Abstract
Autophagy is a cellular process responsible for the degradation of intracellular cargo. Its dynamic nature and the multiple types of autophagy organelles present at a given time make current measurements, such as those done by Western blotting, insufficient to understand autophagy and its roles in aging and disease. Capillary electrophoresis coupled to laser induced fluorescence detection (CE-LIF) has been used previously to count and determine properties of individual organelles, but has never been used on autophagy organelles or for determination of changes of such properties. Here we used autophagy organelles isolated from L6 cells expressing GFP-LC3, which is an autophagy marker, to develop a CE-LIF method for the determination of the number of autophagy organelles, their individual GFP-LC3 fluorescence intensities, and their individual electrophoretic mobilities. These properties were compared under basal and rapamycin-driven autophagy, which showed differences in the number of detected organelles and electrophoretic mobility distributions of autophagy organelles. Vinblastine treatment was also used to halt autophagy and further characterize changes and provide additional insight on the nature of autophagy organelles. This approach revealed dramatic and opposite directions in changes of organelle numbers, GFP-LC3 contents, and electrophoretic mobilities during the duration of the vinblastine treatment. These trends suggested the identity of organelle types being detected. These observations demonstrate that individual organelle analysis by CE-LIF is a powerful technology to investigate the complexity and nature of autophagy, a process that plays critical roles in response to drug treatments, aging, and disease.
2. Introduction
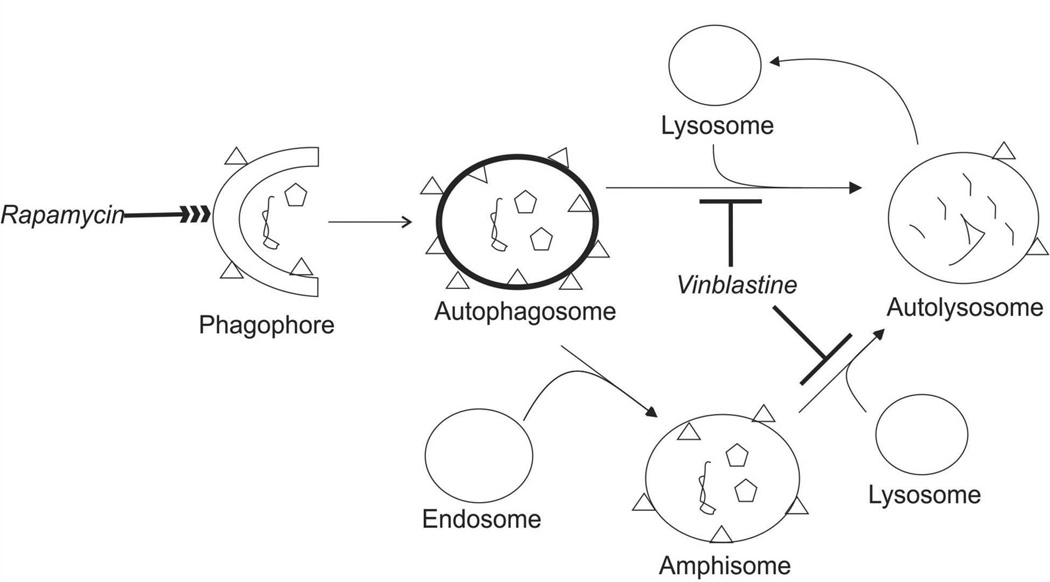
Autophagy is a cellular process responsible for the degradation of intracellular components that involves several organelle types including phagophores, autophagosomes, autolysosomes and amphisomes. These organelles define a commonly accepted autophagy pathway1, 2 and an alternate route involving amphisomes (Figure 1).3, 4 The most commonly accepted autophagy marker is the protein LC3-II, which forms upon lipidation of cytosolic LC3-I with phosphatidylethanolamine.5–7 LC3-II is present in the phagophore, autophagosome, autolysosome, and the amphisome. In autophagosomes LC3-II is an essential protein receptor that when mutated causes loss of autophagy function.6, 8 Not surprisingly, defects in this and other autophagy organelles ultimately links autophagy deficiency to aging, cancer, and neurodegenerative diseases.9
Figure 1.
Autophagy Degradation of Intracellular Contents. Phagophores engulf intracellular components such organelles (pentagons) and have LC3-II localized on their membrane (triangles). Phagophores mature into autophagosomes. Autophagosomes either mature into autolysosomes by direct interaction with lysosomes or into amphisomes by fusing with endosomes. Because amphisomes are a small fraction (<5%), they will not be explicitly mentioned in this report. Autolysosomes degrade the components sequestered in autophagosomes and then are recycled to form lysosomes.47 Treatment with vinblastine halts formation of autolysosomes, while rapamycin enhances the overall autophagy process.4
Understanding autophagy requires characterizing the properties of concomitant organelle types involved in this biological process. Western blotting, fluorescence confocal microscopy, transmission electron microscopy and flow cytometry have been used to monitor autophagy but are inadequate to characterize mixtures of organelles containing LC3-II. Western blots have been used to determine bulk amounts of LC3-II relative to LC3-1 or degradation of GFP-LC3-II in the autolysosome where GFP is cleaved off from GFP-LC3-II.10, 11 Similarly, fluorescence confocal microscopy has been used to determine the number of fluorescently-labeled LC3-II organelles.12–15 Transmission electron microscopy has also been used to evaluate autophagy based on the number of observed autophagosomes.3 Unfortunately microscopies are not high throughput techniques. Flow cytometry has been used to monitor autophagy in single cells by comparing combined levels of GFP-LC3-II and GFP-LC3-I with remaining GFP-LC3-II levels after permeabilization of cells with saponin.16 This technique has not been used for analysis of individual autophagy organelles.
Inhibitors that halt autophagy at specific points of its pathway have been used to evaluate autophagy.3, 4 One of these inhibitors, vinblastine (c.f. Figure 1) binds to tubulin interfering with cytoskeleton polymerization, which is required for scaffolding the fusion of autophagosomes and lysosomes.17–20 As a result, cells treated with vinblastine have increased numbers of autophagosomes compared to non-treated cells.3, 4 Unrelated to vinblastine, rapamycin was used to enhance autophagy. Rapamycin (c.f. Figure 1) activates autophagy by binding to FK-506-binding protein 12, which inactivates the kinase activity of the mammalian target of rapamycin protein (mTOR). When mTOR activity is inhibited, autophagy related proteins such as the Atg1/ULK1 complex, Atg13, FK-200 and Atg101 are activated,25–28 which ultimately causes increased levels of autophagy-related proteins including LC3-II21 and Beclin1.21, 22 It is then not surprising that both LC3-II and Beclin1 have been used to assess the effects of both vinblastine and rapamycin on autophagy.23 Unfortunately, changes in abundance of these proteins after treatments with either vinblastine, rapamycin, or both compounds cannot describe whether such changes result from variations in the numbers of organelles of each type or the individual organelle content of LC3-II or Beclin1.
Capillary electrophoresis coupled to laser induced fluorescence detection (CE-LIF) is a technique previously used to determine the numbers and properties of individual, fluorescently-labeled organelles such as mitochondria,24, 25 nuclei,26 endosomes, and acidic organelles.27, 28 Due to its excellent limits of detection,26, 28, 29 CE-LIF can detect individual organelles tagged with low levels of a fluorescent marker and has the unique capability of providing electrophoretic mobilities, which is strongly dependent on the surface charge of the organelle.25 So far, CE-LIF has not been used to describe the properties of individual autophagy organelles.
This manuscript describes the first analysis of individual autophagy organelles by CE-LIF and the demonstration of its use for comparisons of organelle properties under basal versus rapamycin-enhanced autophagy). Use of vinblastine treatment to halt autophagy provided further evidence on the properties and types of autophagy organelles. Relative to basal autophagy, rapamycin treatment resulted in the detection of a higher number of autophagy organelles, which is expected from previous studies using bulk measurements of autophagy markers.30, 31 Basal and rapamycin-enhanced autophagy displayed opposite trends in GFP-LC3 intensity and electrophoretic mobility distribution changes upon autophagy blockage with vinblastine. Based on these observations autophagosomes and phagophores appear to preferentially contribute to observed changes in the distributions of intensity and electrophoretic mobilities under basal and rapamycin-induced autophagy, respectively, thereby defining properties unique to these organelle types. Future applications of CE-LIF combined with autophagy inhibitors could be used to examine autophagy organelles under conditions prevalent in neurodegenerative diseases32, 33 and aging.32
3. Theory
Various autophagy organelle types are present at a give time in a cell (Figure 1). This composition represents a snapshot of autophagy at a single time point. Sampling of organelles collected under different autophagy conditions (e.g. basal versus rapamycin-enhanced) is commonly used to define differences between these two conditions such that
| Equation 1 |
Where D is the difference in a measured property, iR is the property observed under a given treatment (e.g. rapamycin treatment) and iB is the corresponding basal condition. Bulk differences between two conditions can easily be determined by applying this equation to existing assays (e.g. Western blots). In this study we extend the use of this equation to the number of organelles, individual organelle GFP-LC3-II contents, and individual organelle electrophoretic mobilities.
Assuming steady state conditions, the rates of appearance and disappearance of a given type of autophagy organelle must be equal. That is,
| Equation 2 |
Where Φ is a measure of flux, RA is the rate of appearance and RD is the rate of disappearance. When vinblastine halts autolysosome formation, the rate of accumulation (RA) refers to autophagosome and amphisome formation and rate of disappearance (RD) refer to autolysosome degradation. These rates can be related to the number of detected organelles as follows:
| Equation 3 |
Where No is the number of detected organelles when vinblastine treatment is absent, NT is the number of detected organelles after vinblastine treatment, and Δt is the duration of the treatment. The difference between the rate of accumulation and rate of disappearance of detected organelles can then be calculated as,
| Equation 4 |
When RA – RD is positive, the rate of formation of autophagosomes and amphisomes exceeds the rate of disappearance of autolysosomes. When this difference is negative, the rate of disappearance of autolysosomes exceeds the rate of formation of the other autophagy organelles.
A similar calculation determines changes in properties of organelles,
| Equation 5 |
Where D is the difference of the property of interest it, measured at time t, and the property io measured at the onset of autophagy blockage, divided by the duration of the autophagy blockage (Δt). In this study we used this equation to examine changes in individual organelle contents of GFP-LC3-II and their electrophoretic mobilities before and after vinblastine treatment. A similar interpretation to Equation 4, when D is positive in Equation 5, the property of interest is higher for forming organelles than for disappearing autolysosomes. When D is negative, the reverse is true.
Because individual organelle measurements are commonly represented as distributions, Equation 5 can be modified to compare the xth percentile of two distributions as
| Equation 6 |
Where Dx is the difference at the xth percentile, ix,t and ix,o are values at xth percentile for the property of interest measured at a given time, t, and at the onset of autophagy blockage, respectively. The interpretation of Equation 6 is similar to that of Equation 5, but extends the concept to percentiles of individual organelle measurements.
4. Experimental
Materials, Reagents, and Buffers
Sucrose, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), ethylenediaminetetraacetic acid (EDTA), vinblastine, rapamycin from Streptomyces hygroscopicus, gentamycin, protease inhibitor cocktail, Triton X-100, poly-L-lysine, and poly(vinyl alcohol) (99%+ hydrated, molecular weight 89,000 to 98,0000) were obtained from Sigma Aldrich (Atlanta, GA). Rabbit polyclonal LC3 antibody was obtained from Novus Biologicals (Littleton, CO). Fluorescein, prolong gold antifade reagent with 4’,6’-diamidino-2-phenylindole (DAPI), goat anti-rabbit AlexaFluor568 polyclonal antibody, AlignFlow flow cytometry beads (2.5 µm) and lipofectamine 2000 reagent were obtained from Invitrogen (Carlsbad, CA). Hydrochloric acid was obtained from Mallinckrodt (Phillipsburgh, NJ). Fetal bovine serum was obtained from Omega Scientific (Tarsana, CA). Dubelcco’s Modified Eagle Medium (DMEM) high-glucose cell medium and geneticin were obtained from Gibco (Carlsbad, CA). D-Mannitol was obtained from Riedel-de Haen (Atlanta, GA). Sodium hydroxide was obtained from Fluka (Seelze, Switzerland). Phosphate buffered saline (10× concentration, 1.37 M NaCl, 27 mM KCl, 80 mM Na2HPO4, and 20 mM KH2PO4, pH 7.4) was obtained from BioRad (Hercules, CA). 0.5% trypsin-EDTA (10× concentration, no phenol red) was obtained from Life Technologies (Grand Island, NY). Formaldehyde and sodium chloride were obtained from Fisher Scientific (Pittsburgh, PA). Bovine serum albumin fraction V, heat shock, fatty acid free was obtained from Roche (Basel, Switzerland). Water was purified with a Millipore Synergy UV system (18.2 mΩ/cm, Bedford, MA).
Homogenization buffer was made by adding 11.99 g sucrose (70.0 mM), 19.52 g mannitol (214 mM), 611 mg HEPES (4.31 mM), and 724 mg EDTA (4.94 mM) to 500 mL deionized water and brought to pH 7.2 with 0.1 M HCl and 0.1 M NaOH. Capillary electrophoresis (CE) buffer was made by adding 42.88 g sucrose (250 mM), and 1.18 g HEPES (10.0 mM) to 500 mL deionized water and brought to pH 7.2 with 0.1 M HCl and 0.1 M NaOH. The CE buffer was photobleached with 120-mW LED lights with a λmax at 472 nm for 72 h to reduce background fluorescence intensity.34
Cell Culture
L6 cells (ATCC, Manassas, VA) expressing GFP-LC3 were prepared by transfection with a plasmid containing the EGFP-LC3 gene (plasmid 11546, Addgene)35 and selected by fluorescence activated cell sorting. In brief, both Lipofectamine 2,000 (Life Technologies, Grand Island, NY) transfection reagent (2.4 µL) and plasmid (800 ng) were diluted separately in 50 ML of Opti-MEM medium each, and then mixed. After five minutes, this mixture was added to a well of a 24-well plate containing 70–90% confluent L6 cells. DMEM medium volume was adjusted to 500 µL in each well. After a 48-h treatment, cells were collected by centrifugation at 1,000g for 10 min, and diluted to 2.5 × 106 cells/mL in 1× PBS containing 0.5% w/v bovine serum albumin. Non-transfected L6 cells were used as a control. Cells expressing GFP-LC3 were selected using fluorescence activated cell sorting (FACS) with a BD FACSAria 1 flow cytometer (BD Biosciences, San Jose, CA) using a 130-µm nozzle and a 20 mW, 488 nm-argon ion-laser. Only 32% of the cells were fluorescent. Thus, FACS was used a second time to sort one cell per in each well of 24-well plates. Fluorescent microscopy was used for up to one week following sorting to select fluorescent cell clones. Transfected cells were frozen for future use in liquid nitrogen at −80° C. The studies reported here used the same cell clone.
The original L6 cells and the clone expressing GFP-LC3 were maintained at 37°C, 5% CO2 in DMEM supplemented with 10% fetal bovine serum containing either 100 µg/mL gentamycin or 150 µg/mL of each gentamycin and geneticin, respectively. For maintenance, cells were released with trypsin in PBS (0.5% v/v) from T-flasks and then split 1:40 (v/v) into new flasks. Fluorescence confocal microscopy was used confirm that GFP-LC3 colocalized with an anti-LC3 antibody (see Supplementary Information, S-1, Figure S-1, Table S-1). The high values approaching the maximum correlation and colocalization value for both correlation and colocalization indicate that GFP was bound to LC3 protein (see Supplementary Information, S-2, Table S-1).
Autophagy Treatments
Once cells reached ~90% confluence, cells were treated with vinblastine,3, 4 rapamycin,30, 31 or both. For basal autophagy experiments, two different treatments were performed: One flask was treated with 50 µM vinblastine for two hours, while the second flask was untreated. For rapamycin-induced autophagy experiments, both flasks were treated with 267 µM rapamycin for 3 hours; one hour into this treatment, one flask was treated with 50 µM vinblastine for the remaining two hours, while the second one remained free from vinblastine. For both basal and rapamycin-induced autophagy experiments, cells were harvested by differential centrifugation at 1,000g for 10 minutes and then washed once by suspending in homogenization buffer and differential centrifugation at 1,000g for 10 minutes.
Organelle Isolation and Release
Post-nuclear fractions from GFP-LC3 expressing L6 cells (passages 14–24, 106–107 cells) were prepared. Cell disruption was done in an ice-cooled cell disruption bomb (Parr Instrument Co., Moline, IL) charged to 500–600 psi with nitrogen gas for a minimum of 15 min prior to pressure release. The lysate was collected in a 50-mL falcon tube. Unbroken cells and nuclei were removed by differential centrifugation at 600g for 10 minutes. Organelles were collected by differential centrifugation at 14,000g for 45 minutes. The organelle pellet was reconstituted with homogenization buffer, and centrifuged at 14,000g for 30 minutes. The organelle pellet was reconstituted with CE buffer (100 µL) and was resuspended with a 1.00-mL syringe (Hamilton, Reno NV) to break up the organelle pellet and minimize organelle aggregation. Protein concentrations were determined using the bichinchoninic acid protein assay according to the manufacturer (ThermoScientific, Wilmington, MA).
CE-LIF Instrumentation and Alignment
Poly-vinyl alcohol (PVA) coating of fused silica capillaries (150-µm outer diameter, 30-µm inner diameter, Polymicro, Phoenix, AZ) was done as previously described.36 PVA was used to decrease non-specific binding of organelles to the fused silica capillary.37 The current monitoring method was used to estimate the reduction of electroosmotic flow relative to an uncoated capillary.38 Residual electroosmotic flows were 17 –22% of that of uncoated capillaries. Capillaries were no longer used when GFP-labeled biological material began adhering to the outlet of the capillary. Prior to storage at room temperature, capillaries were flushed with methanol, deionized water, and air.
A previously described custom-built CE-LIF instrument39 was used to determine numbers of autophagy organelles, individual autophagy organelle GFP-LC3-II levels, and individual autophagy organelle electrophoretic mobility. Briefly, a 488-nm argon-ion laser (10 mW) was used for fluorescence excitation. A 530 (± 17.5)-nm filter collected GFP and fluorescein fluorescence. The polyimide coating on the outlet of the capillary was burned to decrease fluorescence from this coating. Fluorescence was detected by a photomultiplier tube (R1477, Hamamatsu Corp., Bridgewater, NJ, 1 kV). Data were collected at 200 Hz and digitized by a NiDaq I/O data board (PCI-MIO-16XE-50, National Instruments, Austin, TX) controlled with Labview 5.1 software (National Instruments). The limit of detection for a fluorescein standard was 11 ± 8 zmol (Ave ± St. Dev., n = 12). Capillary electrophoresis separations were performed at −297 V/cm for ~30 minutes. Hydrodynamic injection (104-cm height, 10 s injection) was used for sample introduction. In between separations, the capillary was washed with methanol and CE buffer for 5 minutes each. Methanol removed any remaining biological material and CE buffer re-equilibrated the capillary.
To align the instrument for fluorescence response reproducibility, AlignFlow beads were continuously flowed through the CE-LIF instrument (see Supplementary Information, S-3, Figure S-2). The %RSD fluorescence matched the manufactorer’s reported levels (see Supplementary Information, Table S-2).
Data Analysis
CE-LIF data was analyzed using Igor Pro (WaveMetrics, Inc., Lake Oswego, OR) as previously described.28 Organelle peaks were selected according to a threshold value defined as follows:
| Equation 7 |
where x̄ is the average background intensity and σ is the standard deviation of the background intensity.
In order to account for variations in contents of each sample, the number of organelle peaks was then normalized to the amount of protein in the sample injected, calculated as follows:
| Equation 8 |
where Vi is the volume injected and [protein] is the protein concentration of the biological sample determined by the BCA assay.
When samples have a large number of organelles, two or more organelle may produce one single detected event due to overlap. Statistical overlap theory (SOT) was used to predict overlap of individual autophagy organelle events (see Supplementary Information, S-4).40, 41 There is a saturation limit above which overlap precludes an accurate count of events, mcritical.41, 42 This saturation limit is based on where σ is the peak width and X is the duration of the bin in seconds.40 When the number of detected organelle events (mdetected), exceeds mcritical, we used mcritical as a conservative value of the number of organelles detected (see Supplementary Information, Table S-3).
Because the detector sensitivity varies from day-to-day, we corrected for variations in instrument sensitivity to allow for comparison of individual autophagy organelle fluorescence levels in separations performed on different days. This correction used the background intensity (c.f. Equation 7), assumed proportional to the LIF detector sensitivity. All the signal intensities of the detected peaks were normalized to that with the lowest background intensity level. This correction is particularly important because each condition (basal autophagy, untreated; basal autophagy, vinblastine-treated; rapamycin-induced autophagy, untreated; and rapamycin-induced autophagy, vinblastine-treated) was done on different days (see Supplementary Information S-3).
The electrophoretic mobilities of detected organelles (µ) are affected by run-to-run fluctuations. We used the observed (µFlu,o) and known (µFlu,r) electrophoretic mobilities of fluorescein to determine the electrophoretic mobilities of observed events (µorganelle) as follows:
| Equation 9 |
| Equation 10 |
where µorganelle is the mobility of a detected organelle, L is the length of the capillary, t is the organelle migration time, V is the separation voltage, µFlu,o is the observed fluorescein electrophoretic mobility, and µFlu,r is the reported electrophoretic mobility of fluorescein in a PVA-coated capillary.42
Quantile-quantile plots (QQ plots) were used to compare, in increments of 5%, the 5th through 95th percentiles of distributions of individual organelle fluorescence levels and electrophoretic mobilities. If the two distributions are similar, their QQ plot approaches a diagonal line with slope equal to one.
Safety Considerations
Biosafety level 1 was observed for all procedures using L6 cells. Biological waste was treated with bleach for 30 min prior to disposal. Used cell culture supplies were autoclaved prior to disposal.
5. Results & Discussion
Detection of Individual Autophagy Organelles by CE-LIF
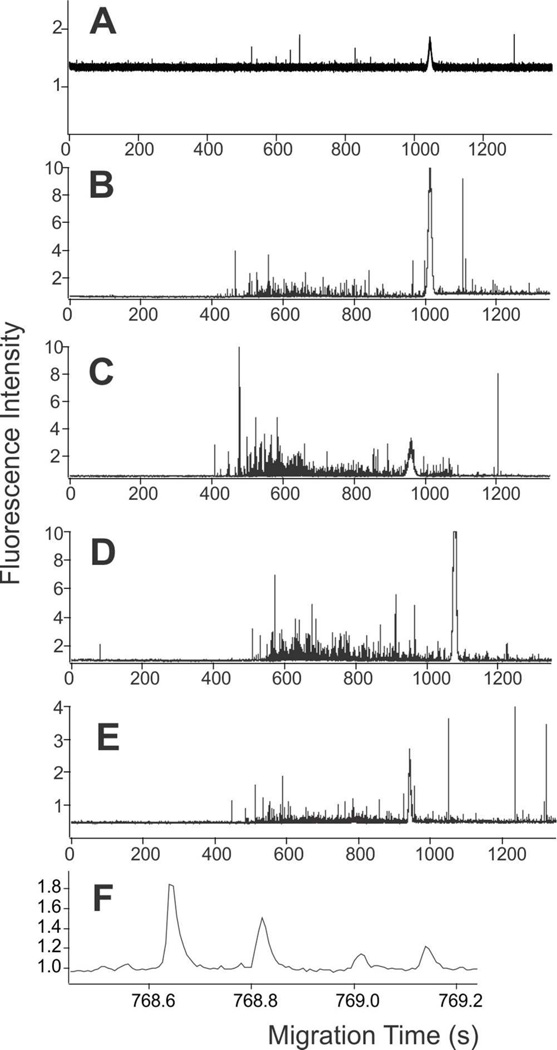
The first focus of this study was to determine the suitability of CE-LIF to detect autophagy organelles and measure their properties at the individual organelle level. Prior work demonstrated the use CE-LIF to determine the numbers and properties of other types of individual organelles.24–28 To evaluate the utility of GFP-LC3 as a label for autophagy organelles, fluorescence confocal microscopy confirmed colocalization of GFP and anti-LC3 antibody at puncta (see Supplementary Information, S-1), which indicated GFP-LC3 is a suitable marker to monitor autophagy organelles by CE-LIF. Treatments with vinblastine and rapamycin also showed suitable colocalization confirming the suitability of GFP-LC3 as a marker out autophagy organelles under treatments relevant to the CE-LIF studies (see Supplementary Information, Table S-1). In agreement, the CE-LIF analysis of organelles isolated from GFP-LC3 expressing L6 cells under conditions of basal autophagy, treated with vinblastine, treated with rapamycin, or treated with both rapamycin and vinblastine demonstrated detection of individual autophagy organelles (Figure 2).
Figure 2.
Electropherograms of L6 cells expressing GFP-LC3. (A) Non-transfected cells. (B)–(F) GFP-LC3 expressing cells. (B) Basal autophagy. (C) Basal autophagy after vinblastine treatment. (D) Rapamycin enhanced autophagy. (E) Rapamycin enhanced autophagy after vinblastine treatment. The broad peaks at ~1,000 s in (A)–(E) are fluorescein peaks. (F) Expansion of individual organelle detected events from (E). CE buffer: 250 mM sucrose, 10 mM HEPES, pH 7.2; −297 V/cm in a PVA coated capillary. Fluorescence detection at 530 ± 18 nm.
Representative electropherograms of individual organelles showed narrow peaks, (FWHM = 36 ± 16 ms; Ave ± St. Dev, n =12,567 peaks), with their width defined by the organelle’s travel time through the laser beam of the LIF detector (Figure 2F). These narrow events are expected when a fluorophore (i.e. GFP-LC3-II) is bound to an organelle. On the other hand, GFP-LC3-I, which is cytosolic, would be free in solution and would be detected as a band with broadening defined by diffusion and other band broadening effects.34 A broad peak from GFP-LC3-I was not detected, because GFP-LC3-I was lost during removal of the cytosol by differential centrifugation during the fractionation procedure.43 Because GFP-LC3-I does not interfere in the CE-LIF results, here we refer to GFP-LC3-II, the organelle bound form, as GF-PLC3.
To assess the presence of random non-specific events among detected events corresponding to GFP-LC3-II labeled organelles, we evaluated the number of peaks in the pre-migration window. This time region (0 to ~ 450 – 500 s) corresponds to the time required by organelles to reach the detector. The number of peaks detected in the pre-migration window was 0.02 ± 0.02 peaks/s (Ave ± St. Dev., n = 12 runs). Therefore, non-specific events do not contribute significantly to the detected organelles observed in Figure 2.
To eliminate the possibility that the observed events were attributed to native fluorescence, we analyzed organelles isolated from L6 cells not expressing GFP-LC3 (Figure 2A). The number of detected events per ng protein in this control was 6 ± 0 (Ave. ±St. Dev., n = 2 runs), while this number was 252 ± 78 events (Ave. ±St. Dev., n = 3 runs) in the sample from GFP-LC3 expressing cells. These results indicate that the bias due to native fluorescence is < 2%. This minimal number of detected organelle peaks in this control (Figure 2A) relative to analyses of GFP-LC3 expressing cells (Figure 2B–E) confirmed that the observed events are mainly due to GFP-LC3 fluorescence and not to native fluorescence. Together, these results confirm that GFP-LC3 labeled organelles can be monitored by CE-LIF.
Next, we assessed the reproducibility of the individual organelle fluorescence intensity and electrophoretic mobility distributions obtained from replicate CE-LIF analysis of the same sample. QQ plots confirmed that both properties had adequate reproducibility (see Supplementary Information, Figure S-3).25, 37
Individual Organelle Comparisons Between Basal and Rapamycin-Enhanced Autophagy
Individual organelle measurements were used to compare snapshots of basal and rapamycin-enhanced autophagy (c.f. Equation 1). These snapshots include: (1) the number of organelles present in the respective samples, (2) the distribution of individual organelle GFP-LC3 intensities, and (3) the distribution of individual organelle electrophoretic mobilities.
Samples from rapamycin-treated cells had higher numbers of autophagy organelles than untreated cells (174 ± 31 and 71 ± 15, events/ng protein, respectively; Avg. ± St. Dev., n = 3 runs). These results are in agreement with previous observations done by confocal fluorescence microscopy in which rapamycin-treatment increased the number of autophagy organelles detected in primary cortical neurons44 and in normal rat kidney cells12 relative to those numbers in untreated cells.
Quantile-quantile plots (QQ plots) were used to compare percentiles of distributions of individual organelle fluorescence levels and electrophoretic mobilities (Figures 3–6). Data from replicate runs of the same sample were pooled as previously done.25, 37 This approach provided the means to compare distributions of individual organelle GFP-LC3 fluorescence intensities and electrophoretic mobility distributions resulting from the CE-LIF of basal and rapamycin enhanced autophagy. Histograms of GFP-LC3-II levels and electrophoretic mobility values are commonly used to compare data sets (see Supplementary Information, Figure S-4). However, the differences between distributions are appreciated better using QQ plots.
Figure 3.
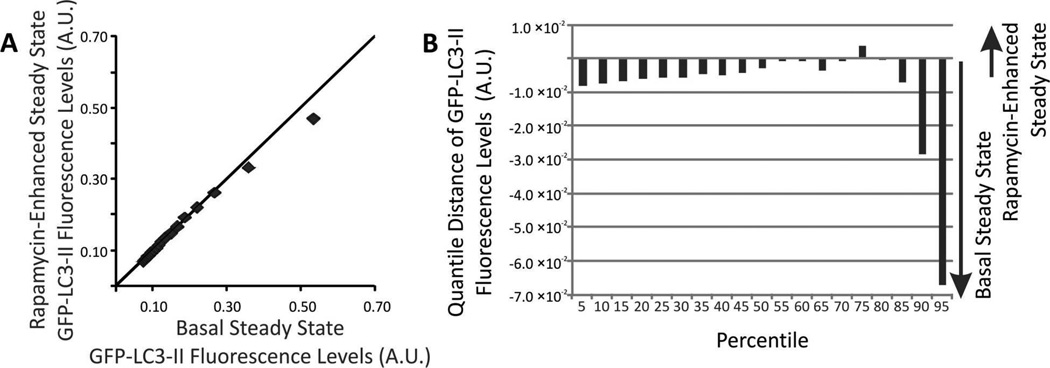
(A) QQ plot of individual GFP-LC3 fluorescent intensity distributions. Comparison of distributions of individual GFP-LC3 levels under basal (x-axis) and rapamycin-enhanced (y-axis). Markers represent 5, 10, 15…95th percentiles. For percentiles with y = x, their intensity value is the same and appear on top of the diagonal line. (B) Changes in the distributions of individual GFP-LC3 levels. The change for a given percentile was calculated according to Equation 6.
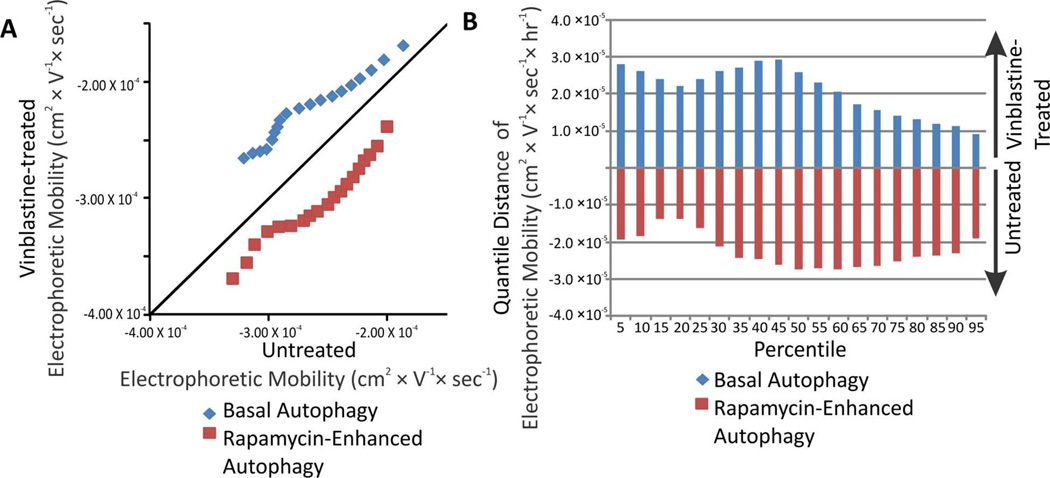
Figure 6.
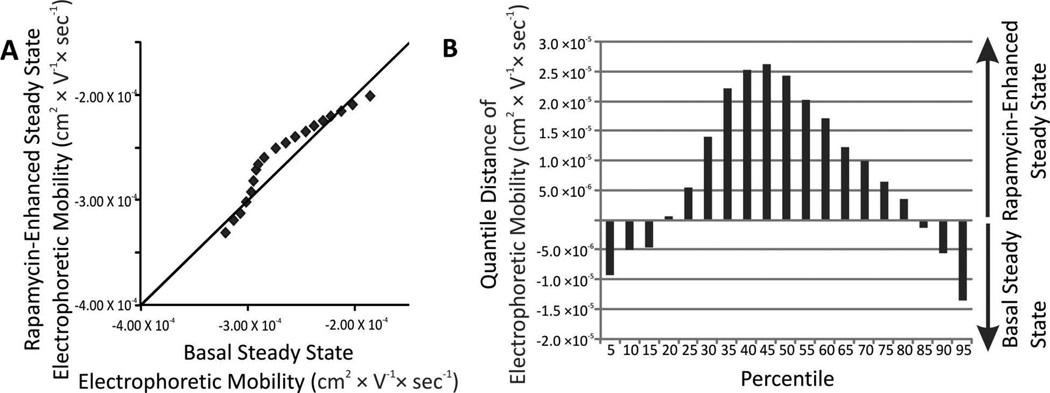
(A) Changes in the distributions of individual electrophoretic mobilities. (B) Changes in the distributions of individual electrophoretic mobilities. The change for a given percentiles was calculated according to Equation 6.
Fluorescence intensity distributions revealed that the contents of GFP-LC3 levels in individual autophagy organelles were remarkably similar between rapamycin-enhanced and basal autophagy (Figure 3). This suggests that, although rapamycin enhances autophagy, the GFP-LC3 contents in autophagy organelles and the proportions of the various types of autophagy organelles remain about the same.
Electrophoretic mobility has been associated with the surface charge of organelles.25 This property is not strongly dependent on organelle size. Indirect evidence from plotting electrophoretic mobilities versus GFP-LC3-II, a modest surrogate of size, does not show any trend (see Supplementary Information, Figure S-5). In agreement, a previous study suggested that surface charge and not size is a major determinant of electrophoretic mobility of organelles in CE-LIF separations.25 In this study, liposomes mimicking mitochondria size and surface composition were compared to theoretical predictions of electrokinetic models of particles, which suggested that the electrophoretic mobilities of these models were dependent primarily on their surface charge density. Given the similarities in size and surface composition between mitochondria and other organelles, it has been accepted that surface charge is mainly responsible for the electrophoretic behavior organelles including nuclei,26 mitochondria,20, 21 and acidic organelles.27, 28 Furthermore, modification of the organelle surface via trypsin digestion led to changes in electrophoretic mobility,34 Thus, it is highly likely that the electrophoretic mobility of autophagy organelles is also a surrogate of their surface composition, primarily determined by lipids and proteins.
The electrophoretic mobilities of autophagy organelles were, in general, more positive when cells were treated with rapamycin relative to untreated cells (25th –80th percentiles in Figure 4.) A maximum difference was observed at the 45th percentile. These observations suggest that rapamycin treatment not only enhances the number of autophagy organelles present at a given time but it is associated with a change in surface composition, which may reflect on changes in proportions of the various organelle types, the subcellular origins of components of the surface of autophagy organelles or on interactions with other organelle types.
Figure 4.
(A) QQ plot of individual autophagy organelle electrophoretic mobility distributions. Comparison of distributions of individual electrophoretic mobility under basal (x-axis and rapamycin-enhanced (y-axis). Markers represent 5, 10, 15…95th percentiles. For percentiles with y = x, their electrophoretic mobility value is the same and should appear on top of the diagonal line. (B) Changes in the distributions of individual autophagy organelle electrophoretic mobility. The change for a given percentiles was calculated according to Equation 6.
Changes in Individual Autophagy Organelle Properties When Halting Autophagy
Treatment with vinblastine disrupts cytoskeletal organization4 that blocks fusion of autophagosomes and lysosomes thereby decreasing formation of autolysosomes. Because degradation of autolysosomes is not interrupted these organelles tend to disappear vinblastine treatment. Comparison of the results of individual autophagy organelle properties in the presence and absence of vinblastine treatments allowed us to assess the overall change in organelle number and organelle properties during the duration of the treatment (c.f. Equations 3 and 4).
For basal autophagy, the numbers of detected organelle events were 107 ± 23 and 230 ± 59 events/ng protein (Avg. ± St. Dev., n = 3 run) for untreated and after a 2-hour vinblastine treatment, respectively. Using Equation 3, the overall change was 62 ± 32 events/(hour × ng protein). Assuming that phagophore is not altered by vinblastine treatment and that amphisomes are secondary contributors to autophagy, the results presented here suggest that the number of autophagosomes forming is higher than the number of autolysosomes disappearing for basal autophagy. The results for rapamycin-enhanced autophagy were surprising. It is not possible to distinguish whether these results are caused by formation of autophagosomes and phagophores with undetectable
Because rapamycin enhances autophagy by inhibition of the mammalian target of rapamycin, leading to increased levels of autophagosome-related proteins such as LC3-II and Beclin1, it was anticipated that the number of accumulating organelles with detectable levels of GFP-LC3-II would be higher after vinblastine treatment. Surprisingly, the results showed the opposite. The number of detected organelle events were 174 ± 31 and 49 ± 20 events/ng protein (Avg. ± St. Dev., n = 3 runs) for untreated and after a 2-hour vinblastine treatment, respectively; the change in the number of detected organelle events was −63 ± 18 events/(hour × ng protein). These results suggest that under rapamycin-treatment, halting autophagy results in faster disappearance of autolysosomes than formation of autophagosomes.
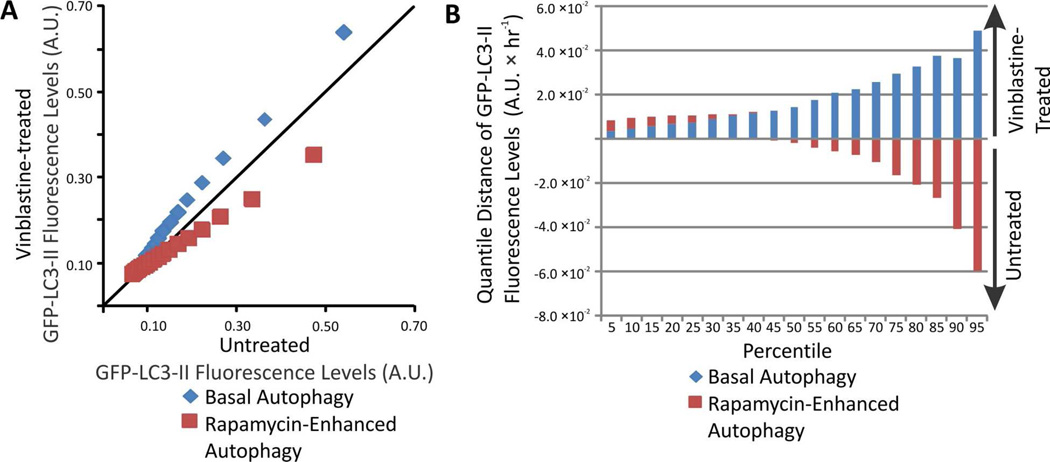
Changes in the distributions of individual autophagy organelle GFP-LC3 levels occurring upon vinblastine treatment were represented as QQ plots (Figure 5A). For basal autophagy, the organelles that accumulate upon vinblastine treatment tend to have higher individual GFP-LC3 fluorescence than autolysosomes that are being degraded. Along the x-axis of the QQ plot, GFP-LC3 levels increased by .10 – 32% when compared to autophagy organelles from untreated cells (Figure 5B). In contrast, for rapamycin-enhanced autophagy, organelles that accumulate have lower individual GFP-LC3 fluorescence than autolysosomes that are being degraded. Along the x-axis of the QQ plot, GFP-LC3 levels decreased for the 50th – 95th percentile, down by 33% at 95th percentile, when compared to autophagy organelles from untreated cells (Figure 5B). Consistently, previous studies also reported an increase in total LC3-II levels upon autophagy blockage under basal conditions,16, 45 and a decrease in GFP-LC3-II levels when rapamycin-treated cells are subject to autophagy blockage.23 Such bulk studies are unable to describe what CE-LIF measurements describe here, that the change in LC3-II or GFP-LC3-II levels are uniquely associated with about 50% of the organelles in the sample.
Figure 5.
(A) Changes in the distributions of individual GFP-LC3 levels. (B) Changes in the distributions of individual GFP-LC3 levels. The change for a given percentiles was calculated according to Equation 6.
QQ plots of individual organelle GFP-LC3-II levels also hints at the types of organelles that change upon vinblastine treatment (c.f. Figure 5). The lower quantiles would represent (1) phagophores because these are the first autophagy structures formed, which suggest lower GFP-LC3 levels and fluorescence than autophagosomes,5 and (2) autolysosomes, which experience cleavage and degradation of GFP-LC3-II as the autolysosome matures and also low quantum fluorescence yield because the GFP fluorescence is decreased at the low pH found in the autolysosomal lumen.46–48 Because below the 50th quantile, changes due to vinblastine treatment were minimal under both basal and rapamycin-induced autophagy, it is likely that phagophores and autolysosomes do not change in GFP-LC3-II contents. This would imply that under basal conditions the increases in GFP-LC3-II contents correspond mainly to individual autophagosomes. In contrast, under rapamycin-enhanced autophagy, organelles with lower GFP-LC3-II contents predominantly accumulate upon vinblastine treatment; these organelles would correspond to phagophores, which continue forming during such treatment.
Changes in the distributions of individual electrophoretic mobilities of autophagy organelles occurring upon treatment with vinblastine were represented as QQ plots (Figure 6A). For basal conditions, vinblastine treatment caused a positive shift in electrophoretic mobilities suggesting that the surface charge of accumulating autophagosomes is relatively more positive than that of the other types of autophagy organelles (Figure 6). This change was 10 – 20% in the quantiles observed (Figure 6B). These results suggest that autophagosomes that accumulate under vinblastine treatment have a more positive charge density relative to the other types of autophagy organelles.
In contrast, for rapamycin-enhanced autophagy conditions, vinblastine treatment caused a negative shift in electrophoretic mobilities suggesting that the surface charge of the accumulated organelles is predominantly more negative than that of other types of autophagy organelles (Figure 6). This change was 8 – 20% in the quantiles observed. Because phagophores continue forming and being transformed into autophagosomes during vinblastine treatment and amphisome numbers are low compared to autophagosomes,4 is it suggested that phagophores are responsible for the negative shift in mobility observed under rapamycin-enhanced autophagy conditions. This would imply that phagophores have a more negative charge density than the other types of autophagy organelles.
Considering the multiple sources of autophagosomes membrane components such as the plasma membrane, Golgi, ER, and mitochondria,49–51 it is not surprising to observe highly heterogeneous electrophoretic mobilities in individual autophagy organelles. Furthermore, in agreement, with rapamycin treatments used here, other studies reported that under starvation-enhanced autophagy, the source of autophagosomes membrane components shifts from the Golgi to the ER,49 Similarly, other studies suggest that starvation induces use of outer mitochondrial membrane as a source autophagosome membrane components.52, 53 Despite of the confounding effects of this heterogeneity, CE-LIF analysis after vinblastine treatment was able to uncover electrophoretic mobility changes after vinblastine treatment. These changes are likely attributed to more positive surface charges of autophagosomes, relative to other types of autophagy organelles under basal conditions (c.f., Figure 6). In contrast, under rapamycin-induced autophagy, phagophores may prevail among the detected organelles, which are characterized by lower levels of detected GFP-LC3-II (c.f. Figure 5) and more negative electrophoretic mobilities (c.f. Figure 6).
6. Conclusions
This study demonstrated the use of CE-LIF to detect individual autophagy organelles based on their GFP-LC3-II contents. Individual organelle detection eliminates biases caused by cytosolic GFP-LC3-I, which is eliminated during sample preparation. Both individual GFP-LC3 fluorescence and electrophoretic mobilities were directly determined from CE-LIF data, which made possible to observe individual organelle properties under basal and rapamycin-enhanced autophagy.
Comparison of basal and rapamycin-enhanced autophagy revealed an increased number of autophagy organelles under rapamycin-enhanced conditions, fairly consistent levels of GFP-LC3 between the two states, and prevalence of organelles with a positive shift in electrophoretic mobilities for rapamycin-enhanced autophagy, which suggest higher relative abundance of organelles with more positive surface charge.
Vinblastine was used to halt autophagy allowing for examination of changes in individual organelle properties during the treatment. These experiments suggest that: (1) the difference in rates of accumulation of autophagosomes and degradation of autolysosomes is positive under basal condition and negative under rapamycin-enhanced autophagy; (2) higher levels of GFP-LC3-II in individual organelles suggest that autophagosomes accumulate under basal conditions, while the lower levels of GFP-LC3 under rapamycin-enhanced conditions points at prevalence of phagophore accumulation; and (3) autophagosomes and phagophores may have more positive and negative surface charge, respectively. These observations could not have been made using currently available technologies such as Western blots, flow cytometry, or microscopy.
Future developments could enhance the description of autophagy via CE-LIF analysis. Fluorescent labels specific for autolysosomes and phagophores could provide a more comprehensive description of the various organelle types. In addition, labeling of other organelle types may aid at monitoring autophagy sub-types such as mitophagy (degradation of mitochondria) and pexophagy (degradation of peroxisomes). These development combined with temporal measurements would respond to a critical need for tools to investigate the role of autophagy flux in disease and aging.32, 33, 54
Supplementary Material
Acknowledgment
The LC3-GFP plasmid was a generous gift from the Professor Do-Hyung Kim laboratory from the University of Minnesota-Twin Cities. Dr. Jonathan Dozier assisted in LC3-GFP plasma preparation and cloning. Dr. Vratislav Kostal assisted in the transformation and selection of transfected cells. Funding was provided by NIH grant AG020866.
Footnotes
Conflict of Interest Disclosure
The authors declare no competing financial interest.
Supplementary Information
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Klionsky DJ. J. Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massey AC, Zhang C, Cuervo AM. Curr. Top. Dev. Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- 3.Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. J. Biol. Chem. 1998;273:21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- 4.Seglen PO, Brinchmann MF. Autophagy. 2010;6:542–547. doi: 10.4161/auto.6.4.11272. [DOI] [PubMed] [Google Scholar]
- 5.Polson HEJ, de Lartigue J, Rigden DJ, Reedijk M, Urbé S, Clague MJ, Tooze SA. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 6.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. EMBO J. 2003;22:4577–4577. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 8.Cherra SJ, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW, Chu CT. J. Cell. Biol. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 11.Welter E, Thumm M, Krick R. Autophagy. 2010;6:794–797. doi: 10.4161/auto.6.6.12348. [DOI] [PubMed] [Google Scholar]
- 12.Jahreiss L, Menzies FM, Rubinsztein DC. Traffic. 2008;9:574–587. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fader CM, Sanchez D, Furlan M, Colombo MI. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 14.Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Mol. Biol. Cell. 2010;21:1001–1010. doi: 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narendra D, Tanaka A, Suen DF, Youle RJ. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eng KE, Panas MD, Hedestam GBK, McInerney GM. Autophagy. 2010;6:634–641. doi: 10.4161/auto.6.5.12112. [DOI] [PubMed] [Google Scholar]
- 17.Gordon PB, Hoyvik H, Seglen PO. Biochem. J. 1992;283:361–369. doi: 10.1042/bj2830361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vacca A, Iurlaro M, Ribatti D, Minischetti M, Nico B, Ria R, Pellegrino A, Dammacco F. Blood. 1999;94:4143–4155. [PubMed] [Google Scholar]
- 19.Olmsted JB, Borisy GG. Annu. Rev. Biochem. 1973;42:507–540. doi: 10.1146/annurev.bi.42.070173.002451. [DOI] [PubMed] [Google Scholar]
- 20.Wilson L. Ann. N. Y. Acad. Sci. 1975;253:213–231. doi: 10.1111/j.1749-6632.1975.tb19201.x. [DOI] [PubMed] [Google Scholar]
- 21.Lee JS, Lee GM. Biotechnol. Bioeng. 2012;109:3093–3102. doi: 10.1002/bit.24567. [DOI] [PubMed] [Google Scholar]
- 22.Sekiguchi A, Kanno H, Ozawa H, Yamaya S, Itoi E. J. Neurotrauma. 2012;29:946–956. doi: 10.1089/neu.2011.1919. [DOI] [PubMed] [Google Scholar]
- 23.Mizushima N, Yoshimori T, Levine B. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strack A, Duffy CF, Malvey M, Arriaga EA. Anal. Biochem. 2001;294:141–147. doi: 10.1006/abio.2001.5148. [DOI] [PubMed] [Google Scholar]
- 25.Duffy CF, Fuller KM, Malvey MW, O'Kennedy R, Arriaga EA. Anal. Chem. 2001;74:171–176. doi: 10.1021/ac010939i. [DOI] [PubMed] [Google Scholar]
- 26.Gunasekera N, Musier-Forsyth K, Arriaga E. Electrophoresis. 2002;23:2110–2116. doi: 10.1002/1522-2683(200207)23:13<2110::AID-ELPS2110>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 27.Fuller KM, Arriaga EA. Anal. Chem. 2003;75:2123–2130. doi: 10.1021/ac026476d. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Arriaga EA. Anal. Chem. 2006;78:820–826. doi: 10.1021/ac051513x. [DOI] [PubMed] [Google Scholar]
- 29.Wu S, Dovichi NJ. J. Chromatogr. A. 1989;480:141–155. doi: 10.1016/s0021-9673(01)84284-9. [DOI] [PubMed] [Google Scholar]
- 30.Ravikumar B, Vacher C, Berger Z, Davies J, Luo S, Oroz L, Scaravilli F, Easton D, Duden R, O'Kane C, Rubinsztein D. Nat. Genet. 2004;36:585. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 31.Webb J, Ravikumar B, Atkins J, Skepper J, Rubinsztein D. J. Biol. Chem. 2003;278:25009. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 32.Cuervo AM. Trends Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuervo AM. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Wolken GG, Kostal V, Arriaga EA. Anal. Chem. 2011;83:612–618. doi: 10.1021/ac102712r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilges M, Kleemiss MH, Schomburg G. Anal. Chem. 1994;66:2038–2046. [Google Scholar]
- 37.Whiting CE, Arriaga EA. Electrophoresis. 2006;27:4523–4531. doi: 10.1002/elps.200600249. [DOI] [PubMed] [Google Scholar]
- 38.Huang X, Gordon MJ, Zare RN. Anal. Chem. 1988;60:1837–1838. [Google Scholar]
- 39.Duffy CF, Gafoor S, Richards DP, Admadzadeh H, O'Kennedy R, Arriaga EA. Anal. Chem. 2001;73:1855–1861. doi: 10.1021/ac0010330. [DOI] [PubMed] [Google Scholar]
- 40.Davis JM, Arriaga EA. J. Chromatogr. A. 2009;1216:6335–6342. doi: 10.1016/j.chroma.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis JM, Arriaga EA. Anal. Chem. 2010;82:307–315. doi: 10.1021/ac901982u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kostal V, Fonslow BR, Arriaga EA, Bowser MT. Anal. Chem. 2009;81:9267–9273. doi: 10.1021/ac901508x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham JM, Rickwood D. Subcellular fractionation : a practical approach. USA: New York, NY: Oxford University Press; 1997. [Google Scholar]
- 44.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. J. Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizushima N, Yoshimori T. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 46.Kimura S, Noda T, Yoshimori T. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y, Yu L. Exp. Cell Res. 2013;319:142–146. doi: 10.1016/j.yexcr.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Han K, Kwon HW, Kang H, Kim J, Lee MS, Choi MY. Physica A. 2012;391:686–692. [Google Scholar]
- 49.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. J. Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cuervo AM. Nat. Cell Biol. 2010;12:735–737. doi: 10.1038/ncb0810-735. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki K, Ohsumi Y. FEBS Lett. 2010;584:1280–1286. doi: 10.1016/j.febslet.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 52.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tooze SA, Yoshimori T. Nat. Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 54.Cuervo AM, Bergamini E, Brunk UT, Dröge W, Ffrench M, Terman A. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.