Abstract
Hypophosphatasia (HPP) is an inherited disorder of mineral metabolism caused by mutations in ALPL, encoding tissue non-specific alkaline phosphatase (TNAP). Here, we report the molecular findings from monozygotic twins, clinically diagnosed with tooth-specific odontohypophosphatasia (odonto-HPP). Sequencing of ALPL identified two genetic alterations in the probands, including a heterozygous missense mutation c.454C>T, leading to change of arginine 152 to cysteine (p.R152C), and a novel heterozygous gene deletion c.1318_1320delAAC, leading to the loss of an asparagine residue at codon 440 (p.N440del). Clinical identification of low serum TNAP activity, dental abnormalities, and pedigree data strongly suggest a genotype-phenotype correlation between p.N440del and odonto-HPP in this family. Computational analysis of the p.N440del protein structure revealed an alteration in tertiary structure affecting the collagen-binding site (loop 422-452), which could potentially impair the mineralization process. Nevertheless, the Probands (compound heterozygous: p.[N440del];[R152C]) feature early-onset and severe odonto-HPP phenotype, whereas the father (p.[N440del];[=]) has only moderate symptoms, suggesting p.R152C may contribute or predispose to a more severe dental phenotype in combination with the deletion. These results assist in defining the genotype-phenotype associations for odonto-HPP, and further identify the collagen-binding site as a region of potential structural importance for TNAP function in the biomineralization.
Keywords: Hypophosphatasia, odontohypophosphatasia, tissue nonspecific alkaline phosphatase, ALPL, collagen-binding site, compound heterozygous mutations, protein 3D modeling
1. INTRODUCTION
Hypophosphatasia (HPP; OMIM ID: 146300, 241500, 241510) is a rare metabolic inherited disorder characterized by defective mineralization of bones and teeth due to deficient enzymatic activity of tissue non-specific alkaline phosphatase (TNAP) [1]. Disease symptoms are highly variable in their clinical expression, and six clinical forms are currently recognized, based on age at diagnosis and severity of features, including: lethal perinatal, benign perinatal, infantile, childhood, adult, and odontohypophosphatasia (odonto-HPP) forms [2]. The birth prevalence of the most severe forms of HPP, i.e. perinatal and infantile, is estimated to be 1:100,000. On the basis of frequency of heterozygotes and proportion of mutations exhibiting a dominant negative effect, it is expected that mild forms of HPP (childhood, adult and odonto-HPP) are more common than severe forms [3].
All clinical isotypes of HPP, including odonto-HPP, share in common reduced serum TNAP activity (ALP), and presence of either one or two pathologic mutations in the ALPL gene [3]. The ALPL gene (OMIM: 171760) is mapped to chromosome 1 (1p36.12) and consists of 12 exons encoding TNAP [4]. Currently, at least 264 distinct mutations and 16 polymorphisms in the ALPL gene have been identified and associated with various forms of HPP. Missense mutations account for 75% of these mutations, while the remaining percentage are represented by small deletions (11%), splicing mutations (5.7%), nonsense mutations (3.8%), small insertions (2.3 %), large deletions (1,1%), insertions or deletions (0.7%), and mutations in regulatory ALPL sequences (0.4%) (http://www.sesep.uvsq.fr/03_hypo_mutations.php#stat). In milder forms, in which one mutant allele is believed to be sufficient to cause disease, mutation detection rate is more difficult to estimate [3].
Deficient TNAP activity is thought to be the major cause for skeletal mineralization defects observed in HPP [1, 5]. TNAP regulates mineralization by hydrolyzing the mineralization inhibitor, inorganic pyrophosphate (PPi), and by increasing inorganic phosphate (Pi) locally which participates in propagation of hydroxyapatite crystals in the extracellular matrix, and in deposition of hydroxyapatite between collagen fibrils [1, 5]. Decrease or loss of TNAP activity leads to accumulation of extracellular PPi, provided in part by nucleotide pyrophosphatase phosphodiesterase 1 (NPP1) and progressive ankylosis protein homolog (ANKH), resulting in inhibition of hydroxyapatite formation [5, 6, 7].
TNAP is reported to be a tetrameric structure on the cell surface, linked to the membrane via glycosylphosphatidylinositol (GPI) anchors, and oriented so that the active sites face the extracellular environment. The enzyme is also active as a homodimer but not as a monomer [8, 9]. Due to the structural properties of the TNAP, some mutations affecting protein structure may exhibit a dominant negative effect. These dominant negative mutations (also called antimorphic mutations) usually result in an altered molecular function due to inhibition of enzymatic activity of the normal monomer by the mutated partner in heterodimers, thus contributing to highly variable clinical phenotypes of HPP [10]. Consequently, genotype-phenotype correlations are difficult to establish, because most patients are compound heterozygous for missense mutations and/or are carriers of mutations exhibiting a dominant negative effect.
Genotype-phenotype correlations have been examined by the use of site-directed mutagenesis and three dimensional (3D) modeling of the enzyme [2, 10-15]. Most of these studies show an excellent correlation between the severity of the phenotype and residual enzymatic activities produced in vitro, and/or localization of mutant residues in the 3D structure, whereas transfection assays may not distinguish structural mutations from functional ones [13]. To date, all clinical forms of HPP have been shown to involve TNAP mutations that compromise the protein structure. According to 3D modeling studies, most mutations affect functional domains of the protein, namely the active site, the calcium binding site, the crown domain, and the homodimer interface, as well as other regions involved in dimerization, tetramerization, or membrane anchoring [13, 16, 17]. In this context, computational approaches for protein 3D modeling may assist in establishing genotype-phenotype and structure-function correlations, as well as predicting the structural and/or functional impact of each mutation.
The goal of the present study was to identify the mutation(s) associated with odonto-HPP affecting monozygotic twin probands, establish a genotype-phenotype association, and use a 3D modeling approach to evaluate the impact of each mutation in the TNAP protein structure. Additionally, we evaluated the expression of mutant protein and its subcellular localization in dental pulp cells from probands by Western blotting and immunocytochemistry.
2. METHODS
2.1. Subjects
The probands were male monozygotic twins of Caucasian descent clinically diagnosed with odonto-HPP. Probands and their biological parents were examined in order to identify potential mutations in the ALPL gene. The family was provided study information and consented to participate (IRB #065/2005). The clinical diagnosis of odonto-HPP in the probands (by physical and dental examinations, radiographs, and blood chemistry assays) and subsequent management of dental symptoms has been reported previously [18, 19]. Briefly, probands (patients A and B), at the age of two, were brought to the Piracicaba Dental School, University of Campinas, Brazil for dental evaluation. Parents reported premature exfoliation of anterior primary teeth, with signs of partial root resorption. Physical examination and radiographs (long bones, joints, and skull) showed age-appropriate growth and development. Routine laboratory testing revealed low serum ALP activity for both probands (patient A: 62 U/L, patient B: 63 U/L; normal range for children 151-471 U/L), while serum phosphate and calcium levels remained within normal limits [18-20].
2.2. Genotype analyses
Genomic DNA of probands and their parents was isolated from peripheral blood leukocytes using a Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA) following the manufacturer's instructions.
Primer sequences were designed to amplify all TNAP coding exons (2-12), as previously reported [21], allowing analysis of the whole coding sequence, including intron-exon borders. Polymerase chain reaction (PCR) was performed in a final volume of 50 μl with 100 ng of DNA, 30 μM forward and reverse primers, 0.2 mM dNTP mix (Invitrogen™, Life Technologies, Carlsbad, CA, USA Life Technologies, Gaithersburg, MD, USA), 0.75 U Gold Tap® Flexi DNA polymerase (Promega), and 1-3 mM MgCl2. Cycle conditions and annealing temperature were optimized for each primer pair.
PCR products were purified from agarose gel using GFX PCR DNA and Gel Band purification kit (GE Healthcare, Piscataway, NJ, USA), according to the manufacturer's instructions. DNA sequence analysis was performed using the BigDye Terminator v3.1 Cycle Sequencing Kit and migrated on capillary 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Sequence similarity was performed using BLASTN [22]. Putative mutations were identified after multiple sequence alignment using Clustal W [23] and electropherogram analysis. The existence of each putative mutation was confirmed by sequencing DNA from both parents, as well as by a secondary validation method, i.e. restriction enzyme digestion of DNA or amplification using specific primers.
2.3. Polymerase Chain Reaction (PCR)
PCR was used additionally to confirm identified gene mutations (described above). Primers were designed to amplify alleles with suspected gene deletion (1318_20delAAC), by using forward primer sequences designed with (native TNAP: 5’-GCCCACAGCTCACAACAAC-3’) or without (1318_20delAAC: 5’-GCCCACAGCTCACAACTAC-3’) the three-base-pair AAC deleted. PCR reactions were performed using 5 ng of DNA template, 0.4 μM each forward and reverse (5’-GTCCACGAGCAGAACTACG -3’) primers and LightCycler® FastStart DNA MasterPLUS SYBER Green I kit 1X (Roche Diagnostics, Penzberg, Germany) in the LightCycler® 2.0 Instrument (Roche Diagnostics).
2.4. Three dimensional (3D) modeling
Three dimensional (3D) models of the native TNAP protein and mutants (p.N440del, p.R152C and p.N440del/p.R152C) were constructed based on the previously determined 3D structure of human placental alkaline phosphatase (PLAP) (PDB ID: 1EW2) [17] using SWISS-MODEL software (http://swissmodel.expasy.org/) [24-26]. These models were aligned, visualized, and analyzed using the open source software PyMOL Graphics System Molecular (Version 1.2r3pre, Schrödinger, LLC) (http:www.pymol.org). Internal contacts for native and mutants residues in the TNAP structure were analyzed using STING Millennium software [27] from Brazilian Enterprise for Agricultural Research (EMBRAPA) (http://www.nbi.cnptia.embrapa.br/SMS/index_s.html).
2.5. Immunofluorescence
Primary dental pulp cells from both probands A and B (genotype: p.[N440del];[R152C]) and four control individuals (native TNAP) were obtained as previously reported [20]. Briefly, extracted teeth were placed in biopsy media, and pulp was harvested by cracking open teeth using a dental chisel and hammer and removing soft tissue with sterile forceps. Pulp cells were obtained by enzymatic digestion with 3 mg/mL collagenase type I and 4 mg/mL dispase (Gibco®, Invitrogen™, Life Technologies) for 1 h at 37 C. Cells at passage four were seeded on coverslips in 24-well cell culture plate (2×104 cells per well) and were cultured in DMEM with FBS 5% for 24 h. After two washes in phosphate-buffered saline (DPBS, Invitrogen™, Life Technologies), cells were fixed in 2% paraformaldehyde in DPBS for 20 min at room temperature (RT). Blocking for nonspecific binding was performed by incubating with blocking buffer solution (10% normal donkey serum in DPBS) for 45 min at RT. Subsequently, coverslips containing fixed cells were incubated overnight at 4 °C with human alkaline phosphatase/ALPL monoclonal rat antibody (R&D Systems, Inc., Minneapolis, MN, USA) in DPBS containing 1% normal donkey serum (Sigma, St. Louis, MO, USA) and 1% bovine serum albumin (BSA, Sigma). Next, coverslips were washed twice in DPBS and incubated with Alexa Fluor 546 goat anti-rat IgG (Invitrogen™, Life technologies) for 2 h at RT in the dark. After washing twice with DPBS, nuclear counterstain was performed by incubation with 0.5 μM de SYTO® 21 green fluorescent nucleic acid stain (Invitrogen™, Life Technologies) in DPBS for 2 min at RT. Coverslips were mounted in the ProLong® Gold antifade reagent (Molecular Probes®, Invitrogen™, Life Technologies). The subcellular localization in dental pulp cells was visualized using the Leica TCS SP5 confocal microscope (Leica Microsystems, Wetzlar, Germany). Negative controls were performed using the incubation buffer with no primary antibody.
2.6. Western blot analysis
For extraction of total proteins, primary dental pulp cells, at passage five, from probands A and B (genotype: p.[N440del];[R152C]) and four control individuals (native TNAP) were seeded in 100 mm tissue culture dishes (40×104 cells per plate) in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen™, Life Technologies) with 10% FBS for 24 h. Next, medium was changed to 5% FBS supplemented with 50 μg/mL ascorbic acid (AA) and 10 mM β-glycerol phosphate (βGP) for 7 days, with medium changed every other day. Cells were washed twice with DPBS, harvested in ice-cold DPBS containing protease inhibitor cocktail (Sigma) and centrifuged at 850 g. The cell pellet was lysed with RIPA buffer (Sigma) containing protease inhibitor cocktail (Sigma). Total protein concentration was determined by the Bradford method. Similar amounts of total protein from each sample (~70 μg) were resolved on 10% SDS-polyacrylamide gel electrophoresis (PAGE) and then transferred to Hybond-ECL nitrocellulose membrane (GE Healthcare). Blots were blocked by incubation with 3% BSA in Tris buffer saline (TBS, pH 7.6) for 1 h. To detect the target protein, blots were probed with human alkaline phosphatase/ALPL rat monoclonal antibody (1:500, R&D Systems, Minneapolis, MN, USA) and secondary antibodies conjugated to horseradish peroxidase (1:30,000, ECL Anti-Rat IgG, GE Healthcare) in TBS containing 0.1% Tween 20. All steps of the incubation were performed for 1h at room temperature with gentle agitation. The antigen-antibody complexes were detected by chemiluminescence using SuperSignal® West Fento Maximum Sensitivity Substrate (Thermo Scientific, Pierce Biotechnology, Rockford, IL, USA) for 1 min. Then, chemiluminescent images were acquired using an acquisition and documentation system (MicroChemi 4.2 from DNR Bio-Imaging Systems, Israel). Blots were re-probed with α-tubulin mouse monoclonal antibody (1:500) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and secondary ECL Anti-Mouse IgG antibodies, (1:20,000). TNAP levels were semi-quantified by densitometry using Scion image analysis software (Scion Corp, Frederick, Maryland) and normalized according to density of α-tubulin protein.
3. RESULTS AND DISCUSSION
3.1. Genotype-phenotype association in odonto-HPP diagnosed subjects
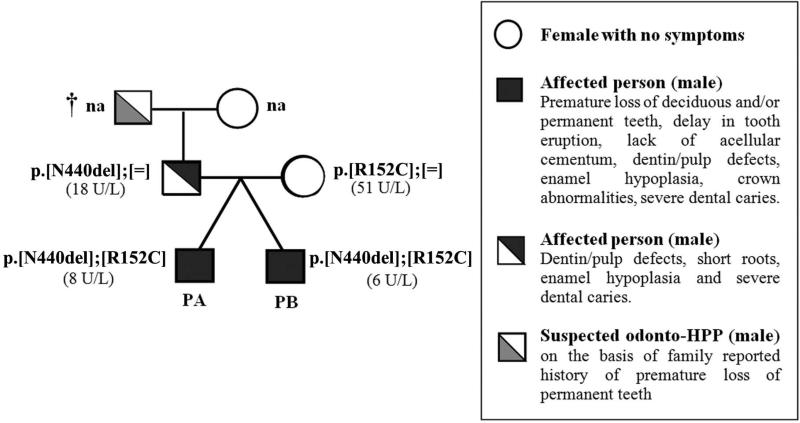
In the present study, clinical symptoms, laboratory assays, pedigree data, and genomic DNA sequence analysis were used in order to establish an odonto-HPP genotype-phenotype association, and these data are summarized in Figure 1. Tracing the family history, the mother of the probands (subjects A and B) was asymptomatic, and her serum ALP activity was found to be normal (51 U/L; normal range 25-100 U/L). However, the father presented clinical signs of dental abnormalities including short roots, mild enamel defects, pulp chamber enlargement, low serum ALP levels (18 U/L; normal range 25-100 U/L), and reported a familial history of early tooth loss in his father. Overall, these findings suggested initially that the disorder was transmitted through autosomal dominant inheritance (Figure 1).
Figure 1. Kindred pedigree: Autosomal dominant inheritance of an odontohypophosphatasia clinical phenotype.
Summarized here for probands (PA and PB) and their biological parents are pedigree data, dental symptoms, and serum ALP activity (U/L), as well as results of genetic analysis (see supplementary data). Serum ALP activities reported were measured at the age of diagnosis, where normal values for the adult range are 25-100 U/L. Dental phenotypes in probands and their father suggest autosomal dominant pattern of inheritance. Genetic analysis revealed the mutation 1318-20delAAC, promoting the deletion of asparagine (N) at position 440 of the TNAP (p.N440del), was paternally inherited, and substitution C > T at position 454-nt leading to the change of arginine 152 to cysteine (p.R152C) was maternally inherited. Abbreviations: (N): normal allele, (na): not clinically or genetically evaluated, (†): deceased person.
Screening for mutations in the ALPL gene revealed two genetic alterations in the probands (Supplementary data). Firstly, there was a heterozygous substitution C → T at position 454-nt, leading to the substitution of cysteine for arginine 152 (p.R152C), an alteration previously described in this family and found in the mother of the probands [18-20]. The p.R152C (c.454C>T) missense mutation, associated to other mutation p.R184W (c.550C>T), has been reported previously in a case of adult HPP (p.[R152C];[R184W]; SESEP – University of Versailles- Saint Quentin), whereas a mutation affecting the same codon p.R152H (c.455G>A) has been described in the mother of a patient with a lethal clinical form of HPP [28]. p.R152H is believed to be a TNAP polymorphism because this alteration was detected in 3 of 168 alleles (frequency of 1.8%) in subjects from a control group with metabolic bone diseases other than HPP, including osteogenesis imperfecta [29]. It is notable that p.R152H would be a conservative missense mutation (also termed a synonymous mutation) in terms of properties of arginine and histidine, while p.R152C would be a non-conservative missense mutation. Notably, studies have indicated that some ALPL polymorphisms, as well as compound heterozygous mutations, may play an important role in HPP severity when associated with additional ALPL mutations [15, 21, 29].
The second alteration identified in the probands was a heterozygous gene deletion of three basepairs in-frame (AAC) at position 1318-1320-nt (c.1318_1320delAAC), leading to deletion of asparagine (Asn, N) at codon 440 (p.N440del). While the missense mutation described above was maternally inherited, the p.N440del was of paternal origin (Figure 1).
The genetic alteration 1318_1320delAAC (p.N440del) has not been reported previously for odonto-HPP or other HPP types. Clinical dental abnormalities and low serum ALP activity in the father, in addition to the shared p.N440del, strongly suggests a genotype-phenotype correlation between p.N440del and odonto-HPP in this family. Notably, the probands (genotype: p.[N440del];[R152C]), who were compound heterozygous for the missense mutation (p.R152C) and deletion (p.N440del), present an early onset and relatively severe odonto-HPP phenotype, whereas the father with only one mutation (genotype: p.[N440del];[=]), presented relatively moderate symptoms with no premature tooth loss and relatively milder enamel phenotype. Thus, our findings suggest that the N440 deletion is a pathological genetic alteration, whereas p.R152C may contribute or predispose to a more severe dental phenotype in combination with the deletion.
3.2. Analysis of 3D models of mutant TNAP proteins
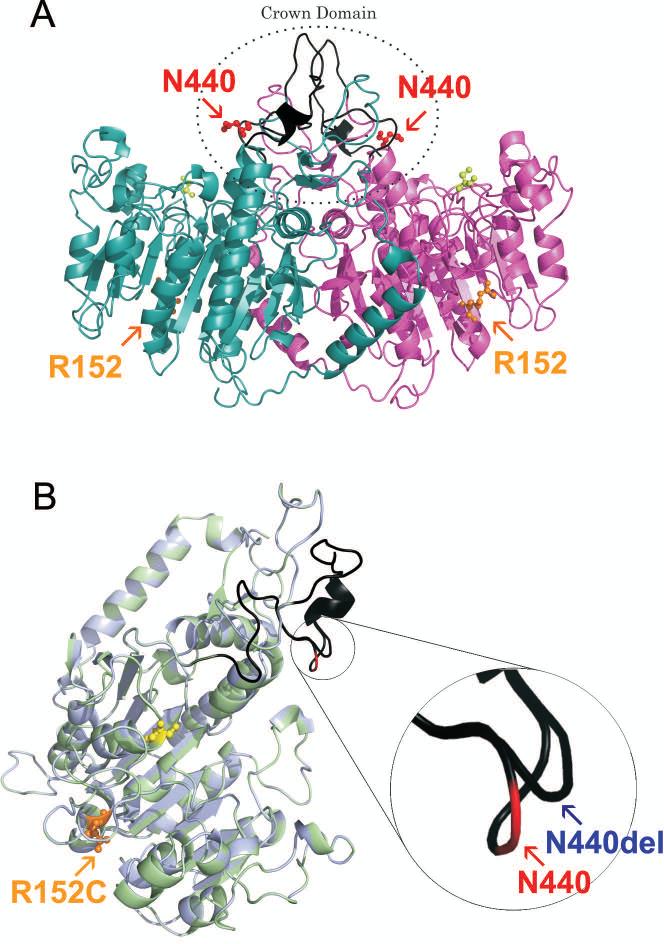
In order to provide insights on potential contribution of each genetic alteration to enzyme function and the odonto-HPP phenotype, 3D protein modeling and computational analysis were used to predict how the identified alterations would affect protein tertiary structure. The alignment of the 3D models of native TNAP protein and mutants revealed that the deletion of the N440 residue was predicted to result in protein conformational changes (Figure 2). The N440 residue is located in the coil structure of loop 422-452 (loop 405-435 excluding the signal peptide), corresponding to a collagen-binding site within the crown domain of TNAP [13]. N440 residue deletion resulted in the change of this coil structure, affecting the protein folding pattern as well as the hydrogen bonding and hydrophobic interactions between neighbor residues (H438, N439, and Y441) and other regions of the molecule (Figures 2 and 3). Residues of this coil structure are located in the highly accessible loop (422-452) within the crown domain that is formed by the insertion of a 60-residue segment (388–448) from each TNAP monomer [13, 17, 30]. The functional and structural importance of the crown domain has been elucidated through building 3D models of the enzyme based on the structure of human PLAP, and localization of residues affected by mutation within the specific domains [13, 17, 30, 31]. Results from these studies demonstrated that the crown domain is critical for isozyme-specific properties such as non-competitive inhibition, heat-stability, and allosteric behavior [17, 30], as well as dimerization and homodimer stability, and interactions between TNAP and extracellular matrix proteins including collagens [13, 31].
Figure 2. Three-dimensional models of TNAP dimer and monomer.
(A) Ribbon representation corresponding to the dimer of native TNAP. The monomer I is shown in purple, monomer II in light blue, the collagen-biding site in black, and the active site highlighted by yellow spheres. Localization of residues R152 (highlighted by orange) and N440 (highlighted by red) are indicated in the model. (B) Ribbon presentation of superimposed structures of native TNAP (cyan) and mutant p.N440del /p.R152C (light green). The model was based on the crystal structure of the PLAP monomer (PDB ID: 1E2W), and signal peptide was not included in the 3D model shown here. The active site is indicated by yellow spheres and the collagen-biding site is highlighted in black. The residue affected by missense mutation (R152C) is highlighted by orange spheres. The native N440 residue is indicated by red, while the mutant residue position (N440del) is indicated by blue arrow. The R152 residue affected by a missense mutation is located in the α-helix on the surface of the molecule, while N440 residue affected by deletion is located in the coil structure of the flexible loop, corresponding to collagen-biding site in the crown domain. N440 residue deletion promotes the change of coil structure, affecting neighbor residues (H438, N439 and Y441), localized into the flexible loop corresponding to collagen-binding site. These figures were assembled with PyMOL Graphics System Molecular.
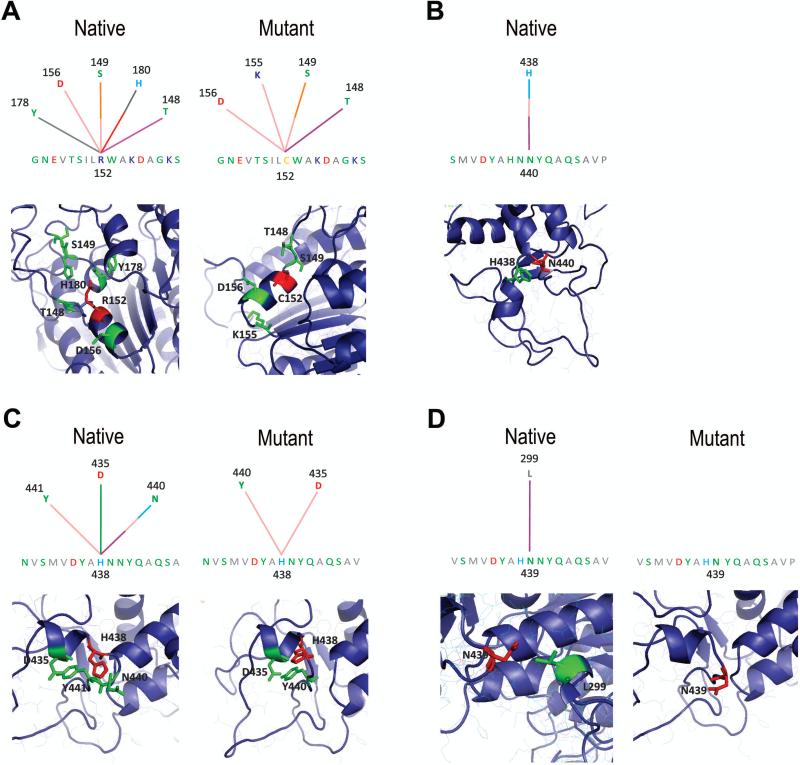
Figure 3. Alterations in amino acid interactions in the p.R152C and p.N440del mutant TNAP proteins.
(A) Graphical representation for internal contacts for R152 (native) and C152 (mutant) residues: interactions with T148, S149, D156, Y178 and H180 are observed in the native protein (R152), however two interactions are abolished (H180 and Y178) and one interaction with a novel residue (K155) is established due to the replacement of R by C residue at 152 position. (B) Graphical representation of internal contacts for N440 residue in the native protein: hydrogen bond and hydrophobic interactions with the H438 residue are observed in the native protein, but these interactions are lost in the protein affected by deletion of N440 residue. (C) Graphical representation of internal contacts for H438 residue in the native and mutant protein: interactions with D435, N440 and Y441 are observed in native TNAP, while in the protein affected by deletion of N440 residue, all interactions with N440 and charge attractive interaction with D435 are abolished. (D) Graphical representation of internal contacts for N439 residue in the native and mutant protein: one hydrophobic interaction with L299 is observed in native protein, but this interaction is abolished in the protein affected by deletion of N440. Part of the modeled TNAP structure showing internal contacts for C152, R152, N440, H438 and N439 residues are presented below each graphical representation. Colored lines represent different types of interactions: salmon = hydrogen bond (main chain – main chain); orange = hydrogen bond (side chain – main chain), light blue = hydrogen bond (side chain – side chain), purple = hydrophobic interaction, gray = aromatic stacking, red = charge repulsive interaction, green = charge attractive interaction. Graphical representation and analysis for internal contacts was performed using the STING Millennium software [27].
The maternally inherited p.R152C missense mutation was not predicted to result in significant conformational changes to the TNAP molecule (Figure 2). On the other hand, differences in internal contacts established for mutant C152 compared to the native R152 residue were observed. In the native protein, the R152 residue interacts with T148, S149, D156, Y178 and H180 residues, however two interactions are abolished (H180 and Y178), and one interaction with a novel residue (K155) is established in the mutated C152 form (Figure 3). R152 is a nonconserved residue located in the α-helix and within a cluster that localizes to the surface of the TNAP. Although surface residues are generally not believed to play an important structural role [13, 14, 17], Monet et al. (2001) pointed to a role for R152 residue in the secondary structure of the protein [13]. For example, mutations affecting residues on the surface of the molecule may alter the tetramer or homodimer formation, leading to dysfunctional interactions with other important players for biological function [8, 9].
TNAP enzyme activity may be directly or indirectly affected by genetic alterations, depending on the nature of interactions between altered residues and nearby residues and/or critical domains, such as the active site, ligand-biding site, and homodimer interface [14]. In the present study, both genetic alterations (p.N440del and p.R152C) were distant from active site (Figure 2) and did not appear to directly affect the catalytic properties of TNAP. In addition, based in localization these residues in 3D model of homodimer (Figure 2A), we observed that neither p.R152C nor p.N440del appear to affect the dimer formation. On the other hand, TNAP activity also could be indirectly affected by incorrect biosynthesis, loss the molecule stability, impaired trafficking of TNAP to cell surface, or abnormal interactions with other cellular proteins [31-35], therefore we performed additional studies to gain further insights as to the mechanism for loss of ALP activity in these probands.
3.3. Expression and localization of mutant TNAP in dental pulp cells
Mutations mapped to different domains of the ALPL, affecting alkaline phosphatase activity (e.g. p.D306V, p.E235G, p.N170D, p.A179T, and p.G334D, have been described to exhibit improper folding and incorrect assembly [32, 33, 35, 36]. Misfolded and incorrectly assembled proteins are generally recognized and degraded by the endoplasmic reticulum (ER) quality-control system [33, 36]. The ER quality-control system is crucial for securing the fidelity of gene expression at the posttranslational level and permits only correctly folded and completely assembled proteins to proceed to the Golgi, subsequently to be transported to cell membrane [33]. Therefore, decreased expression of TNAP mutants on the cell surface may be due to improper protein folding and incorrect assembly, resulting in the defective transport, accumulation in the early stages of the secretory pathway and degradation of mutant proteins in a proteasome-dependent manner [32, 33, 35, 36].
To attempt to define how molecular defects of TNAP (p.R152C and p.N440del) may indirectly affect TNAP activity, immunofluorescence staining and western blotting analysis were performed in dental pulp cells obtained from probands and control individuals. Based on previous studies, TNAP is synthesized as a 66-kDa immature form with high mannose-type oligosaccharides, which is then converted to the ~80-kDa mature form bearing complex-type carbohydrate chains, which after being properly trimmed and terminally glycosylated through the Golgi apparatus, finally reaches the plasma membrane and is exposed on the cell surface via a GPI anchor [32, 35]. Studies have demonstrated that the 80-kDa mature form, but not the 66-kDa one, is predominantly expressed on the cell surface. Incorrect post-translational modification of 66-kDa immature form may lead to formation of disulfide-bonded aggregates in the endoplasmic reticulum, that ultimately do not proceed to the cell surface [32, 34, 35].
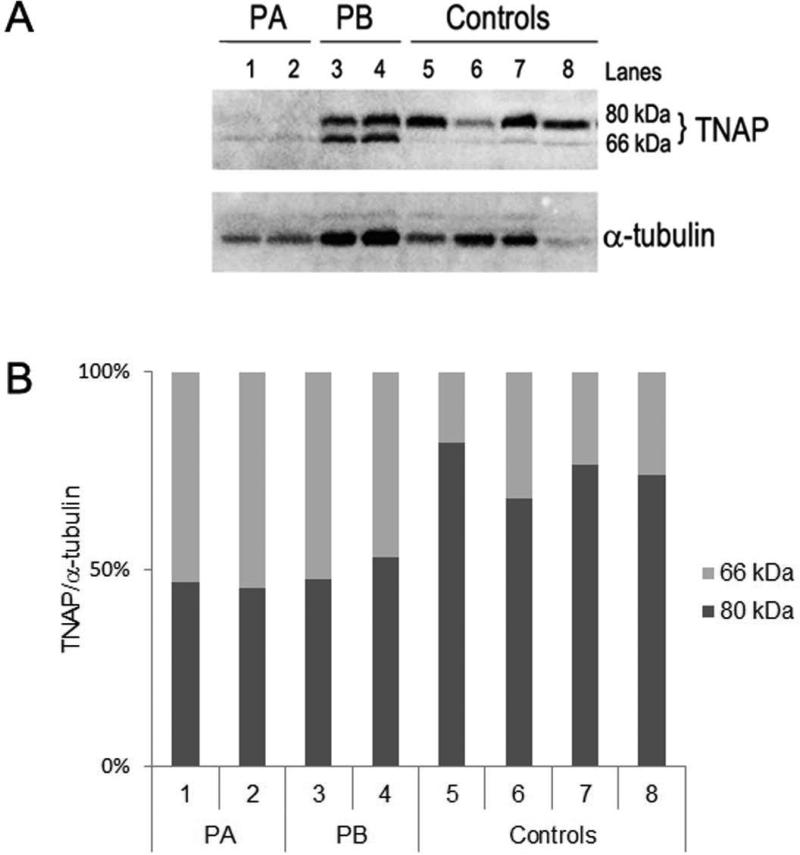
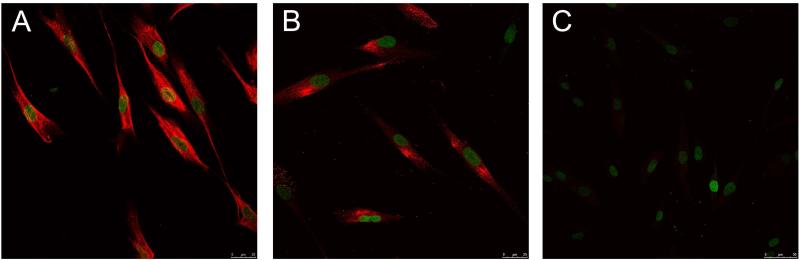
Here, we analyzed dental pulp cells from probands carrying both a heterozygous missense mutation (p.R152C) and a heterozygous deletion (p.N432del) in different alleles. Western blotting analysis revealed the presence of both the 66-kDa and ~80-kDa forms of TNAP in total protein extracts from probands and control cells (Figure 4A), however the ~80-kDa form predominated in control cells, whereas there was increased ratio of the 66-kDa form (non-glycosylated, immature form of TNAP) to the 80-kDa (glycosylated or mature form of TNAP) in probands compared to control cells (Figure 4B). Furthermore, immunocytochemistry revealed that TNAP was localized to the cell surface (and cytoplasm) in control dental pulp cells (with native TNAP), whereas mutated TNAP protein was more predominantly localized to the perinuclear region and cytoplasm in cells from the probands (Figure 5).
Figure 4. Mutant TNAP protein expression in dental pulp cells.
(A) Representative image showed the TNAP protein expression in primary dental pulp cells from patient A (PA: lanes 1 and 2) and patient B (PB: lanes 3 and 4) and four controls (Control individuals: lanes 5, 6, 7, and 8) by Western Blotting. Two bands, corresponding to the major ~80-kDa (glycosylated) and ~66-kDa (non-glycosylated) form of TNAP, were detected in dental pulp cells from probands and control individuals. However, an increased level of the 66-kDa form of TNAP versus the 80-kDa was observed in extracts of total protein from probands’ cells, expressing mutant TNAP proteins (PA: 1, 2 and PB: 3, 4; Genotype: p.[R152C];[N440del]), compared to control cells, expressing the native TNAP protein (Controls: 5, 6, 7 and 8). (B) The graph shows the percentage of expression values normalized for each ~80 kDa and ~66 kDa form of TNAP in each sample. TNAP protein levels (~80 kDa and ~66 kDa) were estimated using Scion Image software and normalized by α-tubulin protein levels (~55 kDa). Similar results were obtained in additional experiments.
Figure 5. Immunolocalization of TNAP protein in the dental pulp cells.
Subcellular localization of TNAP protein in dental pulp cells from a (A) control individual and (B) a proband. Negative control was performed in the absence of primary antibody and is shown in panel (C). TNAP expression was visualized by red fluorescent staining (Alexa Fluor 546) using a Leica TCS SP5 confocal microscope (A and B bar = 25 μM; in C bar = 50 μM). Nuclear counterstain was performed with SYTO® 21 Green Fluorescent Nucleic Acid Stain and was visualized by green fluorescent staining.
We have demonstrated previously that primary periodontal ligament and dental pulp cells harvested from the same probands exhibited increased ALPL mRNA, whereas residual ALP activity and ability to promote mineralization in vitro were markedly reduced (40% and 50%, respectively) [18, 20]. Here, we showed that increased ALPL mRNA concentration in these cells does not result in increased protein expression, suggesting that regulatory mechanisms, such as the ER quality-control system, may be intervening and resulting in defective intracellular transport of mutant TNAP, likely due to retention of a fraction of mutant TNAP molecules in the intracellular compartment.
Mutations affecting TNAP trafficking have been described previously. Shibata et al. [35] showed that a homozygous missense mutation in TNAP affecting the 179 residue (p.A179T), associated with a lethal hypophosphatasia, exhibited defective folding that affected trafficking of TNAP molecules, causing only a small fraction of mutated TNAP protein to reach the cell membrane, presumably due to the formation of disulfide-bonded high-molecular mass aggregates. Intriguingly, cells expressing mutant TNAP (p.A179T) exhibited residual TNAP activity, suggesting the mutation did not lead to complete inactivation of the enzyme [35]. Conversely, Numa et al. [34] showed that the TNAP mutation (p.V423A), mapped to the collagen binding domain, had a markedly reduced alkaline phosphatase activity, but this was not the result of defective transport, rather suggesting a structural importance of the crown domain (more specifically of collagen binding domain) with respect to the catalytic function of TNAP.
A novel mutation, p.N440del, localized to the TNAP crown domain was identified in the probands in this study. A number of TNAP missense mutations affecting amino acids residues localized in flexible loop corresponding to the collagen-binding region have been identified in individuals diagnosed with HPP (Table 1). Moreover, Mornet et al. [13], showed that among 10 mutations localized the crown domain and associated with HPP, at least six were located to this loop, including p.V423A, p.G426C, p.Y436H, p.S445P, p.R450C, and p.R450H. Interestingly, genetic alterations affecting amino acid residues in the collagen binding loop correspond to homozygous severe forms of HPP or heterozygous mild phenotypes (Table 1), indicating that this region play an important, but presently unclear, role in TNAP function.
Table 1.
Summary of reported ALPL gene mutations affecting amino acid residues located in the collagen binding loop 422-452 within the crown domain.
| Clinical form in patient | Base change | Amino acid change |
Genotype of patient | Reference | |
|---|---|---|---|---|---|
| Non-standardized nomenclature | Standardized nomenclature | ||||
| Perinatal | c.1268T>C | V406A | p.V423A | p.[V423A];[A116T] * | [28] |
| Adult | c.1270G>A | V407M | p.V424M | p.[V424M];[V424M] | Versailles, 2007 |
| Infantile | c.1276G>T | G409C | p.G426C | p.[K224A];[G426C] * | [37] |
| Infantile | c.1276G>A | G409S | p.G426S | p.[G426S];[G426S] | [13] |
| Childhood | c.1277G>A | G409D | p.G426D | p.[G426D];[E191K] * | [29] |
| Perinatal | c.1282C>T | R411X | p.R428X | p.[R428X];[R428X] | [38] |
| Perinatal | c.1283G>C | R411P | p.R428P | p.R428P ; c.997+2T>A * | [39] |
| Odonto-HPP | c.1285G>A | E412K | p.E429K | p.[E429K];[?]* | Versailles, 2006 |
| Infantile | c.1306T>C | Y419H | p.Y436H | p.[A33V];[Y436H] * | [40] |
| Infantile | c.1313A>T | H421L | p.H438L | p.[H438L];[H438L] | Versailles, 2009 |
| Odonto-HPP | c.1318_20delAAC | N423del | p.N440del | p.[N440del];[R152C] * | Present study |
|---|---|---|---|---|---|
| Adult | c.1328C>T, | A426V | p.A443V | p.[A443V];[=] * | [11] |
| Infantile/perinatal | c.1333T>C | S428P | p.S445P | p. [S445P];[Y263]a * | [14, 21] |
| Odonto-HPP | c.1349G>A | R433H | p.R450H, | p.[D406G];[R450H] * | [41] |
| Infantile/perinatal | c.1348C>T | R433C | p.R450C | p.[R450C];[R450C] | [14, 21] |
Normal allele or uncharacterized mutation (N), Heterozygous (*), previously described as a polymorphism (a), SESEP Laboratory and the Human Molecular Genetics laboratory of the University of Versailles-Saint Quentin en Yvelines, France (Versailles).
Nucleotide numbering is according to the international recommendation and Weiss et al. 1988 the first nucleotide (+1) corresponds to the A of the ATG initiation codon. Non-standardized nomenclature: Amino acid numbering is according to Weiss et al. 1988 (the nomenclature takes into account of a 17-residues signal peptide). Standardized nomenclature: Amino acid numbering is according to the recommendations to the international recommendation. The first amino acid corresponds to the ATG initiation codon.
It is difficult to assign specific dysfunctions to each heterozygous mutation in these probands. Considering the functional and structural importance of the crown domain, and based on the pedigree, reduced ALP activity in vivo and in vitro, and predicted alterations in mutant TNAP structures, we hypothesize that the odonto-HPP phenotype is associated with the heterozygous deletion of residue N440, which may indirectly affect the enzyme activity, possibly from failureto reach a correct conformation, or via alterations in interactions with collagen. On the other hand, based on TNAP protein expression and immunolocalization, analysis of internal contacts, and reports in the literature, we propose that the consequences of heterozygous N440 deletion are intensified by association with the heterozygous missense mutation, p.R152C. However, because the p.R152C mutation was of maternal heritance and the mother was asymptomatic, we propose that when heterozygous and in the absence of other mutations, this alteration is not in itself sufficient to significantly and deleteriously affect TNAP function.
It remains unclear how particular ALPL mutations cause more severe clinical forms of HPP, in contrast to mild forms, including odonto-HPP. Dental tissues have been reported to be highly sensitive to dysregulation of phosphate and pyrophosphate metabolism [42-44], perhaps indicating that less deleterious ALPL mutations may preferentially affect the dentition, but not the broader skeleton.
4. CONCLUSIONS
We characterized a novel genetic alteration (c.1318_1320delAAC, p.N440del) in the ALPL gene resulting in odonto-HPP in the probands, monozygotic twins. Based on pedigree information, clinical symptoms, genetic analysis, and residual ALP activity, a genotype-phenotype association was established for p.N440del and odonto-HPP in this case. The heterozygous gene deletion was paternally inherited, and predicted to alter the loop harboring a collagen-binding site in the TNAP protein crown domain. In addition to this gene deletion, the probands feature an p.R152C missense mutation of maternal inheritance, which was not predicted to alter protein structure, but may contribute to the more aggressive dental phenotype and reduced ALP activity observed in probands (p.[N440del];[R152C]) compared to their father (heterozygous p.N440del). Therefore, we propose that the molecular basis of odonto-HPP phenotype described here is associated with both p.N440del and p.R152C heterozygous compound mutations.
Supplementary Material
Highlights.
- Identification of a novel heterozygous gene deletion (p.N440del) in ALPL gene.
- Genotype-phenotype relationships of compound heterozygous mutations in Odonto-HPP.
- p.R152C heterozygous missense mutation contributes to a more severe dental phenotype.
- Structural consequences of p.N440del and p.R152C mutations in the TNAP by 3D modeling.
- N440 deletion is predicted to alter the collagen-binding site in the TNAP enzyme.
ACKNOWLEDGMENTS
This research was supported in part by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health. A portion of this research was performed while MJS and BLF were affiliated with the University of Washington School of Dentistry, Seattle, WA, USA. The present study was supported by the São Paulo State Research Foundation (FAPESP, Brazil, grant #07/08192-5 and 08/00534-7), Coordination for the Improvement of the Higher Level Personnel (CAPES): 02426/09-9, National Council for Scientific and Technological Development (CNPq): 553386/2008-5, National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR) DE15109, and NIH Fogarty International Research Collaboration Award (FIRCA) grant 5R03TW007590-03.
Footnotes
Conflict of interest: The authors declare no conflict of interest related to this study.
REFERENCES
- 1.Whyte MP. Hypophosphatasia and the role of alkaline phosphatase in skeletal mineralization. Endocr Rev. 1994;15:439–61. doi: 10.1210/edrv-15-4-439. [DOI] [PubMed] [Google Scholar]
- 2.Mornet E. Hypophosphatasia. Orphanet J Rare Dis. 2007;2:40. doi: 10.1186/1750-1172-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mornet E, Nunes ME. Hypophosphatasia. In: Pagon TDB RA, Dolan CR, Stephens K, Adam MP, editors. GeneReviews™ [Internet] 2010/03/20 ed. University of Washington, Seattle; Seattle (WA): 2007. 1993-2007. [Google Scholar]
- 4.Weiss MJ, Ray K, Henthorn PS, Lamb B, Kadesch T, Harris H. Structure of the human liver/bone/kidney alkaline phosphatase gene. J Biol Chem. 1988;263:12002–10. [PubMed] [Google Scholar]
- 5.Orimo H. The mechanism of mineralization and the role of alkaline phosphatase in health and disease. J Nippon Med Sch. 2010;77:4–12. doi: 10.1272/jnms.77.4. [DOI] [PubMed] [Google Scholar]
- 6.Boskey AL. Mineral-matrix interactions in bone and cartilage. Clin Orthop Relat Res. 1992:244–74. [PubMed] [Google Scholar]
- 7.Whyte MP. Physiological role of alkaline phosphatase explored in hypophosphatasia. Ann N Y Acad Sci. 2010;1192:190–200. doi: 10.1111/j.1749-6632.2010.05387.x. [DOI] [PubMed] [Google Scholar]
- 8.Fedde KN, Michell MP, Henthorn PS, Whyte MP. Aberrant properties of alkaline phosphatase in patient fibroblasts correlate with clinical expressivity in severe forms of hypophosphatasia. J Clin Endocrinol Metab. 1996;81:2587–94. doi: 10.1210/jcem.81.7.8675582. [DOI] [PubMed] [Google Scholar]
- 9.Seetharam B, Tiruppathi C, Alpers DH. Hydrophobic interactions of brush border alkaline phosphatases: the role of phosphatidyl inositol. Arch Biochem Biophys. 1987;253:189–98. doi: 10.1016/0003-9861(87)90651-5. [DOI] [PubMed] [Google Scholar]
- 10.Lia-Baldini AS, Muller F, Taillandier A, Gibrat JF, Mouchard M, Robin B, Simon-Bouy B, Serre JL, Aylsworth AS, Bieth E, Delanote S, Freisinger P, Hu JC, Krohn HP, Nunes ME, Mornet E. A molecular approach to dominance in hypophosphatasia. Hum Genet. 2001;109:99–108. doi: 10.1007/s004390100546. [DOI] [PubMed] [Google Scholar]
- 11.Fauvert D, Brun-Heath I, Lia-Baldini AS, Bellazi L, Taillandier A, Serre JL, de Mazancourt P, Mornet E. Mild forms of hypophosphatasia mostly result from dominant negative effect of severe alleles or from compound heterozygosity for severe and moderate alleles. BMC Med Genet. 2009;10:51. doi: 10.1186/1471-2350-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herasse M, Spentchian M, Taillandier A, Keppler-Noreuil K, Fliorito AN, Bergoffen J, Wallerstein R, Muti C, Simon-Bouy B, Mornet E. Molecular study of three cases of odontohypophosphatasia resulting from heterozygosity for mutations in the tissue non-specific alkaline phosphatase gene. J Med Genet. 2003;40:605–9. doi: 10.1136/jmg.40.8.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mornet E, Stura E, Lia-Baldini AS, Stigbrand T, Menez A, Le Du MH. Structural evidence for a functional role of human tissue nonspecific alkaline phosphatase in bone mineralization. J Biol Chem. 2001;276:31171–8. doi: 10.1074/jbc.M102788200. [DOI] [PubMed] [Google Scholar]
- 14.Zurutuza L, Muller F, Gibrat JF, Taillandier A, Simon-Bouy B, Serre JL, Mornet E. Correlations of genotype and phenotype in hypophosphatasia. Hum Mol Genet. 1999;8:1039–46. doi: 10.1093/hmg/8.6.1039. [DOI] [PubMed] [Google Scholar]
- 15.Chang KC, Lin PH, Su YN, Peng SS, Lee NC, Chou HC, Chen CY, Hsieh WS, Tsao PN. Novel heterozygous tissue-nonspecific alkaline phosphatase (TNAP) gene mutations causing lethal perinatal hypophosphatasia. J Bone Miner Metab. 2011;30:109–13. doi: 10.1007/s00774-011-0282-8. [DOI] [PubMed] [Google Scholar]
- 16.Le Du MH, Millan JL. Structural evidence of functional divergence in human alkaline phosphatases. J Biol Chem. 2002;277:49808–14. doi: 10.1074/jbc.M207394200. [DOI] [PubMed] [Google Scholar]
- 17.Le Du MH, Stigbrand T, Taussig MJ, Menez A, Stura EA. Crystal structure of alkaline phosphatase from human placenta at 1.8 A resolution. Implication for a substrate specificity. J Biol Chem. 2001;276:9158–65. doi: 10.1074/jbc.M009250200. [DOI] [PubMed] [Google Scholar]
- 18.Rodrigues TL, Foster BL, Silverio KG, Martins L, Casati MZ, Sallum EA, Somerman MJ, Nociti FH., Jr. Correction of hypophosphatasia-associated mineralization deficiencies in vitro by phosphate/pyrophosphate modulation in periodontal ligament cells. J Periodontol. 2012;83:653–63. doi: 10.1902/jop.2011.110310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodrigues TL, Giorgetti AP, Martins L, Pereira Neto JS, Foster B, Nociti FH., Jr. Clinical correlate: Cementum and periodontal defects resulting from odontohypophosphatasia predispose for premature tooth loss. In: McCauley LK, Somerman MJ, editors. Mineralized Tissues in Oral and Craniofacial Science. John Wiley & Sons, Inc; Hoboken, NJ: 2011. [Google Scholar]
- 20.Rodrigues TL, Foster BL, Silverio KG, Martins L, Casati MZ, Sallum EA, Somerman MJ, Nociti FH., Jr. Hypophosphatasia-associated deficiencies in mineralization and gene expression in cultured dental pulp cells obtained from human teeth. J Endod. 2012;38:907–12. doi: 10.1016/j.joen.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mornet E, Taillandier A, Peyramaure S, Kaper F, Muller F, Brenner R, Bussiere P, Freisinger P, Godard J, Le Merrer M, Oury JF, Plauchu H, Puddu R, Rival JM, Superti-Furga A, Touraine RL, Serre JL, Simon-Bouy B. Identification of fifteen novel mutations in the tissue-nonspecific alkaline phosphatase (TNSALP) gene in European patients with severe hypophosphatasia. Eur J Hum Genet. 1998;6:308–14. doi: 10.1038/sj.ejhg.5200190. [DOI] [PubMed] [Google Scholar]
- 22.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 25.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 26.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neshich G, Togawa RC, Mancini AL, Kuser PR, Yamagishi ME, Pappas G, Jr., Torres WV, Fonseca e Campos T, Ferreira LL, Luna FM, Oliveira AG, Miura RT, Inoue MK, Horita LG, de Souza DF, Dominiquini F, Alvaro A, Lima CS, Ogawa FO, Gomes GB, Palandrani JF, dos Santos GF, de Freitas EM, Mattiuz AR, Costa IC, de Almeida CL, Souza S, Baudet C, Higa RH. STING Millennium: A web-based suite of programs for comprehensive and simultaneous analysis of protein structure and sequence. Nucleic Acids Res. 2003;31:3386–92. doi: 10.1093/nar/gkg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taillandier A, Lia-Baldini AS, Mouchard M, Robin B, Muller F, Simon-Bouy B, Serre JL, Bera-Louville A, Bonduelle M, Eckhardt J, Gaillard D, Myhre AG, Kortge-Jung S, Larget-Piet L, Malou E, Sillence D, Temple IK, Viot G, Mornet E. Twelve novel mutations in the tissue-nonspecific alkaline phosphatase gene (ALPL) in patients with various forms of hypophosphatasia. Hum Mutat. 2001;18:83–4. doi: 10.1002/humu.1154. [DOI] [PubMed] [Google Scholar]
- 29.Mumm S, Jones J, Finnegan P, Henthorn PS, Podgornik MN, Whyte MP. Denaturing gradient gel electrophoresis analysis of the tissue nonspecific alkaline phosphatase isoenzyme gene in hypophosphatasia. Mol Genet Metab. 2002;75:143–53. doi: 10.1006/mgme.2001.3283. [DOI] [PubMed] [Google Scholar]
- 30.Hoylaerts MF, Manes T, Millan JL. Mammalian alkaline phosphatases are allosteric enzymes. J Biol Chem. 1997;272:22781–7. doi: 10.1074/jbc.272.36.22781. [DOI] [PubMed] [Google Scholar]
- 31.Bossi M, Hoylaerts MF, Millan JL. Modifications in a flexible surface loop modulate the isozyme-specific properties of mammalian alkaline phosphatases. J Biol Chem. 1993;268:25409–16. [PubMed] [Google Scholar]
- 32.Ishida Y, Komaru K, Ito M, Amaya Y, Kohno S, Oda K. Tissue-nonspecific alkaline phosphatase with an Asp(289)-->Val mutation fails to reach the cell surface and undergoes proteasome-mediated degradation. J Biochem. 2003;134:63–70. doi: 10.1093/jb/mvg114. [DOI] [PubMed] [Google Scholar]
- 33.Ito M, Amizuka N, Ozawa H, Oda K. Retention at the cis-Golgi and delayed degradation of tissue-non-specific alkaline phosphatase with an Asn153-->Asp substitution, a cause of perinatal hypophosphatasia. Biochem J. 2002;361:473–80. doi: 10.1042/0264-6021:3610473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Numa N, Ishida Y, Nasu M, Sohda M, Misumi Y, Noda T, Oda K. Molecular basis of perinatal hypophosphatasia with tissue-nonspecific alkaline phosphatase bearing a conservative replacement of valine by alanine at position 406. Structural importance of the crown domain. FEBS J. 2008;275:2727–37. doi: 10.1111/j.1742-4658.2008.06414.x. [DOI] [PubMed] [Google Scholar]
- 35.Shibata H, Fukushi M, Igarashi A, Misumi Y, Ikehara Y, Ohashi Y, Oda K. Defective intracellular transport of tissue-nonspecific alkaline phosphatase with an Ala162-->Thr mutation associated with lethal hypophosphatasia. J Biochem. 1998;123:968–77. doi: 10.1093/oxfordjournals.jbchem.a022032. [DOI] [PubMed] [Google Scholar]
- 36.Fukushi M, Amizuka N, Hoshi K, Ozawa H, Kumagai H, Omura S, Misumi Y, Ikehara Y, Oda K. Intracellular retention and degradation of tissue-nonspecific alkaline phosphatase with a Gly317-->Asp substitution associated with lethal hypophosphatasia. Biochem Biophys Res Commun. 1998;246:613–8. doi: 10.1006/bbrc.1998.8674. [DOI] [PubMed] [Google Scholar]
- 37.Mochizuki H, Saito M, Michigami T, Ohashi H, Koda N, Yamaguchi S, Ozono K. Severe hypercalcaemia and respiratory insufficiency associated with infantile hypophosphatasia caused by two novel mutations of the tissue-nonspecific alkaline phosphatase gene. Eur J Pediatr. 2000;159:375–9. doi: 10.1007/s004310051290. [DOI] [PubMed] [Google Scholar]
- 38.Taillandier A, Zurutuza L, Muller F, Simon-Bouy B, Serre JL, Bird L, Brenner R, Boute O, Cousin J, Gaillard D, Heidemann PH, Steinmann B, Wallot M, Mornet E. Characterization of eleven novel mutations (M45L, R119H, 544delG, G145V, H154Y, C184Y, D289V, 862+5A, 1172delC, R411X, E459K) in the tissue-nonspecific alkaline phosphatase (TNSALP) gene in patients with severe hypophosphatasia. Mutations in brief no. 217. Online. Hum Mutat. 1999;13:171–2. doi: 10.1002/(sici)1098-1004(1999)13:2<171::aid-humu16>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 39.Spentchian M, Brun-Heath I, Taillandier A, Fauvert D, Serre JL, Simon-Bouy B, Carvalho F, Grochova I, Mehta SG, Muller G, Oberstein SL, Ogur G, Sharif S, Mornet E. Characterization of missense mutations and large deletions in the ALPL gene by sequencing and quantitative multiplex PCR of short fragments. Genet Test. 2006;10:252–7. doi: 10.1089/gte.2006.10.252. [DOI] [PubMed] [Google Scholar]
- 40.Henthorn PS, Raducha M, Fedde KN, Lafferty MA, Whyte MP. Different missense mutations at the tissue-nonspecific alkaline phosphatase gene locus in autosomal recessively inherited forms of mild and severe hypophosphatasia. Proc Natl Acad Sci U S A. 1992;89:9924–8. doi: 10.1073/pnas.89.20.9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taillandier A, Cozien E, Muller F, Merrien Y, Bonnin E, Fribourg C, Simon-Bouy B, Serre JL, Bieth E, Brenner R, Cordier MP, De Bie S, Fellmann F, Freisinger P, Hesse V, Hennekam RC, Josifova D, Kerzin-Storrar L, Leporrier N, Zabot MT, Mornet E. Fifteen new mutations (−195C>T, L-12X, 298-2A>G, T117N, A159T, R229S, 997+2T>A, E274X, A331T, H364R, D389G, 1256delC, R433H, N461I, C472S) in the tissue-nonspecific alkaline phosphatase (TNSALP) gene in patients with hypophosphatasia. Hum Mutat. 2000;15:293. doi: 10.1002/(SICI)1098-1004(200003)15:3<293::AID-HUMU11>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 42.Foster BL, Nagatomo KJ, Bamashmous SO, Tompkins KA, Fong H, Dunn D, Chu EY, Guenther C, Kingsley DM, Rutherford RB, Somerman MJ. The progressive ankylosis protein regulates cementum apposition and extracellular matrix composition. Cells Tissues Organs. 2011;194:382–405. doi: 10.1159/000323457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foster BL, Nagatomo KJ, Nociti FH, Jr., Fong H, Dunn D, Tran AB, Wang W, Narisawa S, Millan JL, Somerman MJ. Central role of pyrophosphate in acellular cementum formation. PLoS One. 2012;7:e38393. doi: 10.1371/journal.pone.0038393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foster BL, Nagatomo KJ, Tso HW, Tran AB, Nociti FH, Jr., Narisawa S, Yadav MC, McKee MD, Millan JI, Somerman MJ. Tooth root dentin mineralization defects in a mouse model of hypophosphatasia. J Bone Miner Res. 2013;28:271–82. doi: 10.1002/jbmr.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.