Abstract
Single-molecule fluorescence spectroscopy of freely diffusing molecules in solution is a powerful tool used to investigate the properties of individual molecules. Single-Photon Avalanche Diodes (SPADs) are the detectors of choice for these applications. Recently a new type of SPAD detector was introduced, dubbed red-enhanced SPAD (RE-SPAD), with good sensitivity throughout the visible spectrum and with excellent timing performance. We report a characterization of this new detector for single-molecule fluorescence resonant energy transfer (smFRET) studies on freely diffusing molecules in a confocal geometry and alternating laser excitation (ALEX) scheme. We use a series of doubly-labeled DNA molecules with donor-to-acceptor distances covering the whole range of useful FRET values. Both intensity-based (μs-ALEX) and lifetime-based (ns-ALEX) measurements are presented and compared to identical measurements performed with standard thick SPADs. Our results demonstrate the great potential of this new detector for smFRET measurements and beyond.
Keywords: SPAD, FRET, single-molecule, confocal, diffusion, ALEX, lifetime, TCSPC
1. INTRODUCTION
Single-molecule fluorescence spectroscopy methods are used by an increasing number of investigators to study fundamental questions in fields as diverse as polymer physics1, DNA or protein interactions and conformations2 or enzyme activity3 at the single-molecule level. Current technical developments are focusing mostly on expanding the number of simultaneously recorded spectral channels4–5, improving the sophistication of data acquisition and analysis methods6 or combining single-molecule spectroscopy with other experimental systems7–8. All these progresses rely on low-noise, high-sensitivity detectors, which have attained maturity during the past decade9–10. In the specific case of point detection used in confocal detection of molecules diffusing in solution, SPADs have become a de facto standard. They are by design limited to a point-like detection volume but have single-photon counting capabilities and exquisite temporal resolution allowing time-resolved studies down to the picosecond (ps) time scale.
In the context of this work, it is worth noting that the generic term “SPAD” hides a surprising variety of architectures and fabrication processes, which have direct implications for single-molecule spectroscopy applications11. Among them, SPADs manufactured using a thick reach-through structure (henceforth referred to as “thick SPADs”), have been around for over a decade12 and have exceptional sensitivity throughout the visible and near-infrared (NIR) range. Arguably their introduction has been the main reason why single-molecule spectroscopy has been possible13. However, they suffer from several drawbacks: possible damage by high photon flux, relatively slow instrument response function (IRF), count rate-dependent IRF walk14 and in general performance depending on the size and location of the illuminated area. Although some of these problems can be partially fixed by implementing additional electronic circuitry15 or have been improved upon in a recent device16, they remain less robust and reliable than shallow junctions (or “thin”) SPADs11, fabricated using a custom-technology, which do not suffer from these issues.
Thin or custom-technology SPADs, on the other hand, had until recently a lower sensitivity in the red region of the spectrum, but very good temporal resolution (<100 ps)11, which explains that a number of groups have implemented “dual” setup architectures, in which “green” photons are detected using thin SPADs (to take advantage of their excellent temporal resolution), whereas “red” photons are detected using thick SPADs (preferred despite their lower temporal resolution because of their better sensitivity)17–18. The recent development of “red-enhanced” thin SPADs19 (RE-SPAD) thus promises a significant simplification in single-molecule setup design, improvements in temporal resolution and even more importantly, opens up the perspective of arrays of SPADs developed with this technology for high-throughput single-molecule studies20–25.
This work examines the suitability of the single-pixel RE-SPAD for smFRET experiments and compares its performance with that of a standard thick SPAD. To this effect, we performed smFRET experiments on freely-diffusing short double-stranded DNA (dsDNA) molecules. DNA molecules were doubly-labeled with a FRET pair, separated by variable distances covering the range of low to high FRET efficiencies. We used a single-spot confocal geometry with either continuous-wave lasers (for intensity-based FRET measurements using a μs-ALEX approach26–27) or pulsed lasers (for lifetime-based measurements using a ns-ALEX approach28) and compared results obtained in identical conditions with thick SPADs and custom technology SPADs.
This paper is organized as follows: Section 2 describes the molecules and setup used in this work as well as our data acquisition and analysis protocols. Results are reported in the next two sections: intensity-based smFRET results in Section 3 and lifetime-based results in Section 4. Comparison of both sets of results is presented in Section 5. In both cases, the RE-SPAD performed as expected from its theoretical specifications and appeared as a reliable alternative to existing detectors, with a few definite advantages discussed in Section 6.
2. MATERIALS AND METHODS
2.1. Samples
A set of 5 different FRET samples and their corresponding singly-labeled counterparts was used. All samples consisted of a common 40 base-pair (bp) long doubly-labeled double-stranded DNA (dsDNA) with the donor (ATTO 550) on one strand and the acceptor (ATTO 647N) on the other. The acceptor dye was bound to the 5′ end of the top strand (Fig. 1), while the donor dye was attached to the bottom strand at different positions from the 3′ end (7, 12, 17, 22 and 27 bp away from the acceptor, respectively). The sequence is identical to that used in previous work27 and is designed in such a way that the environment of the donor dye is similar for all molecules, in order to minimize variations in donor quantum yield between samples. Dyes were attached to a dT residue through a C6 linker using NHS-ester chemistry. Dual HPLC purified singly-labeled ssDNA samples were purchased from IDT (Coralville, IO) and used without further purification. ssDNA molecules were hybridized to their complementary strand in a 1:1 stoichiometry to form doubly-labeled samples, and with two-fold excess of unlabeled complementary strand for singly-labeled samples. dsDNA samples were prepared with filtered, freshly prepared buffer (Tris-EDTA 50) and kept on ice until observed. 5 μl of sample at single-molecule concentration (<100 pM) were deposited in a sealed chamber consisting of a polymer gasket sandwiched between two glass coverslips. 10 to 30 minutes measurements were performed at room temperature.
Figure 1.
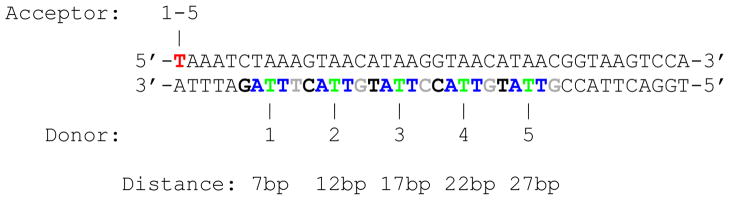
DNA sequence used in this work, with location of the dyes indicated for each sample (A: acceptor, D: donor).
2.2. μs-ALEX setup
A custom-made single-molecule setup was modified to incorporate different types of detectors and lasers in such a way as to make switching between different configurations simple, as schematically indicated in Fig. 2. Briefly, two continuous wave lasers (D-excitation: 532 nm, 100 mW, MLL-III-532–100 mW, Changchun, China; A-excitation: 638 nm, 22 mW, RedFlame, Coherent Inc., Auburn, CA) were alternated using a computer-controlled acousto-optical modulator (AOM, model PCAOM 48058–2.5-.55, Gooch & Housego, Melbourne, FL). A period of 50 μs, equally divided into a 25 μs D-excitation period and a 25 μs A-excitation period was created using a TTL pulse-generating computer board (PCI 6602, National Instruments, Austin, TX) programmed in LabVIEW as previously described27, 29. The two alternated laser beams were coupled into a single-mode fiber and coupled into a water-immersion microscope objective lens (60X, NA 1.2, Olympus, Piscataway, NJ) after expansion to overfill the back aperture. Laser power was measured before the objective lens and was typically 40 μW for A-excitation (peak power: 80 μW) and 100 μW for D-excitation (peak power: 200 μW), in order to achieve a mean S value of ~ 0.5 for doubly-labeled samples27.
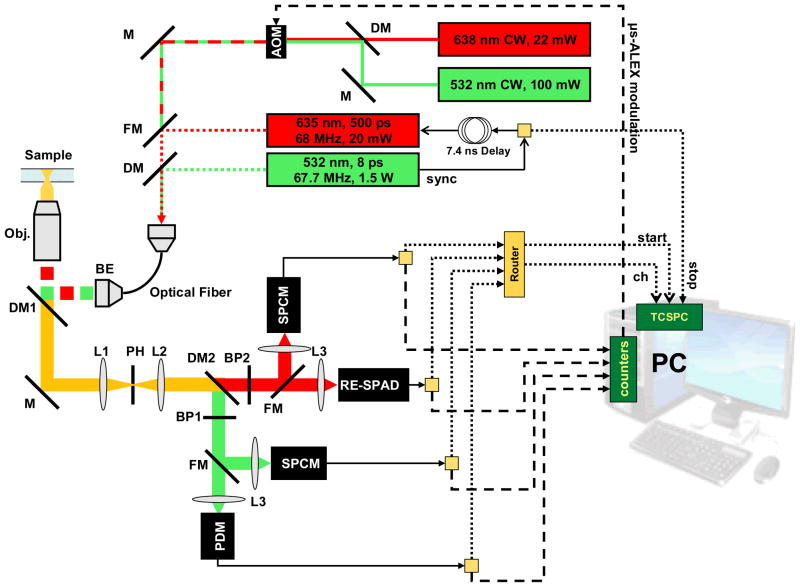
Figure 2.
Setup schematics showing the excitation paths (top right) and the emission path (bottom left) as well as the data acquisition system (bottom right). A detailed description of the setup can be found in the main text. Abbreviations: M = mirror; DM = dichroic mirror; FM = flippable mirror; BP = band-pass filter; L = lens; BE = beam expander; AOM = acousto-optic modulator; PDM = standard custom-technology SPAD; RE-SPAD = red-enhanced custom-technology SPAD; SPCM = Perkin-Elmer SPAD.
In the emission path, two flippable mirrors (FM in Fig. 2) were installed in front of the detectors in order to be able to switch from detection with the thick SPADs (SPCM in Fig. 2) to detection with the custom technology SPADs† (PDM and RE-SPAD in Fig. 2). A non red-enhanced custom technology SPAD (PDM) was used for the donor channel, the red-enhanced SPAD being reserved for the acceptor channel. Because of the smaller detection area of the custom technology SPADs (50 μm diameter), the flippable mirrors were used to redirect light to the larger area SPCM SPADs (~180 μm diameter) for reproducible alignment from one switch to the next. Appropriate bandpass filters (BP) and dichroic mirrors (DM) for each dye were used. A pinhole (PH, diameter: 69 μm) was used after the microscope tube lens (L1), and a pair of demagnifying lenses (L2, L3) was used to relay the light emerging from the pinhole to the detector. The size of the imaged light spot at the detector planes was measured with a CCD camera (CoolSnap cf, Photometrics, Tucson, AZ) and found to have a full-width at half-maximum of ~12 μm, ensuring that both detectors’ sensitive areas were not a limiting factor for collection efficiency.
Measurements series were performed within a single day, to avoid possible alignment drift. Each single sample measurement lasted 10 min, in order to accumulate enough single-molecule bursts. Measurements were repeated using different new batches of samples and found to be reproducible.
2.3. ns-ALEX setup
For ns-ALEX experiments, two pulsed lasers were used. A 532 nm, 67.7 MHz repetition rate, 6.6 ps FWHM pulsed laser (IC-1000–532, High Q Laser Production GmbH, Hohenems, Austria) was used for D-excitation. A 635 nm, <20 mW, triggerable pulsed laser diode (FWHM ~ 350 ps) was used for A-excitation (LDH-P-635, PicoQuant, Berlin, Germany). The sync pulse output of the 532 nm laser was amplified and fed (i) to the pulsed red diode controller (PDL 800-B, PicoQuant) with a 7.4 ns delay and (ii) to the Stop input of a time-correlated single-photon counting (TCSPC) acquisition board (SPC-630, Becker & Hickl GmbH, Berlin, Germany).
Both lasers were coupled to the same single-mode fiber used for μs-ALEX experiments, a single flippable mirror allowing switching from CW to pulsed excitation experiments without needing any realignment of the excitation or detection paths (see Fig. 2).
The TCSPC board receives single-photon TTL pulses output by SPAD detectors via a “router” unit (HRT-82, B&H), which accepts up to 8 different inputs and outputs a single Start pulse (sent to the CFD, or constant fraction discriminator input of the SPC-630 board). The identity of the source detector is transmitted separately by the router to the TCSPC board via a serial cable. In order to take advantage of the fast NIM output of the custom technology SPADs, the following strategy was used for those detectors: the TTL output of each detector was connected to one of the router input, while the NIM output of each detector was connected to one of the output port of a fast signal splitter (ZFCS-2–1, Mini-Circuits, Brooklyn, NY). The splitter input was then connected to the CFD input of the TCSPC board. For the thick SPAD detectors, which only output a TTL signal, the standard CFD output of the router was connected to the TCSPC board CFD input.
The B&H software provided with the SPC-630 board was used for data acquisition (SPCM version 8.72). Data was saved in the .spc format, which contains a macrotime stamp (50 ns resolution) and a nanotime stamp (7.3 ps resolution for the 30 ns TAC window used in these experiments) as well as channel number for each photon.
Data for IRF measurement was acquired by focusing the lasers onto a bare glass coverslip. Ambient light was used to acquire TAC non-linearity calibration data.
2.4. μs-ALEX Data Analysis
Raw photon time stamp data files were analyzed using software written in LabVIEW. Briefly, data analysis consisted in first determining the location of the D-excitation and A-excitation periods in order to be able to identify the different photon streams . This was done by building a histogram of all photon time stamps modulo the 50 μs alternation period and graphically defining the beginning and end of each period. Next, background rates were computed for all four photons streams for later background correction. Bursts were detected using all photons and using a simple probabilistic criterion based on the assumption of Poisson background30. The exact parameters (common to all data sets) used in this procedure do not affect the final FRET measurement results, but of course have a direct influence on burst-size histograms. In particular, we used an identical min burst size parameter Smin = 50.
A background-corrected ALEX two dimensional histogram was constructed for each sample as described in ref. 27. The donor leakage parameter was estimated from a vs burst scatter plot fit for a donor-only population, while proximity ratios (EPR(i)) and uncorrected stoichiometry ratios (S(i)) were computed27 for all samples (i = 1, …, 5). The detection correction factor γ was computed27 from the slope of the best linear fit to the set of data points (EPR(i), S(i)). γcorrected FRET efficiency values E(i) for the FRET populations were then computed after construction of background-, leakage- and γ-corrected ALEX histograms for each sample i.
2.5. ns-ALEX Data Analysis
Raw B&H .spc files were analyzed using the same LabVIEW software as for μs-ALEX. Briefly, the excitation period definition of μs-ALEX analysis was replaced by an emission period definition step based on the nanotime histograms for each spectral channel. The TAC window of 30 ns comprised one full laser repetition period and a second distorted period which was not used for analysis. The first period was graphically split into two adjacent periods: the D-emission period comprised the first half-period (7.4 ns) starting shortly before the beginning of the D-channel decay, whereas the A-emission period comprised the remainder of the 14.8 ns period. Nanotime histograms were corrected for TAC nonlinearity using a calibration file acquired with uncorrelated ambient light.
Background rates calculation for each photon stream, burst detection and ALEX histogram computation were performed exactly as for μs-ALEX data. After a population of interest was selected within the ALEX histogram, the corresponding photon nanotimes were histogrammed to form a population-specific nanotime histogram, which was fitted with the appropriate decay model. Fitting was performed using a simplex downhill minimization algorithm31 using a weighted least square cost function in which the model consisted in a sum of exponential decays with time and intensity offsets, convolved with the measured IRF. In some cases, a component with a fixed time constant corresponding to the D-only labeled molecule was incorporated in the fit, as discussed later in the text. Parameter uncertainties were evaluated using the χ2 curvature matrix32–33.
3. INTENSITY-BASED SINGLE-MOLECULE FRET MEASUREMENTS
μs-ALEX histograms for all 5 samples and both sets of detectors allowed straightforward identification of the doubly-labeled molecule bursts. In practice, the samples turned out to contain very little D-only and A-only labeled molecules. From the selected bursts, a FRET population-specific FRET efficiency histogram was built after photon-stream specific background-correction, donor-leakage correction and γ-correction factor calibration.
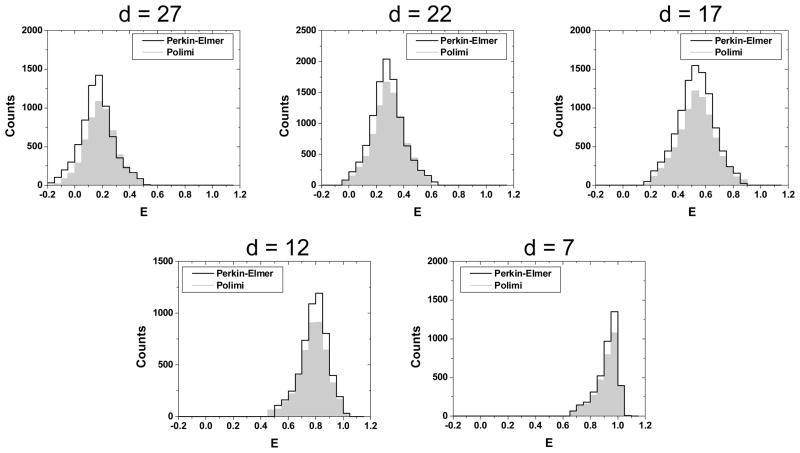
Comparison of histograms obtained for both sets of detectors and all 5 samples is presented in Fig. 3 and results are summarized in Table 1. The mean FRET efficiency values extracted with both sets of detectors were in excellent agreement with one another, as expected after setup calibration. The main difference was a slightly smaller number of bursts in the case of the Polimi detectors, as expected from their lower photon detection efficiency (PDE) in the visible range of the spectrum10. Interestingly, this had little to no effect on the width of the FRET histograms, as reported in Table 1 and in Fig. 3. Since the width of FRET histograms is mainly governed by the effect of shot-noise34–35, this result emphasizes that the small PDE deficit of the custom technology SPAD has no noticeable effect on the quality of the smFRET results.
Figure 3.
Comparison of the FRET histograms obtained with the two different sets of detectors (Polimi: filled gray histogram and Perkin-Elmer: black outline) for all 5 samples (d = 7–27 bp). Both sets of detectors provide identical results, although the Polimi detectors collect slightly less bursts than the Perkin-Elmer ones during the same amount of time, due to their slightly lower PDE.
Table 1.
Summary of μs-ALEX smFRET measurements on the 5 dsDNA samples. For each set of detector and sample, the position E of the peak and standard deviation σ of the best Gaussian fit (histogram width) to the corresponding histogram shown in Fig. 4 is reported as E ± σ.
| Sample | d = 27 bp | d = 22 bp | d = 17 bp | d = 12 bp | d = 7 bp |
|---|---|---|---|---|---|
| E (Perkin-Elmer) | 0.13 ± 0.10 | 0.25 ± 0.10 | 0.51 ± 0.13 | 0.78 ± 0.09 | 0.93 ± 0.05 |
| E (Polimi) | 0.16 ± 0.10 | 0.27 ± 0.11 | 0.51 ± 0.13 | 0.77 ± 0.09 | 0.93 ± 0.06 |
4. LIFETIME-BASED SINGLE-MOLECULE FRET MEASUREMENTS
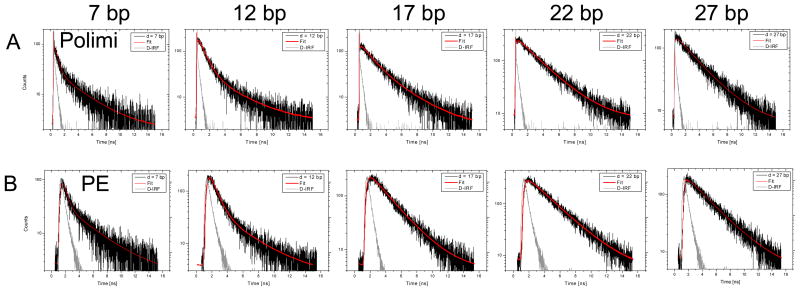
ns-ALEX experiments were performed as described previously28. Due to the high frequency (67.7 MHz) of the D-excitation laser, this technique requires slightly more sophisticated analysis than the similar Pulsed Interleaved Excitation (PIE) technique introduced by Lamb and colleagues36. In particular, the short repetition period (~14.8 ns) of the D-excitation laser is interrupted by the A-excitation laser pulse after only ~7.4 ns, resulting in significant donor decay “wrapping” from one period to the next and overlap of the acceptor decay due to FRET into the acceptor decay due to direct excitation by the A-excitation laser (data not shown). To compound this problem, the pulse red diode laser used in this study had a far from ideal temporal profile. For this reason, we only studied the donor decay of the ALEX histogram-selected FRET population, recorded with the standard custom-technology SPAD, whose IRF FWHM is similar (22 ps) to that of the RE-SPAD (70 ps) and much shorter than that of the Perkin-Elmer SPAD (~500 ps). The corresponding nanotime histograms (or lifetime decays) are shown in Fig. 4.
Figure 4.
Nanotime histograms of the FRET population for the 5 samples (d = 7, 12, 17, 22 and 27 from left to right) as collected with the Polimi detectors (top row) and Perkin-Elmer detectors (bottom row). A single laser period of 14.8 ns is shown (first time axis tick: 0 ns, last tick: 16 ns). The black curves (in units of counts per TAC bin) are the calibrated decays, shown with the corresponding multi-exponential fits in red. The IRF is shown in light gray (arbitrary units). The fitted lifetimes are reported in Table 2.
As briefly mentioned in Section 2, some of the resulting nanotime histograms were best fit using two or three exponential components. When the mean square error (MSE) of a fit with n + 1 components was larger than that of a fit with n components, the former was rejected as unnecessary. In general, we observed that high FRET samples (d = 7 and 12 bp) required one more component than low FRET samples (d = 22 and 27 bp), with the intermediate FRET sample case depending on the set of detectors used (3 components needed with the Polimi detectors but only 2 with the Perkin-Elmer detectors).
The shortest component was easily attributed to Raman or Rayleigh scattering due to our choice of spectral filters. Its amplitude was in general negligible, but its presence was more noticeable when using the Polimi detectors, due to their very narrow IRF. In the absence of any other plausible source, the longest component needed in the case of high FRET samples was fixed to that of the D-only lifetime, measured on a D-only labeled sample with the donor located at the same position as that of the studied doubly-labeled sample. We hypothesized that the combination of high acceptor excitation rate due to FRET followed by direct A-excitation 7.4 ns later might lead to increased acceptor triplet-state shelving (i.e. blinking) and or bleaching, resulting in a molecule with a temporarily dark acceptor, i.e. a D-only molecule. Similar unwanted photophysics effects have been reported in the past37 and will require further studies to be validated.
The fitted lifetimes and the corresponding FRET efficiency values are reported in Table 2. The FRET efficiency was computed using the standard relation:
| (1) |
where τD/A is the fitted lifetime component (reported in Table 2) and τ0 is the measured lifetime of a D-only labeled dsDNA sample with the donor located in the same position as the doubly-labeled sample. Table 2 shows a reasonable agreement between results obtained with both pairs of detectors, with the exception of the d = 17 bp sample, which shows up to 20% discrepancy with the μs-ALEX calculated FRET efficiency, and a significant difference between results obtained with the Polimi and Perkin-Elmer SPADs. Further studies will be necessary to better understand the origin of this discrepancy and difference.
Table 2.
Fitted FRET lifetime component and corresponding FRET efficiency for the 5 dsDNA samples and the two pairs of detectors. The typical uncertainty on fitted lifetimes was 0.01–0.02 ns.
| Sample | d = 27 bp | d = 22 bp | d = 17 bp | d = 12 bp | d = 7 bp |
|---|---|---|---|---|---|
| τ in ns (Perkin-Elmer) | 3.33 | 2.96 | 2.44 | 0.75 | 0.41 |
| E (Perkin-Elmer) | 0.20 | 0.19 | 0.43 | 0.81 | 0.90 |
| τ in ns (Polimi) | 3.31 | 2.92 | 1.59 | 0.82 | 0.47 |
| E (Polimi) | 0.17 | 0.19 | 0.61 | 0.78 | 0.88 |
5. COMPARISON OF THE RE-SPAD DETECTOR WITH OTHER SPADS
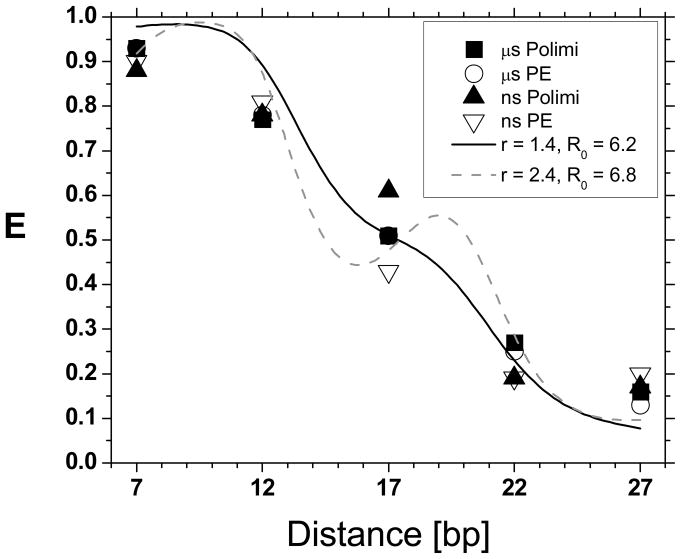
Fig. 5 summarizes the FRET efficiency results obtained in both sets of measurements (μs-ALEX and ns-ALEX measurements using custom-technology and thick SPADs). The μs-ALEX results were reasonably consistent with a simple geometrical model38 in which the dyes attached n-base pairs away from one another were located:
Figure 5.
FRET efficiencies measured in this work using different techniques and detectors. The two sets of μs-ALEX measurements performed with different detectors (black squares: Polimi detectors, open circles: Perkin-Elmer detectors) are in excellent agreement with one another and in reasonable agreement with the values obtained using ns-ALEX (black triangles: Polimi detectors, open triangles: Perkin-Elmer detectors), except for the 17 bp sample, where ns-ALEX FRET efficiencies exhibit a significant dispersion. The plain and dashed curves correspond to the prediction of a simple geometric model for the dsDNA molecule and its attached dyes, discussed in the text, for different parameter values r and R0 (values in nm).
at a radial distance r away from the axis of a straight cylinder representing the dsDNA molecule;
at a distance n×Δ from one another along the axis of the cylinder (Δ = 0.334 nm);
pointed away from the cylinder at an angle n×π/5 from one another;
and using a fixed Förster radius R0 for all dye positions. Fixing r = 1.4 nm, yielded a best fit R0 = 6.2 nm (plain curve), while a two-parameter fit yielded a less plausible distance r = 2.4 nm and R0 = 6.8 nm. These discrepancies are not surprising in light of recent work having elucidated the complex interplay between linker flexibility, dye anisotropy and local DNA deformations in FRET measurements39–40 and go beyond the scope of this simple comparison.
As mentioned, the ns-ALEX measurements closely followed the μs-ALEX results except for the 17 bp distance sample, where a larger discrepancy was observed. Future measurements using two detectors for time-resolved polarization anisotropy measurements will be performed to investigate its origin, and in particular, the potential role of donor anisotropy variation as a function of dye position along the dsDNA strant. Narrow IRF detectors such as the Polimi detectors used in this study will be essential for this study.
6. CONCLUSION AND PERSPECTIVES
We have performed smFRET measurement on a set of 5 dsDNA samples covering the whole range of FRET efficiencies, comparing the results obtained with standard thick-junction SPADs (Perkin-Elmer) and custom-technology SPADs, in particular a new red-enhanced custom-technology SPAD (RE-SPAD) developed by the Politecnico di Milano group.
For intensity-based measurements (μs-ALEX), we have verified that, provided the emission PSF size is matched to the sensitive area of the detectors, the detected count rates scale according to the measured PDE. This requirement is not very demanding, considering the relatively large size of the RE-SPAD (50 μm diameter, compared to ~200 μm diameter for the Perkin-Elmer SPAD). Since the difference in PDE between the Perkin-Elmer and RE-SPAD is of the order of 20% at most in the visible spectrum, there is no difficulty detecting single-molecules, even when most photons are emitted in the red region of the spectrum, as is the case for large FRET samples. The main effect of the difference in PDE is visible in the total number of bursts collected during a given acquisition time, or alternatively, in the maximum burst size. This difference is negligible in practice, and moreover there is no measurable effect on the measured FRET value or the shot-noise broadening of FRET histograms.
For time-resolved measurements, the much narrower IRF of the custom-technology SPADs did not provide any significant advantage in the range of FRET values studied in this work. This is because the effect of the IRF shape is well taken into account by convolution in the fitting process. However, we were able to verify that for very short fluorescence lifetimes such as that of Erythrosin B (95 ps in aqueous buffer), the narrow IRF of custom technology SPADs is essential for accurate extraction of the correct decay constant (data not shown). These characteristics of the RE-SPAD could be critical for time-resolved anisotropy measurements40 or photoinduced electron-transfer measurements41.
Most importantly, the improved spectral sensitivity of RE-SPADs should be compatible with custom-technology SPAD arrays24, 42, which opens up exciting prospects for high-throughput smFRET experiments using multispot excitation22, 25.
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number 5R01 GM095904. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Also referred to as Polimi detectors or Polimi SPADs in the text.
References
- 1.Vacha M, Habuchi S. Conformation and physics of polymer chains: a single-molecule perspective. NPG Asia Mater. 2010;2(4):134–142. [Google Scholar]
- 2.Michalet X, Weiss S, Jäger M. Single-molecule fluorescence studies of protein folding and conformational dynamics. Chem Rev. 2006;106(5):1785–1813. doi: 10.1021/cr0404343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Min W, English BP, Luo GB, Cherayil BJ, Kou SC, Xie XS. Fluctuating enzymes: Lessons from single-molecule studies. Accounts Chem Res. 2005;38(12):923–931. doi: 10.1021/ar040133f. [DOI] [PubMed] [Google Scholar]
- 4.DeRocco VC, Anderson T, Piehler J, Erie DA, Weninger K. Four-color single-molecule fluorescence with noncovalent dye labeling to monitor dynamic multimolecular complexes. Biotechniques. 2010;49(5):807–816. doi: 10.2144/000113551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heilemann M, Kasper R, Tinnefeld P, Sauer M. Dissecting and Reducing the Heterogeneity of Excited-State Energy Transport in DNA-Based Photonic Wires. J Am Chem Soc. 2006;128(51):16864–16875. doi: 10.1021/ja065585x. [DOI] [PubMed] [Google Scholar]
- 6.Sisamakis E, Valeri A, Kalinin S, Rothwell PJ, Seidel CAM. Accurate single-molecule FRET studies using multiparameter fluorescence detection. Methods Enzymol. 2010;475:455–514. doi: 10.1016/S0076-6879(10)75018-7. [DOI] [PubMed] [Google Scholar]
- 7.Lipman EA, Schuler B, Bakajin O, Eaton WA. Single-Molecule Measurement of Protein Folding Kinetics. Science. 2003;301(5637):1233–1235. doi: 10.1126/science.1085399. [DOI] [PubMed] [Google Scholar]
- 8.Kim S, Streets AM, Lin RR, Quake SR, Weiss S, Majumdar DS. High-throughput single-molecule optofluidic analysis. Nature Meth. 2011;8:242–245. doi: 10.1038/nmeth.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michalet X, Siegmund OHW, Vallerga JV, Jelinsky P, Millaud JE, Weiss S. Detectors for single-molecule fluorescence imaging and spectroscopy. Eur Biophys J. 2007;54(2–3):239–282. doi: 10.1080/09500340600769067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michalet X, Colyer RA, Scalia G, Ingargiola A, Lin R, Millaud JE, Weiss S, Siegmund OHW, Tremsin AS, Vallerga JV, Cheng A, Levi M, Aharoni D, Arisaka K, Villa F, Guerrieri F, Panzeri F, Rech I, Gulinatti A, Zappa F, Ghioni M, Cova S. Development of new photon-counting detectors for single-molecule fluorescence microscopy. Philos Trans R Soc B-Biol Sci. 2013;368(1611):20120035. doi: 10.1098/rstb.2012.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghioni M, Gulinatti A, Rech I, Zappa F, Cova S. Progress in silicon single-photon avalanche diodes. IEEE J Sel Top Quantum Electron. 2007;13(4):852–862. [Google Scholar]
- 12.Spinelli A, Davis LM, Dautet H. Actively quenched single-photon avalanche diode for high repetition rate time-gated photon counting. Rev Sci Instrum. 1996;67(1):55–61. [Google Scholar]
- 13.Li LQ, Davis LM. Single-Photon Avalanche-Diode for Single-Molecule Detection. Rev Sci Instrum. 1993;64(6):1524–1529. [Google Scholar]
- 14.Felekyan S, Kuhnemuth R, Kudryavtsev V, Sandhagen C, Becker W, Seidel CAM. Full correlation from picoseconds to seconds by time-resolved and time-correlated single photon detection. Rev Sci Instrum. 2005;76(8):083104. [Google Scholar]
- 15.Rech I, Labanca I, Ghioni M, Cova S. Modified single photon counting modules for optimal timing performance. Rev Sci Instrum. 2006;77(3):033104. [Google Scholar]
- 16.Kell G, Bülter A, Wahl M, Erdmann M. τ-SPAD: a new red sensitive single-photon counting module. Proc SPIE. 2011;8033:803303. [Google Scholar]
- 17.Andrecka J, Lewis R, Brückner F, Lehmann E, Cramer P, Michaelis J. Single-molecule tracking of mRNA exiting from RNA polymerase II. Proc Natl Acad Sci USA. 2008;105:135–40. doi: 10.1073/pnas.0703815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milles S, Lemke EdwardA. Single Molecule Study of the Intrinsically Disordered FG-Repeat Nucleoporin 153. Biophys J. 2011;101(7):1710–1719. doi: 10.1016/j.bpj.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulinatti A, Rech I, Maccagnani P, Ghioni M, Cova S. Improving the performance of Silicon Single Photon Avalanche Diodes. Proc SPIE. 2011;8033:803302. [Google Scholar]
- 20.Colyer RA, Scalia G, Kim T, Rech I, Resnati D, Marangoni S, Ghioni M, Cova S, Weiss S, Michalet X. High-throughput multispot single-molecule spectroscopy. Proc SPIE. 2010;7571:75710G. doi: 10.1117/12.841398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colyer RA, Scalia G, Rech I, Gulinatti A, Ghioni M, Cova S, Weiss S, Michalet X. High-throughput FCS using an LCOS spatial light modulator and an 8×1 SPAD array. Biomed Opt Express. 2010;1(5):1408–1431. doi: 10.1364/BOE.1.001408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingargiola A, Colyer RA, Kim D, Panzeri F, Lin R, Gulinatti A, Rech I, Ghioni M, Weiss S, Michalet X. Parallel multispot smFRET analysis using an 8-pixel SPAD array. Proc SPIE. 2012;8228:82280B. doi: 10.1117/12.909470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michalet X, Colyer RA, Scalia G, Kim T, Levi M, Aharoni D, Cheng A, Guerrieri F, Arisaka K, Millaud J, Rech I, Resnati D, Marangoni S, Gulinatti A, Ghioni M, Tisa S, Zappa F, Cova S, Weiss S. High-throughput single-molecule fluorescence spectroscopy using parallel detection. Proc SPIE. 2010;7608:76082D. doi: 10.1117/12.846784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gulinatti A, Rech I, Cammi C, Labanca I, Maccagnani P, Ghioni M. Silicon single-photon avalanche diodes for high performance parallel photon timing. Proc SPIE. 2012;8375:8375N. [Google Scholar]
- 25.Ingargiola A, Panzeri F, Sarkosh N, Gulinatti A, Rech I, Ghioni M, Weiss S, Michalet X. 8-spot smFRET analysis using two 8-pixel SPAD arrays. Proc SPIE. 2013;8590 doi: 10.1117/12.2003704. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapanidis AN, Lee NK, Laurence TA, Doose S, Margeat E, Weiss S. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci U S A. 2004;101(24):8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee NK, Kapanidis AN, Wang Y, Michalet X, Mukhopadhyay J, Ebright RH, Weiss S. Accurate FRET Measurements within Single Diffusing Biomolecules Using Alternating-Laser Excitation. Biophys J. 2005;88:2939–2953. doi: 10.1529/biophysj.104.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurence TA, Kong XX, Jager M, Weiss S. Probing structural heterogeneities and fluctuations of nucleic acids and denatured proteins. Proc Natl Acad Sci U S A. 2005;102(48):17348–17353. doi: 10.1073/pnas.0508584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapanidis AN, Laurence TA, Lee NK, Margeat E, Kong XX, Weiss S. Alternating-laser excitation of single molecules. Accounts Chem Res. 2005;38(7):523–533. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- 30.Fries JR, Brand L, Eggeling C, Kollner M, Seidel CAM. Quantitative identification of different single molecules by selective time-resolved confocal fluorescence spectroscopy. J Phys Chem A. 1998;102(33):6601–6613. [Google Scholar]
- 31.Nelder JA, Mead R. A simplex method for function minimization. Comput J. 1965;7(4):308–313. [Google Scholar]
- 32.Brumby S. Exchange of comments on the simplex algorithm culminating in quadratic convergence and error estimation. Anal Chem. 1989;61(15):1783–1786. [Google Scholar]
- 33.Phillips GR, Eyring EM. Error estimation using the sequential simplex method in nonlinear least squares data analysis. Anal Chem. 1988;60(8):738–741. [Google Scholar]
- 34.Nir E, Michalet X, Hamadani KM, Laurence TA, Neuhauser D, Kovchegov Y, Weiss S. Shot-noise limited single-molecule FRET histograms: Comparison between theory and experiments. J Phys Chem B. 2006;110(44):22103–22124. doi: 10.1021/jp063483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antonik M, Felekyan S, Gaiduk A, Seidel CAM. Separating structural heterogeneities from stochastic variations in fluorescence resonance energy transfer distributions via photon distribution analysis. J Phys Chem B. 2006;110(13):6970–6978. doi: 10.1021/jp057257+. [DOI] [PubMed] [Google Scholar]
- 36.Muller BK, Zaychikov E, Brauchle C, Lamb DC. Pulsed interleaved excitation. Biophys J. 2005;89(5):3508–3522. doi: 10.1529/biophysj.105.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong XX, Nir E, Hamadani K, Weiss S. Photobleaching pathways in single-molecule FRET experiments. J Am Chem Soc. 2007;129(15):4643–4654. doi: 10.1021/ja068002s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 39.Sindbert S, Kalinin S, Hien N, Kienzler A, Clima L, Bannwarth W, Appel B, Muller S, Seidel CAM. Accurate Distance Determination of Nucleic Acids via Forster Resonance Energy Transfer: Implications of Dye Linker Length and Rigidity. J Am Chem Soc. 2011;133(8):2463–2480. doi: 10.1021/ja105725e. [DOI] [PubMed] [Google Scholar]
- 40.Kudryavtsev V, Sikor M, Kalinin S, Mokranjac D, Seidel CAM, Lamb DC. Combining MFD and PIE for Accurate Single-Pair Forster Resonance Energy Transfer Measurements. Chem Phys Chem. 2012;13(4):1060–1078. doi: 10.1002/cphc.201100822. [DOI] [PubMed] [Google Scholar]
- 41.Doose S, Neuweiler H, Sauer M. Fluorescence Quenching by Photoinduced Electron Transfer: A Reporter for Conformational Dynamics of Macromolecules. Chem Phys Chem. 2009;10(9–10):1389–1398. doi: 10.1002/cphc.200900238. [DOI] [PubMed] [Google Scholar]
- 42.Gulinatti A, Rech I, Maccagnani P, Ghioni M. A 48-pixel array of single photon avalanche diodes for multispot single molecule analysis. Proc SPIE. 2013;8631:86311D. doi: 10.1117/12.2003984. [DOI] [PMC free article] [PubMed] [Google Scholar]