoding regions (Figures 2B and 2E). An evolutionary model previously proposed (Tsao et al., 2000) suggests that HAS events are more likely to have functional impact than LAS events. Thus, a substantially higher LAS/HAS ratio provides additional evidence for negative selection of MSI events in coding regions.
Figure 2. The distribution of allelic shift in MSI events and the properties of recurrent coding MSI.
(A) For MSI events occurring at the mononucleotide MS (y-axis; each row) in the CRC genomes, the deviations in the allele lengths (−10bp to +5bp) compared to the germline counterparts are shown as normalized allelic fractions in a heatmap (the values in each row add up to 1), clustered by their similarity. The locations of the corresponding MS (coding, noncoding and 5′/3′ UTR) are shown on the right. (B) MSI events are classified into low- and high-allelic shift (MSI-LAS and MSI-HAS) cases. The graph shows the different frequencies of the two MSI types for the four categories in CRC genomes. (C) MSI events in the coding sequences (CDS) and non-CDS regions are further classified into frameshift and in-frame mutations for CDS (non-triplet and triplet for non-CDS). The frameshift-to-inframe ratio increases with respect to the level of recurrence (% of MS-unstable genomes harboring the mutation; the width of each bar is proportional to the number of MSI) for CDS MSI events. (D–F) Similar to the above for EC genomes. (G) The distribution of A10 homopolymer length on TGFBR2 locus is shown for one CRC genome with positive MSI calls as measured by Sanger- (upper) and exome-sequencing data (below). (H) Similar to AA-2676 as an MSI-negative example. (I–J) The MSI events per sample are compared to those made after local realignment by GATK or by global realignment by Novoalign for 27 MSI-H CRC (I) and 30 EC genomes (J). Overlap and specific calls are distinguished to those overlapped with BWA-based calls or not, respectively. See also Figure S2.
MSI events on trinucleotide repeats were primarily observed in coding sequences and showed comparable numbers of LAS and HAS events for coding MSI (Figure S2), probably due to their relatively neutral nature (i.e., in-frame) in coding sequences (Metzgar et al., 2000). Thus, we further categorized coding MSI into nontriplet (frameshift) and triplet (in-frame) events, similar to the distinction between nonsynonymous and synonymous SNV mutations (Greenman et al., 2007). The percentage of frameshift and in-frame MSI events is shown with respect to the level of recurrence for CRC and EC genomes (Figures 2C and 2F). MSI events of mononucleotide repeats are largely responsible for this relationship given the predominance of mononucleotide-MSI events (92.4% and 93.0% of total MSI events in CRC and EC genomes). For both tumor types, non-recurrent coding MSI events show a lower frequency of frameshift MSI events compared to those occurring in non-coding or UTR regions (non-CDS) at a similar recurrence level, concordant with the negative selection of frameshifting MSI events on coding sequences. Importantly, highly recurrent coding MSI events show a higher frameshift-to-inframe ratio compared to non-recurrent coding MSI (Figures 2C and 2F). This suggests that these non-neutral genomic events may provide selective advantages to the affected clones to overcome the purifying selection on mutations involving coding sequences. Thus, we hypothesize that the genes inactivated by the recurrent frameshift MSI may have tumor suppressive roles in CRC and EC genomes.
We evaluated the performance of our sequencing-based method by comparing its MSI calls on one of the Bethesda markers (TGFBR2, A10 homopolymer) with those from the fragment length assay by Sanger sequencing. Sequencing-based MSI screening identified 20 out of 22 TGFBR2 MSI calls made from Sanger sequencing in 126 CRC genomes without false positives (sensitivity and specificity of 91% and 100%, respectively). For EC genomes, exome and Sanger calls for TGFBR2 were made only on 3 of 130 genomes and they were concordant in every case. Examples of one positive and one negative TGFBR2 MSI calls are shown in Figures 2G and 2H (See also Figure S2). These results strongly support the robustness of our sequencing-based MSI calls. In the two false negative cases, the differences observed in the distributions of MS lengths were not statistically significant due to low read coverage. Refinement on the significance threshold may improve sensitivity for the exome-based approach.
Next, we calculated the frequency of frameshift MSI for each gene in the MSI-H tumors (Figure 3). The frequencies in the two tumor types were moderately correlated (R = 0.470), with some loci such as ASTE1 and CASP5 showing comparable MSI frequency in both. But we also discovered a substantial number of genes targeted by recurrent frameshift MSI with tumor type-specificity. These genes include a few well-known ones such as ACVR2A and TGFBR2 (Markowitz et al., 1995; Parsons et al., 1995; Wang et al., 1995), as well as MSH3. Various molecular functions are perturbed by CRC-specific recurrent MSI events, e.g., SLC22A9 and TMEM22 encode transport-related molecules; SREK1IP1, LTN1 and SEC63 are related to protein metabolism. Among the novel loci with frequent frameshift MSI such as SMAP1 and AIM2, the potential apoptotic role of AIM2 has been reported (Fernandes-Alnemri et al., 2009).
Figure 3. The genes harboring frameshift MSI in CRC and EC genomes and tumor type specificity.
A scatter plot shows the distribution of genes with respect to their frequency of frameshift MSI in CRC and EC genomes. The 27 genes with frameshift MSI in >30% of CRC or in >15% of EC MSI-H genomes are noted. The color gradient indicates the extent of tumor type-specificity (red and blue for CRC- and EC-specificity, respectively). The size of the circles indicates the number of genes with the corresponding MSI frequencies. See also Figure S3 and Table S4.
Among the novel genes with EC-specific frameshift MSI, MSI events on JAK1 coding sequences were observed in 30% of MSI-H EC genomes but none in CRC genomes. Although the protein tyrosine kinase encoded by JAK1 has been reported as an upstream component of the oncogenic JAK-STAT signaling pathway, whether the locus is frequently subject to MSI or what its functional implication might be was largely unknown. Gene set enrichment analysis (GSEA) revealed that MS-unstable EC genomes harboring the JAK1 frameshift MSI may have suppression of JAK-STAT signaling, as evidenced by the repressed transcript levels of genes belonging to the pathway and the transcriptional activation of cell cycle-related genes (Figure S3). TFAM also showed EC-exclusive frameshift MSI. mtTFA (mitochondrial transcription factor A) encoded by TFAM has a role in apoptosis and DNA repair (Larsson et al., 1998), and expression of mtTFA was associated with cancer prognosis (Nakayama et al., 2012). EC-specific frameshift MSI events were also observed in PDS5B, whose interaction with BRCA2 is required for BRCA2-RAD51-mediated DNA damage repair process (Brough et al., 2012), and in ESRP1, whose underexpression is involved in the aberrant splicing pattern during TGFβ-induced epithelial-mesenchymal transformation (Horiguchi et al., 2012). In addition, immune and apoptosis-related gene functions are enriched in genes frequently targeted by frameshift MSI in both tumor types (Table S4).
Bias in allelic expression due to MSI events
To investigate the potential influence of MSI events on the expression level of the affected genes, we compared the differential allelic read counts (wild type versus mutant alleles) from RNA-seq with those from exome data. A statistically significant bias (FDR < 0.05, Fisher’s exact test) was observed for 223 and 131 MSI calls in the MS-unstable CRC and EC genomes, corresponding to 16% and 11% of the total MSI calls with a minimum of 10 RNA-seq reads (Table S5). When we categorized these biases into overexpressed MSI (RNAseqmutant/RNAseqwildtype > Exomemutant/Exomewildtype) and underexpressed MSI, most of the frameshift MSI were in the underexpressed group in both tumor types (Figures 4A and 4B).
Figure 4. Association between MSI and changes in expression level.
(A) The MSI events in CRC genomes accompanied by a significant deviation in expression levels between the wildtype versus mutant alleles are classified into ‘MSI-overexpressed’ and ‘MSI-underexpressed’ in each of four regions. The asterisk indicates significant differential counts (binomial test; P < 0.05) for frameshift coding (P = 0.0009), in-frame coding (P = 0.0462) and 3′ UTR MSI (P = 0.0002). (B) Similarly for EC genomes with significant differential counts for 5′ UTR (P = 0.0110) and frameshift coding MSI (P = 0.0027). (C) The 37 MS loci showing MSI-overexpression or MSI-underexpression in two or more CRC genomes are shown (x-axis; left), along with 14 such MS loci from EC genomes (right). The associated gene symbols and the location of the MS (‘C’, ‘N’, ‘5’, and ‘3’ for coding, noncoding, 5′ UTR, and 3′ UTR MSI) are shown. For each MS locus, the number of samples showing differential expression (over- or under-expressed) is plotted (y-axis). (D) The log2 ratio of the expression levels is shown (y-axis). A higher ratio indicates that the gene showed higher expression in the genomes with the corresponding MSI than those without. An asterisk indicates significant (T-test, P < 0.05) difference in the expression level. See also Table S5.
For genes with significant allelic expression biases in multiple samples, the expression changes for transcripts with the mutant alleles were generally in the same direction (33 of 37 genes for CRC and 8 of 14 for EC showed perfect concordance; Figure 4C). For example, the MS alleles with DNA slippage events in the 3′UTR of ANTXR1 showed significantly lower transcript levels than the wildtype alleles in all eight CRC genomes. We also compared the transcript levels between the genomes with and without the corresponding MSI (Figure 4D). The expression changes were concordant with the within-sample ratios of over- or under-expression of the mutant allele, with 13 genes showing significant differences.
Genome-wide landscape of MSI
We extended our analysis to genome-wide using whole-genome sequencing data from 7 CRC and 10 EC genomes (4 and 5 MSI-H genomes, respectively). The number of MSI events for MSI-H genomes ranged from 11,380 to 332,565 (excluding one EC outlier with 162, which is likely to be a misclassification by the Bethesda panel), in contrast to 5 to 7446 observed in MSS cases (Figure 5A). For subsequent analyses, we selected the six MSI-H genomes (4 CRC and 2 EC genomes) with the largest number of MSI events. The genome-wide distribution of MS loci targeted by MSI reveals a strong depletion at coding sequences and 5′ UTRs, similar to the exome-wide mutational spectrum (Figure 5B). After normalizing for the MS counts in each category, the frequency of MSI in 3′ UTR is comparable to those in intronic or intergenic regions (Figure 5C). Analysis of MSI calls with respect to nucleotide composition and repeat length reveals high variability of mutation frequency depending on the MS length. For instance, up to 50% and 40% of A/T and C/G mononucleotide MS with germline length 12–14 bp can have MSI in some samples, but the MSI frequency of di- and trinucleotide repeats tends to increase with longer repeats (Figure S4). Although variable genomic abundances of different MS repeat types have been reported (Subramanian et al., 2003), our results further suggest that the preference of DNA slippage events largely depends on the sequence composition and length of the repeats.
Figure 5. Genome-wide landscape of MSI.
(A) The number of MSI events genome-wide is shown for the 17 samples with whole-genome sequencing data. Six genomes (4 CRC and 2 EC genomes) with >60,000 MSI events are shaded grey and used for subsequent analyses. (B) The MSI events are classified into five categories based on their genomic location. (C) The number of MSI calls is normalized by the background MS abundance in their respective regions of the genome to obtain MSI frequency. See also Figure S4.
Next, we employed correlative analysis to identify genomic features associated with the occurrences of MSI. First, we find that the local MSI frequency (measured in 1 Mb bins) is inversely correlated with SNV density in four human cancer types (Figure 6A). Second, MSI frequencies are positively correlated with H3K4me3, H3K9ac, H3K36me3, and others that mark open chromatin and transcriptionally active regions, but are negatively correlated with repressive histone modifications such as H3K9me2, H3K9me3, and H3K27me2 (Figure 6B). Figure S5 also shows the correlation of MSI frequency with other genomic features. The preference of DNA slippage events toward open chromatin-like domains is consistent regardless of the bin sizes used (100 kb to 10 Mb; Figure S5); when the MSI frequency across the genome was compared with the chromatin state map defined in nine human cell lines (Ernst et al., 2011), the same pattern was observed (Figure S5). Similarly, genomic segments with early, intermediate, and late DNA-replicating timing have high-to-low MSI frequencies (Figure 6C). Multiple linear regression models (Schuster-Bockler and Lehner, 2012) were adopted to examine the extent of variations in MSI frequencies that can be predicted by a combination of multiple genomic features in CRC and EC genomes (Figure S5).
Figure 6. Correlation with epigenomic features.
(A) The Pearson correlation between MSI frequency and SNV density (measured using 1Mb bins) is shown for four human cancer types. For the ‘Total’ category, SNV densities from the cancer types were combined. (B) The same correlation analysis was performed between the frequency of MSI and enrichment of various histone modifications. (C) MSI frequencies in the early-, intermediate- and late-replicating timing regions are shown. See also Figure S5.
The over-representation of cancer-specific somatic SNVs in heterochromatin-like (Schuster-Bockler and Lehner, 2012) and late-replicating domains (Koren et al., 2012) may be explained by the limited accessibility of DNA repair complexes on closed, heterochromatin-like domains (Peterson and Cote, 2004). However, this assumption is not applicable to MSI in MS-unstable genomes with a deficient MMR system. Further investigation is required to determine whether the increased MSI frequency in open chromatin-like domains arises during DNA replication or is a post-replication event. We also observed that MSI frequency is higher in introns than intergenic regions (Figure 5C; P = 0.002), which is the opposite of SNV (Bass et al., 2011). The depletion of SNVs in introns is probably due to transcription-coupled repair (Pleasance et al., 2010a); elevated MSI frequency introns suggests that MSI in MMR-deficient cancer genomes may undergo different evolutionary or fixation processes.
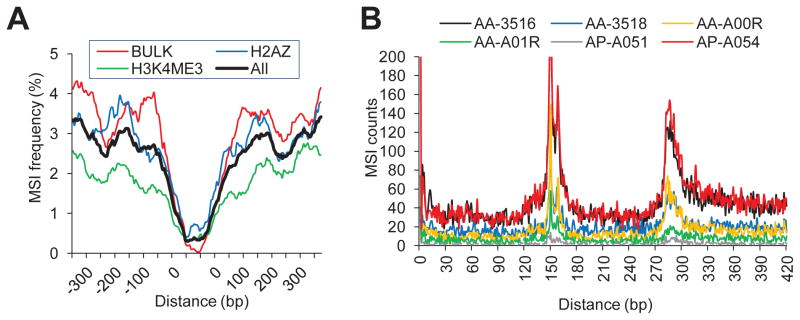
Finally, high-resolution analysis of the MSI frequency with respect to nucleosome occupancy demonstrated that the depletion of MSI events around the positions of bulk nucleosomes as well as epigenetically modified nucleosomes H2A.Z and H3K4me3 (Figure 7A and Figure S6). Analysis of the distances between adjacent MSI events (a pair of MSI calls separated by <500 bp) showed two pronounced peaks at ~150bp and ~285bp (Figure 7B). This periodicity agrees well with the known core nucleosome size of 147bp. Neither a depletion around nucleosomes nor a local periodicity was observed for somatic SNVs from four cancer types (Figure S6).
Figure 7. Depletion of MSI around stable nucleosome positions.
(A) MSI frequency around stable nucleosome positions is shown for one CRC genome (AA-3516; also see Figure S6). (B) The distribution of distances between adjacent MSI pairs indicates periodicity associated with the nucleosome size. See also Figure S6.
Discussion
Our comprehensive survey of genomic loci with MSI has allowed us to gain insights on functional consequences of DNA slippage events on coding sequences and their associations with various genomic and epigenomic features. The classification of samples into the traditional MSI-H, MSI-L and MSS categories based on the number of MSI events agreed well with the benchmark results based on the Bethesda guidelines, but the number of MSI calls was highly variable across the genomes. Besides categorizing the cancer genomes into MS-unstable and -stable ones, the number of MSI events and the related features can be useful in the evolutionary study of cancer genomes (Tsao et al., 2000).
We observed that the MSI-L and MSS categories do not show significant differences in the number of MSI events (Figure S1). Although down-regulation of transcript levels or allelic loss of MSH3 has been reported for MSI-L CRC genomes (Plaschke et al., 2012), our analysis of MSI-L and MSS genomes does not show significant differences in MSH3 expression levels (Figure S1). Most studies of clinical correlates for CRC have observed little or no differences between MSI-L and MSS tumors, and usually collapse these two groups into one (Jass, 2007). In light of our finding of similar numbers of MSI events in MSS and MSI-L tumors, we recommend discontinuation of the use of MSI-L as a distinct classification of CRC and EC tumors.
A gene-level analysis of recurrent events revealed that the ratio of frameshift-to-inframe mutations can be informative in distinguishing driver mutations from passenger ones, and both CRC and EC genomes showed a substantial level of tumor type-specificity in the genes targeted by MSI. The MSI events on the TGFβ pathway genes such as ACVR2A and TGFBR2 in MS-unstable CRC genomes may represent pathway-level equivalent of recurrent SNVs at other TGFβ pathway genes (e.g., SMAD2 and SMAD4) in MS-stable CRC genomes (Cancer Genome Atlas Network, 2012). It was previously shown that some MS loci with recurrent MSI events in CRC genomes are not frequently altered in EC genomes (Kuismanen et al., 2002). Consistent with this, our exome-wide MSI screening clearly demonstrates tumor type-specificity in recurrent MSI targets, with some novel candidates in EC genomes such as JAK1 and TFAM. JAK1 MSI may be functional given its level of recurrence (30% in MSI-H EC genomes) and its association with transcriptional downregulation of multiple gene members in the JAK-STAT pathway. The genetic perturbation of the JAK-STAT pathway was shown to decrease cellular survival of colon cancer cells in vitro (Xiong et al., 2008), which may explain the absence of the JAK1 MSI in MS-unstable CRC genomes. Tumor type-specific MSI targets often involve a similar molecular function, such as MSH3 (CRC) and PDS5B (EC) in DNA repair processes and AIM2 (CRC) and TFAM (EC) in apoptotic pathways. Elucidation of the mechanisms for tumor-type specific targeting of MSI as well as potential molecular functions of the common and tumor type-specific mutations will require further investigation.
Alteration of transcription levels due to an MSI event in the 3′UTR has been attributed to the disruption of the nearby binding sites for microRNA or RNA-binding proteins (Paun et al., 2009; Yuan et al., 2009), but the impact of the MSI mutations in the coding regions and the subsequent changes on expression has not been reported previously. Our result on allele-specific expression combining transcriptome- and exome-sequencing data suggests that frameshift MSI events are often accompanied by lower transcript levels of the corresponding alleles. Increased frequency of SNVs in low-expressed genes has been reported for some cancer types (Nik-Zainal et al., 2012; Pleasance et al., 2010a; Pleasance et al., 2010b). The preference of underexpression for frameshift MSI may be associated with a known RNA surveillance pathway that eliminates mRNA containing a premature stop codon (e.g., nonsense-mediated decay) (Chang et al., 2007), consistent with the negative selection of the non-neutral mutations in the coding region.
In spite of the similarities between MSI and SNV such as their preference on 3′ UTR (Pleasance et al., 2010a) and depletion on coding sequences (Bass et al., 2011), our correlative analysis revealed that their frequencies are largely anti-correlated with major differences in their regional frequencies. First, MSIs and SNVs are overrepresented in euchromatin- and heterochromatin-like domains, respectively. Second, MSIs are more enriched in introns than intergenic regions as opposed to SNVs. Third, the depletion of MSI associated with nucleosome occupancy was not observed for SNVs. For SNVs, it has been proposed that the inaccessibility of DNA repair machineries and the transcription-coupled repair are responsible for the overrepresentation of SNVs in heterochromatin-like domains and intergenic regions, respectively. However, in the context of MMR dysfunction in MS-unstable genomes, the regional preference of MSIs might have arisen during DNA replication not as post-mutational events like SNVs. One hypothesis to explain the increased MSI frequency in open chromatin is that the proofreading capabilities of DNA polymerases may be dependent on the accessibility of the chromatin. For example, the replication fork can move more rapidly in open chromatin with increased DNA slippage errors but in closed chromatin, the slower movement of the replication fork may enhance the proofreading capabilities of DNA polymerase subunits of POLE and POLD1 (Preston et al., 2010). This chromatin states-dependent fidelity of DNA polymerases hypothesis may also explain the decreased MSI frequencies in the nucleosome-occupied DNA segments.
The performance of our MSI-calling algorithm depends on the ability to accurately measure the length of a given MS allele. One problem with current sequencing technology is the frequent error in measuring the length of homopolymers (i.e., mononucleotide MS repeat). In this study, we used data from the Illumina platform, which uses reversible terminators that allows incorporation of just a single nucleotide at a time and is currently the most reliable platform with respect to the homopolymer issue (Dohm et al., 2008). High concordance between exome- and Sanger-sequencing data for a A10 homopolymer (the Bethesda marker TGFBR2) suggests that our method performs well (Figures 2G and 2H; Figure S2). Illumina sequencing is still prone to a higher error rate for longer homopolymers (Minoche et al., 2011), but its impact on our analysis is minimal because ours is based on tumor versus normal comparison.
We used read alignment by BWA (Burrows-Wheeler Aligner) (Li and Durbin, 2009) to associate the intra-read MS repeats to the corresponding genomic loci. We have also tested additional local realignment by Genome Analysis Toolkit (GATK) (DePristo et al., 2011) or indel-sensitive Novoalign (Krawitz et al., 2010), but the number of MSI calls was very similar (Figures 2I and 2J) and the sensitivity in detecting MSI events on TGFRB2 remained exactly the same. Local or global realignment may improve MSI calling, but a systematic evaluation will be required to delineate its platform- or software-dependencies.
In this study, we demonstrated that conventional exome sequencing of tumor and matched normal genomes is able to capture the exon-centric MSI events; however, there may also be some intronic and intergenic MSI events with functional significance. It was reported that MSI events near splicing sites may alter the transcript level or splicing pattern of the target genes as shown for MRE11 (Giannini et al., 2004) and HSP110 (Dorard et al., 2011), respectively. In addition, the quasimonomorphic allelic nature of a Bethesda marker (BAT-26 located at the 3′ splice site of MSH2 exon 5) in the normal population (Zhou et al., 1997) has suggested potential functional significance of MS repeats around splice sites. The availability of a larger cohort with whole-genome sequencing data will be needed to facilitate the identification of functionally important, recurrent non-coding MSI events in intronic or intergenic regions.
Experimental Procedures
Datasets
TCGA data were downloaded from dbGaP (http://www.ncbi.nlm.nih.gov/gap, accession: phs000178.v8.p7). We obtained exome data for 147 CRC and 130 EC patients as well as whole-genome data for 7 CRC and 10 EC patients (tumor and matched normal genomes). All reads were 100 bp paired-end reads. We confined our analysis to those generated on the Illumina platforms.
MSI annotation of TCGA genomes
The MSI status (MSI-H, MSI-L and MSS) and the clinicopathological parameters were obtained from the TCGA website (https://tcga-data.nci.nih.gov). MSI status was evaluated by TCGA using a panel of four mononucleotide repeats (BAT25, BAT26, BAT40, and TGFBRII) and three dinucleotide repeats (D2S123, D5S346 and D17S250), except for a subset of CRC genomes evaluated by five mononucleotide markers (BAT25, BAT26, NR21, NR24, and MONO27). Tumors were classified as MSI-H (> 40% of markers altered), MSI-L (< 40% of markers altered) and MSS (no marker altered). The methylation data of MLH1 promoter was available for 141 CRC and 115 EC genomes.
Identification of a reference set of MS repeats
To generate an exome-wide reference set of MS repeats, we downloaded the mRNA sequences of 39,496 RefSeq genes (USCS Genome Browser; hg18). We used Sputnik (http://espressosoftware.com/sputnik/) to identify MS repeats with different unit length (mono-, di-, tri-, and tetra-nucleotide). We limited our analysis to MS with the size 7–60 bp, as those MS could be detected accurately with the 100 bp reads, and the statistical power to detect longer repeats is lower. The frequency of MS repeats decreases logarithmically with the length of the repeats (e.g., >99% of repeats in the final set of exome- and genome-reference MS are smaller than 40bp), suggesting that the vast majority of MS repeats are examined in our analysis. We found 265,862 MS in total RefSeq sequences. The repeats that encompass splice sites, have undetermined genomic coordinates, or are redundant due to multiple isoforms were removed. The filtered 146,447 MS repeats were categorized into four groups: 50,910 coding, 14,648 5′ UTR, 65,502 3′ UTR and 15,387 noncoding (without reported coding sequences) MS, as annotated in the UCSC Genome Browser. For a genome-wide reference set of MS repeats, a total of 7,894,295 MS repeats were obtained (chromosome 1 to Y) and categorized into five groups (coding, 68,856; 5′ UTR, 15,093; 3′ UTR, 64,849; intronic, 3,193,265; intergenic, 4,552,232).
Detection of a DNA slippage event
The reads were aligned to NCBI build 36 (hg18) using BWA. After filtering reads with low mapping quality, intra-read MS repeats were identified with the same method used to identify reference MS repeats and then intersected with the reference MS repeats by their coordinates. We required the 2bp flanking sequences (both 5′ and 3′) of the intra-read MS repeats to be identical to those of matching reference repeats, ignoring truncated MS repeats. In each genome, the distribution of the repeat allelic length at an MS locus was obtained by collecting the lengths of all intra-read MS repeats mapped to that locus. We compared the distributions of MS lengths from tumor and matched normal genomes at each locus using the Kolmogorov-Smirnov statistic. A false discovery rate (FDR) of < 0.05 was used as a threshold for statistical significance, with a minimum of five tumor and five matched normal reads. We note that the number of MSI “events” refers to the absolute MSI counts per sample, while MSI “frequency” refers to the number of events normalized by the background MS numbers in the reference sets.
Categorization of MS based on allelic length shift
The length of MS repeat measured from each read was compared to the length of the corresponding germline MS repeat (+/− for insertion and deletion, respectively) in the reference set. The differential read counts among different lengths were normalized to obtain relative fractions for each MSI event. Hierarchical clustering was used to group the MSI events with similar profiles. We distinguished MSI events at coding sequences into frameshift and in-frame events depending on whether the allelic length corresponding to the mode of the distribution is non-triplet or not. MSigDB v3 c5 GO categories were used for GSEA (Subramanian et al., 2005). For genes with recurrent frameshift MSI, we used the pre-ranked version of GSEA using the level of recurrence as the weighting parameter for genes.
Allele-specific expression using RNA-seq
RNA-seq reads from MS-unstable CRC and EC tumor genomes were aligned on the RefSeq sequences using BWA. For the 1,143 and 1,224 MSI calls supported by >10 RNA-seq reads with intra-read MS repeats, the differential RNA-seq read counts from wildtype and mutant alleles (depending on whether the allelic MS length is equal to that of germline or not) were compared with those from exome sequencing using Fisher’s exact test. For the 37 and 14 MS loci with significant expression bias in more than one genome, the extent of differential expression was compared between the cancer genomes with or without the MSI at each locus. For gene-level expression, we used log2(RPKM + 1) from RNA-seq data (RPKM, reads per kb per million mapped reads).
Correlative analysis with epigenomic features
Genome-wide features were obtained as previously described (Schuster-Bockler and Lehner, 2012). We limited the analysis to autosomal features. SNV of four human cancer types (leukemia, lung, melanoma, and prostate cancers) were downloaded from the Supplementary Data sections of the respective studies (Berger et al., 2011; Pleasance et al., 2010a; Pleasance et al., 2010b; Puente et al., 2011). Germline polymorphisms (dbSNP build 130), GC contents, genomic coordinates of CpG islands, recombination rates, and conservation scores (all in hg18) were downloaded from the UCSC Genome Browser. Hi-C data (Lieberman-Aiden et al., 2009) were obtained from the Gene Expression Omnibus database (accession: GSE18199). For comparison with the 18 histone acetylation and 17 methylation markers as well as the occupancy of RNA polII, CTCF, and H2AZ, the reads were downloaded as instructed in the original studies (Barski et al., 2007; Wang et al., 2008). The coordinates of the chromatin state map defined in nine human cell lines were downloaded from the UCSC genome browser. To annotate the chromatin states with respect to DNA replication timing, Repli-Seq data were obtained (Hansen et al., 2010). For GM12878, for which both chromHMM and RepliSeq datasets were available, we calculated the ratio of the early-vs-late replication timing (G1B and S1 vs S4 and G2) for each of the 15 chromatin states. Three chromHMM states with the highest and lowest early-vs-late ratio were annotated as ‘Early’ and ‘Late’ replication, with the remaining segments annotated as ‘Intermediate’. To examine the extent of variations in MSI frequencies that can be predicted by a combination of multiple genomic features, we adopted a multiple linear regression model (Schuster-Bockler and Lehner, 2012). Fifty genomic features including the gene expression level were tested in an iterative manner and the models with minimal Bayesian information criterion (BIC) were selected. The genomic occupancy profiles of nucleosomes were obtained from our previous study (Tolstorukov et al., 2011).
Supplementary Material
Highlights.
Novel genes with recurrent MSI, many with tumor type-specificity, are identified
Recurrent coding MSI are distinguished by higher frameshift-to-inframe ratios
Coding MSI has functional consequences, such as lower transcript levels on average
MSI frequency is associated with chromatin organization and nucleosome positioning
Acknowledgments
We thank The Cancer Genome Atlas Research Network for generating the data used in this work. We also thank the members of the Park laboratory (especially Drs. Eunjung Lee, Nils Gehlenborg, and Semin Lee) and Dr. Peter Kharchenko for providing comments on the manuscript, Dr. David Wheeler for helpful discussions, and the Research Information Technology Group at Harvard Medical School for providing computational resources. This work was supported by grants from the National Institutes of Health (R01 GM082798 and U24CA144025 to PJP) and the National Research Foundation of Korea (2012R1A5A2047939 to TMK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, Sougnez C, Voet D, Saksena G, Sivachenko A, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43:964–968. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brough R, Bajrami I, Vatcheva R, Natrajan R, Reis-Filho JS, Lord CJ, Ashworth A. APRIN is a cell cycle specific BRCA2-interacting protein required for genome integrity and a predictor of outcome after chemotherapy in breast cancer. EMBO J. 2012;31:1160–1176. doi: 10.1038/emboj.2011.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Integrated Genomic Characterization of Endometrial Carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012;22:398–406. doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Chapelle A, Hampel H. Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol. 2010;28:3380–3387. doi: 10.1200/JCO.2009.27.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del AG, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorard C, de TA, Collura A, Marisa L, Svrcek M, Lagrange A, Jego G, Wanherdrick K, Joly AL, Buhard O, et al. Expression of a mutant HSP110 sensitizes colorectal cancer cells to chemotherapy and improves disease prognosis. Nat Med. 2011;17:1283–1289. doi: 10.1038/nm.2457. [DOI] [PubMed] [Google Scholar]
- Duggan BD, Felix JC, Muderspach LI, Tourgeman D, Zheng J, Shibata D. Microsatellite instability in sporadic endometrial carcinoma. J Natl Cancer Inst. 1994;86:1216–1221. doi: 10.1093/jnci/86.16.1216. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini G, Rinaldi C, Ristori E, Ambrosini MI, Cerignoli F, Viel A, Bidoli E, Berni S, D’Amati G, Scambia G, et al. Mutations of an intronic repeat induce impaired MRE11 expression in primary human cancer with microsatellite instability. Oncogene. 2004;23:2640–2647. doi: 10.1038/sj.onc.1207409. [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurin CC, Federici MG, Kang L, Boyd J. Causes and consequences of microsatellite instability in endometrial carcinoma. Cancer Res. 1999;59:462–466. [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci U S A. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi K, Sakamoto K, Koinuma D, Semba K, Inoue A, Inoue S, Fujii H, Yamaguchi A, Miyazawa K, Miyazono K, et al. TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene. 2012;31:3190–3201. doi: 10.1038/onc.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- Jung B, Doctolero RT, Tajima A, Nguyen AK, Keku T, Sandler RS, Carethers JM. Loss of activin receptor type 2 protein expression in microsatellite unstable colon cancers. Gastroenterology. 2004;126:654–659. doi: 10.1053/j.gastro.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Koren A, Polak P, Nemesh J, Michaelson JJ, Sebat J, Sunyaev SR, McCarroll SA. Differential Relationship of DNA Replication Timing to Different Forms of Human Mutation and Variation. Am J Hum Genet. 2012;91:1033–1040. doi: 10.1016/j.ajhg.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz P, Rodelsperger C, Jager M, Jostins L, Bauer S, Robinson PN. Microindel detection in short-read sequence data. Bioinformatics. 2010;26:722–729. doi: 10.1093/bioinformatics/btq027. [DOI] [PubMed] [Google Scholar]
- Kuismanen SA, Moisio AL, Schweizer P, Truninger K, Salovaara R, Arola J, Butzow R, Jiricny J, Nystrom-Lahti M, Peltomaki P. Endometrial and colorectal tumors from patients with hereditary nonpolyposis colon cancer display different patterns of microsatellite instability. Am J Pathol. 2002;160:1953–1958. doi: 10.1016/S0002-9440(10)61144-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen LA, Nystrom-Lahti M, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- Metzgar D, Bytof J, Wills C. Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 2000;10:72–80. [PMC free article] [PubMed] [Google Scholar]
- Minoche AE, Dohm JC, Himmelbauer H. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol. 2011;12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama Y, Yamauchi M, Minagawa N, Torigoe T, Izumi H, Kohno K, Yamaguchi K. Clinical significance of mitochondrial transcription factor A expression in patients with colorectal cancer. Oncol Rep. 2012;27:1325–1330. doi: 10.3892/or.2012.1640. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van LP, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- Paun BC, Cheng Y, Leggett BA, Young J, Meltzer SJ, Mori Y. Screening for microsatellite instability identifies frequent 3′-untranslated region mutation of the RB1-inducible coiled-coil 1 gene in colon tumors. PLoS One. 2009;4:e7715. doi: 10.1371/journal.pone.0007715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. doi: 10.1101/gad.1182704. [DOI] [PubMed] [Google Scholar]
- Plaschke J, Preussler M, Ziegler A, Schackert HK. Aberrant protein expression and frequent allelic loss of MSH3 in colorectal cancer with low-level microsatellite instability. Int J Colorectal Dis. 2012;27:911–919. doi: 10.1007/s00384-011-1408-0. [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010a;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010b;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- Preston BD, Albertson TM, Herr AJ. DNA replication fidelity and cancer. Semin Cancer Biol. 2010;20:281–293. doi: 10.1016/j.semcancer.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, Escaramis G, Jares P, Bea S, Gonzalez-Diaz M, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed JC, Perucho M. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstorukov MY, Volfovsky N, Stephens RM, Park PJ. Impact of chromatin structure on sequence variability in the human genome. Nat Struct Mol Biol. 2011;18:510–515. doi: 10.1038/nsmb.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao JL, Yatabe Y, Salovaara R, Jarvinen HJ, Mecklin JP, Aaltonen LA, Tavare S, Shibata D. Genetic reconstruction of individual colorectal tumor histories. Proc Natl Acad Sci U S A. 2000;97:1236–1241. doi: 10.1073/pnas.97.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veigl ML, Kasturi L, Olechnowicz J, Ma AH, Lutterbaugh JD, Periyasamy S, Li GM, Drummond J, Modrich PL, Sedwick WD, et al. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci U S A. 1998;95:8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun L, Myeroff L, Wang X, Gentry LE, Yang J, Liang J, Zborowska E, Markowitz S, Willson JK, et al. Demonstration that mutation of the type II transforming growth factor beta receptor inactivates its tumor suppressor activity in replication error-positive colon carcinoma cells. J Biol Chem. 1995;270:22044–22049. doi: 10.1074/jbc.270.37.22044. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Zhang ZG, Tian XQ, Sun DF, Liang QC, Zhang YJ, Lu R, Chen YX, Fang JY. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia. 2008;10:287–297. doi: 10.1593/neo.07971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Shin J, Wilson A, Goel S, Ling YH, Ahmed N, Dopeso H, Jhawer M, Nasser S, Montagna C, et al. An A13 repeat within the 3′-untranslated region of epidermal growth factor receptor (EGFR) is frequently mutated in microsatellite instability colon cancers and is associated with increased EGFR expression. Cancer Res. 2009;69:7811–7818. doi: 10.1158/0008-5472.CAN-09-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XP, Hoang JM, Cottu P, Thomas G, Hamelin R. Allelic profiles of mononucleotide repeat microsatellites in control individuals and in colorectal tumors with and without replication errors. Oncogene. 1997;15:1713–1718. doi: 10.1038/sj.onc.1201337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.