Abstract
During inflammation, local tissue responses are augmented by complement and acute phase proteins that exude into the tissue because of increased blood vessel permeability mediated by bradykinin, which is proteolytically released from kininogen. Moreover, proteases govern inflammatory responses by processing extracellular matrix proteins and soluble bioactive mediators; however, the consequences of their complex interplay in inflamed mammalian tissues are largely unknown. We quantified changes in the proteome and the nature of protein N-termini (the N-terminome) and the altered abundance of murine proteases and inhibitors during skin inflammation. Through analysis of the N-terminome by iTRAQ-terminal amine isotopic labeling of substrates (TAILS), we identified cotranslational and posttranslational αN-acetylation motifs, quantitative increases in protein abundance, and qualitative changes in the proteolytic signature during inflammation. Of the proteins identified in normal skin, 50% were cleaved, which increased to 60% during inflammation caused by phorbol esters, including chemokines and complement in which we identified previously uncharacterized cleavage sites. In mice deficient in matrix metalloproteinase 2 (MMP2), exudation of serum proteins was diminished compared to that in wild-type mice, and their proteolytic networks differed. We found that the complement 1 (C1) inhibitor was a key regulator linking these inflammatory responses. Cleavage and inactivation of the C1 inhibitor by MMP2 increased complement activation and bradykinin generation by plasma kallikrein in wild-type mice, leading to increased vessel permeability during inflammation. Thus, our degradomics analysis dissected proteolysis in skin inflammation and demonstrated perturbance of the proteolytic signaling network and its functional consequences arising from lack of a single protease.
Introduction
Inflammation is a complex, well-orchestrated response that ultimately restores tissue function and homeostasis. Proteolysis is an important aspect of the inflammatory response. Proteases remove damaged tissue and extracellular matrix proteins, which releases growth factors and bioactive fragments that enable tissue remodeling and healing (1). Proteases also irreversibly alter the function of many bioactive mediators during inflammation by limited, specific, and efficient processing. The exact nature of the N-terminus of a protein and its modification by proteolysis and αN-acetylation often alters protein bioactivity and stability (2, 3), such as within networks of interdependent proteases and inhibitors during complement activation and blood coagulation (4, 5). Thus, what has been termed the protease web (6), which is formed by the interconnected activities of the 578 proteases and inhibitors in humans and 664 in mice (7), sculpts the inflammatory proteome and regulates the activity of inflammatory mediators. Vasoactive peptides are crucial for increasing vascular permeability to enable the exudation of serum proteins. The vasodilator bradykinin is excised from kininogen by plasma kallikrein, which is itself activated by factor XIIa (8) and inhibited by serpin G1, the complement 1 (C1) inhibitor (9). N-terminal processing of chemokines by diprolylpeptidase 4 and matrix metalloproteinases (MMPs) regulates chemotaxis, increasing, and then attenuating, the extent of leukocyte recruitment during inflammation (10). A prime example of this is mitigation of the attraction of macrophages to the site of injury by MMP2 (11, 12), which shows increased abundance in response to transforming growth factor–β1 during wound healing (13). However, the global changes in protease and inhibitor abundances during inflammation and the extent of their proteolytic modification of the inflammatory proteome are unknown.
The ultimate goal of large-scale proteomics experiments is to generate hypotheses as a basis for more detailed studies that aim to confirm in vivo mechanisms. However, for this strategy to be effective, we require functional information, which is missing without quantification, as well as the (patho)physiologically relevant context of an in vivo model. Protease substrates in cells can be identified by degradomics (14) with isotopic labels (15-17). Protein N-termini, including protein neo-N-termini that are formed by proteolytic cleavage, can now be enriched by several approaches to reveal both substrates and their cleavage sites in the same experiment (2, 18, 19). One such technique, terminal amine isotopic labeling of substrates (TAILS) (20, 21), enables quantitative analysis of both proteomes and N-terminomes from in vitro and cellular samples, and can be extended to multiplex assays through the use of isobaric tags for relative and absolute quantitation (iTRAQ), a variant of the technique referred to as iTRAQ-TAILS (22, 23). However, analysis of tissues, in which cells are in their natural microenvironments and where they respond to stresses with all of the influencing factors present at appropriate concentrations, has yet to be reported by any terminomics approach.
Although a massive body of in vitro studies implicates individual proteins, proteases, and pathways as being active during inflammation, studies globally analyzing inflamed tissue protein and protease networks in vivo have so far proven intractable. The functional status and interconnectivity of the molecular mediators of inflammation is hard to ascertain in vivo because many are present at nanomolar concentrations. Quantitative proteomic analysis of tissues is further complicated by the exudation of serum proteins, including acute response proteins released from the liver during inflammation, and inflammatory cells from the blood stream into the tissue. Protein degradation and proteolytic processing also limit the functional insight that can be derived by conventional proteomics.
Here, we mapped the proteolytic fingerprint of a complex tissue that reflects net proteolytic activity in murine skin. With DNA microarrays, we described the repertoire of genes encoding proteases and inhibitors that were responsible for a diverse and large number of stable protein cleavage products, some of which were posttranslationally acetylated after cleavage, which we identified in skin with iTRAQ-TAILS. We dissected this repertoire further in an unbiased global approach aiming to identify how MMP2, which is immunomodulatory (11) and is associated with tissue repair (13) and angiogenesis (24), alters the proteolytic signature of skin inflammation at the levels of intact and cleaved protein abundance and activity. We report that the proteolytic fingerprint changes in the inflammatory response, and we identified previously undescribed cleavage events in the complement cascade in vivo. Further, we suggest that by cleavage inactivation of the protease C1 inhibitor, MMP2 enhances complement activation and controls vascular permeability and acute phase protein exudation by increasing the extent of bradykinin release.
Results
Experimental design
We used the well-established 12-O-tetradecanoylphorbol-13-acetate (TPA) model of murine skin inflammation (25), and administered a single dose of TPA to the shaved back skin of animals (Fig. 1A). We collected skin samples 48hours later, and analyzed their transcriptome and proteome by CLIP-CHIPTM DNA microarray (26) and 4plex-iTRAQ-TAILS (22), respectively. At the 48-hour time-point, the accumulation of neutrophils is still high, and mast cells are depleted, whereas macrophages start entering the site of inflammation (27), which enabled us to monitor several overlapping acute inflammatory responses. The epidermis already showed marked hyperproliferation (28) (Fig. 1B), thus revealing TPA-induced changes to epidermal differentiation that are also associated with proteolysis (29).
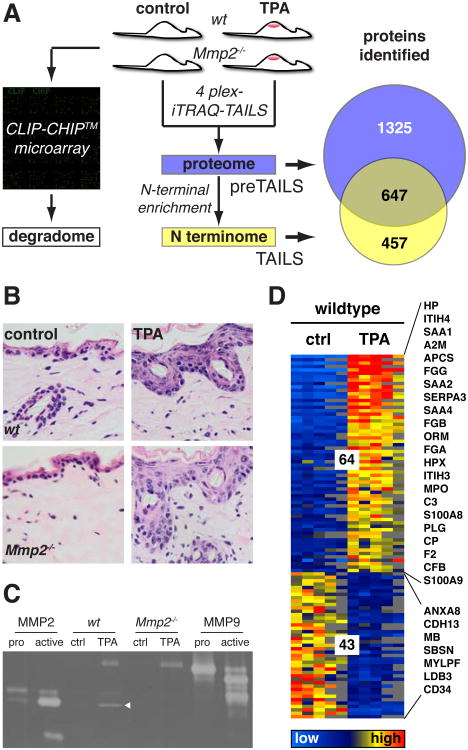
Fig. 1.
Proteomics and degradomics analysis of skin and skin inflammation. (A) The experimental strategy used to determine the transcriptome of the expressed proteases and inhibitors (also known as the protease degradome), the proteome, and the N-terminome in normal and inflamed skin of wild-type and Mmp2-/- mice. Analyzed were two animals per treatment type and genotype (8 mice in total) in five experiments (fig. S1). The total number of proteins identified by both iTRAQ-TAILS and iTRAQ-preTAILS is their union (2429), that is, 1325 proteins were identified only by preTAILS, 457 only by TAILS, and 647 by both iTRAQ-preTAILS and iTRAQ-TAILS. (B) Hematoxylin and eosin staining of back skin sections from wild-type and Mmp2-/- mice 48 hours after treatment with TPA or vehicle control. Multiple layers demonstrate hyperproliferation of epidermal keratinocytes and a higher cell density in the dermis as a result of the infiltration of immune cells, but no significant differences between wild-type and Mmp2-/- mice occurred. Back skins from all 8 animals (two per treatment type and genotype) were analyzed and representative images for each treatment type and genotype are shown. Scale bar: 20 μm. (C) Increased abundance of active MMP2 in inflamed skin. Gelatin zymography shows the increased abundance of active MMP2 (white triangle) in samples from TPA-treated wild-type compared to that in TPA-treated Mmp2-/- skin. ProMMP9 was present in the inflamed skin of mice of both genotypes. Recombinant pro- and active forms of MMP2 and MMP9 proteins were loaded as standards. (D) Differential abundance of proteins in response to TPA in the skin from wild-type mice. 976 proteins were quantified in at least three of five experiments (fig. S2A) and were analyzed for statistically significant changes in abundance between vehicle and TPA-treated skin from wild-type mice. The color code indicates the extent of change; thus, 64 proteins were increased ≥2- fold in abundance and 43 were reduced ≥ 2-fold in abundance (P ≤ 0.05, Limma moderated t-statistic, Benjamini-Hochberg multiple testing correction). Heatmap columns represent mice from each of five experiments. For details and a complete list of differentially abundant proteins see table S2.
We enriched for extracellular proteins to increase the sensitivity of detection of low-abundance secreted bioactive mediators, such as cytokines and chemokines, some of which are prone to functional modification by extracellular proteases (10). We found that MMP2 was increased in abundance and present in its activated form in inflamed skin from TPA-treated wild-type mice compared to that in vehicle-treated animals (Fig. 1C); thus, we studied its effects by including Mmp2-/- and wild-type mice in the same multiplex proteomic experiments. We analyzed 8 animals (2 for each condition) in 5 independent 4plex-iTRAQ-TAILS experiments, in which either individual mice or iTRAQ labels were swapped (fig. S1). iTRAQ-TAILS enriches the naturally blocked and α-amine iTRAQ-labeled original protein N-termini and protease-generated neo-N-termini, while excluding internal trypsin-generated peptides, thus increasing the dynamic range of the abundance of proteins identified (20, 21). We also identified and quantified internal tryptic peptides by analyzing samples before N-terminal peptide enrichment, similar to a typical shotgun approach, which were designated as iTRAQ-preTAILS peptides. Large numbers of tryptic peptides harbored an iTRAQ-labeled lysine, which trypsin skips, enabling ∼85% of the internal tryptic peptides and their corresponding proteins to be quantified.
Global skin inflammatory responses
In the iTRAQ-preTAILS analysis of inflamed skin, we identified 1972 proteins (Fig. 1A and fig. S2A), of which 1147 were found in at least three (of five) replicates as follows: 1183 proteins (ProteinProphet probability corresponding to an error rate of <0.8%), 1048 proteins (error rate <0.9%), 1388 proteins (error rate <0.7%), 1291 proteins (error rate <0.7%), and 1132 proteins (error rate <0.8%) for experiments 1 to 5, respectively (fig. S2A and table S1). Of these proteins, 647 were also identified by their N-terminus in the subsequent iTRAQ-TAILS analysis (Fig. 1A). Gene ontology (GO) analysis revealed the specific enrichment of inflammatory enzymes, secreted proteins, and proteins reflecting a high metabolic activity in the inflamed tissue (fig. S2B). Proteins involved in “response to stimulus” by GO analysis confirmed an acute inflammatory molecular response, including the influx of complement proteins with other acute phase proteins from plasma, as well as the enrichment of proteases and inhibitors.
Of the 1147 proteins identified in at least 3 of 5 iTRAQ-preTAILS analyses, 976 (85%) could be quantified, 64 of which were significantly (Benjamini-Hochberg corrected P <0.05) and greater than two-fold increased in abundance in inflamed skin compared to that in normal skin (Fig. 1D and table S2). Among those proteins whose amounts increased in the exudate were acute phase proteins including haptoglobin (HP), ceruloplasmin (CP), serum amyloid A (SAA), α2-macroglobulin (A2M), plasminogen (PLG), and complement and coagulation proteins. In addition, we detected increased amounts of the pro-inflammatory S100A8/A9 complex components in TPA-treated skin compared to those in untreated skin, as well as the increased abundance of myeloperoxidase (MPO), a marker of neutrophil influx. On the other hand, TPA led to the decreased abundance (≥ two-fold, Benjamini-Hochberg corrected P <0.05) of 43 proteins, including annexin A8 (ANXA8), cadherin-13 (CDH13), suprabasin (SBSN), and filaggrin 2 (FLG2) (table S2), indicating a hyperproliferative response of the epidermis (30, 32).
Protein N-terminal annotation
We identified with high confidence 1813 N-terminal peptides that could be assigned to 1104 proteins (Fig. 2A and tables S3 to S8). Of these, 647 were also identified in the iTRAQ-preTAILS sample (Fig. 1A), thereby facilitating protein isoform assignment (23). By omitting detergent, the skin sample preparation was designed to enrich for extracellular proteins, and by using iTRAQ-TAILS, we enriched for N-terminal peptides only. Thus, both high- and low-abundance proteins were identified from one peptide, or in the case of proteolytic fragmentation, a few peptides; thus, the high-abundance proteins did not excessively mask the identification of low-abundance proteins. Indeed, only 11% of all N-termini were from the 10 most abundant serum proteins or structural components, such as actin, collagen, myosin, and keratin (Fig. 2A). This broad dynamic range of iTRAQ-TAILS led to the uncovering of many very low abundance proteins that were identified by their protein N-termini only (Fig. 2A) and not their tryptic peptides in the iTRAQ-preTAILS shotgun analyses. These included chemokines and growth factors, such as small inducible cytokine A6 (CCL6), macrophage inflammatory protein 2 (CXCL2), small inducible cytokine B5 (CXCL5), platelet basic protein (CXCL7), stromal cell-derived factor 2 (SDF2), hepatocyte growth factor–like protein (MSP), as well as their inhibitory proteins, including insulin-like growth factor binding protein 3 (IGFBP3) and IGFBP4, and latent TGF-β–binding protein 4 (LTBP4).
Fig. 2.
Characterization of the N-terminome in inflamed skin. (A) Union of N-terminal peptides identified in all five experiments (fig. S1) and their assigned proteins. Examples of low-abundance proteins that were not identified by shotgun-like iTRAQ-preTAILS analysis but were identified after iTRAQ-TAILS are shown by the left side arrow. (B) Frequency distribution of N-termini based on annotation in the UniProtKB/Swiss-Prot database. Natural N-termini include proteins starting with or without an initiator methionine (Met), a signal peptide (Sig), a propeptide (Pro), or a mitochondrial transit peptide (Trans). Data for normal skin was from 298 N-termini identified by 2plex CLIP-TRAQ-TAILS analysis of untreated skin samples from wild-type and Mmp2-/- mice. (C) Original mature N-terminus of MPO as identified by iTRAQ-TAILS. MPO was identified in the TPA-treated skin of wild-type and Mmp2-/- mice by an N-terminal peptide (underlined) of the processed precursor one amino acid residue upstream of a “potential” start site predicted in UniProtKB/Swiss-Prot. Inset shows equivalent high-intensity iTRAQ reporter ions only in the 114 (WT-TPA) and 117 (KO-TPA) channels. (D) Yellow inset and upper yellow graph: Acetylation status and amino acid frequency distribution for methionine removal and αN-acetylation for N-termini starting at positions 1 and 2; after initiator methionine (1M) removal with or without acetylation (Ac) [1M.(Ac)P1’-X(n)] (367); all acetylated after removal of initiator methionine (1M.AcP1’-X(n)) (241); and following an acetylated intact initiator methionine (1AcM.P2’-X(n)) (69). Green inset and lower graph: Positional distribution, acetylation status, and amino acid frequency distribution for potential methionine removal and αN-acetylation for N-termini starting at positions ≥3. Forty N-terminal peptides started after a Met (XM.(Ac)X). Seven of these were acetylated (XM.Ac.X), and together with 8 acetylated peptides starting with a Met (X.AcMX) are candidates for cotranslational acetylated alternative start sites. Nineteen more were internal and acetylated (X.AcXX), representing posttranslational αN-acetylation. Asterisks show distribution patterns for 39 N-termini that are similar to patterns from known translation start sites and thus indicate that these are alternative start sites. Inset shows the different amino acid distributions for posttranslational αN-acetylation of internal cleaved peptides vs. cotranslational patterns. (E) Quantitative categorization of N-termini in normal and inflamed wild-type skin. The distribution of quantitative ratios for natural N-termini in TPA-treated and control skin was used to define cutoffs for statistically significant changes in the abundances of N-terminal peptides in inflamed vs. normal skin. Roman numerals I to V indicate quantitative categories for N termini based on log2 wt-TPA/wt-ctrl)-ratios. I: present only in TPA-treated skin; II: significantly more abundant in TPA-treated skin compared to normal skin but present in both conditions; III: not significantly changed in abundance by TPA; IV: significantly less abundant in TPA-treated skin compared to normal skin but present in both conditions; V: absent from TPA treated skin.
Extensive numbers of stable cleaved proteins in normal and inflamed skin
Of the 1813 N-termini identified by iTRAQ-TAILS in normal and inflamed skin, 1292 had reviewed annotation data in the UniProtKB/Swiss-Prot database, thus enabling us to assign their position in the mature protein (Fig. 2B). Of these, 39% were from an original mature protein N-terminus (labeled Natural). Notably, 61% of all identified N-terminal peptides (labeled Internal) were derived either from proteolytic cleavage or from alternative translation initiation at non-AUG codons. However, because of the isotopic contributions across iTRAQ reporter channels, even after isotopic normalization, the absolute determination of expressed proteomes for individual discrete conditions in iTRAQ experiments was difficult. Therefore, we analyzed non-treated, non-shaved skin samples from wild-type and MMP2-deficient mice in a duplex CLIP-TRAQ-TAILS experiment with in-house CLIP-TRAQ isobaric labels (23). Again, in untreated skin, almost half of all N-terminal peptides were internal (44%) indicative of stable cleavage products (Fig. 2B and table S9 and table S10). Hence, this revealed a high level of proteolysis in normal skin that is increased upon inflammation. Moreover, iTRAQ-TAILS enabled annotation of important protein maturation events that are predicted but lack supporting experimental data. As an example, MPO was identified by a peptide starting with glycine (Fig. 2C), but that was one amino acid upstream of the proposed cleavage site that removes the propeptide during MPO maturation (33).
Methionine aminopeptidase and αN-acetylation specificity in vivo
To comprehensively describe the proteolytic fingerprint of the skin, we first studied N-terminal acetylation. The distributions of unblocked and acetylated N-terminal peptides followed previously described patterns (20, 22, 23, 34, 35) with 50% of all natural N-termini being acetylated (Fig. 2B). This result is statistically significantly below the 90% of intracellular proteins that undergo acetylation, and is consistent with our extraction procedure, which was designed to enrich for extracellular proteins. We also found acetylation of proteins (from 620 acetylated N-termini) upon removal of the methionine and signal or propeptides, prompting a detailed analysis of these cotranslational and posttranslational acetylation events, respectively.
We first determined the specificity of methionine aminopeptidase activity in vivo by calculating the distribution of amino acids in the P1’ position for 367 N-terminal peptides that started at position 2 after removal of the initiator methionine in the mature protein (Fig. 2D). We found a high prevalence of alanine, serine, proline, valine and glycine in P1’ in vivo as found previously in eukaryotic in vitro or cell-based systems (20, 22, 36). Of the 367 N-terminal peptides, 241 were cotranslationally acetylated, whereby the preferred acetylated amino acids were alanine, serine, and threonine (n = 229) (Fig. 2D). Nonetheless, 10, 9, and 40% of the terminal peptides commencing with alanine, serine, and threonine, respectively, were not acetylated (fig. S3A). In these cases, but particularly for alanine, the N-terminal residues were followed by a proline, which prevents alanine acetylation (fig. S3A, inset) (37). In 69 proteins, the intact initiator methionine carried an acetyl modification when the following amino acid (P2’) was glutamate, aspartate, phenylalanine, asparagine, leucine, lysine, or glutamine, which is in agreement with previous in vitro studies (35). Here, 1Met residues followed by glutamate, aspartate, or phenylalanine were almost exclusively acetylated, which suggested their crucial contribution to the specificity of the responsible N-terminal acetyltransferase (fig. S3B).
Alternate start site identification, posttranslational acetylation, and N-terminal cyclization
To seek alternative translation start sites in the 1344 peptides commencing at a position ≥3 (where the start is 1Met), we calculated the amino acid frequency distributions of 40 peptides starting after an “internal” methionine, of which 7 were acetylated, and of an additional 8 protein chains starting with an acetylated methionine (Fig. 2D). Indeed, the methionine removal and acetylation sequence dependencies of 39 of these 48 N-terminal peptides were similar to those for known original mature protein N-termini—suggesting that these are alternative start sites (table S11). To confirm this, we could assign 10 of these 39 N-terminal peptides to the N-termini of protein chains predicted from alternative splicing transcripts in the ECGene Alternative Splicing Database (38). Notably, we also found 19 acetylated internal peptides with no methionine as a preceding or starting amino acid (table S12), which is suggestive of posttranslational N-terminal acetyltransferase activity and not cotranslational modification (Fig. 2D inset). Indeed, their sequence dependencies differed from that for cotranslational acetylation, strengthening the concept of an alternative N-terminal acetyltransferase enzyme complex for this type of protein N modification (39). Furthermore, these peptides included all known posttranslational N-acetylation events at 3Glu or 3Asp for cardiac, skeletal, and smooth muscle α-actin that are formed after removal of a posttranslationally acetylated 2Cys (also identified) by acetyl-aminopeptidase after removal of the initiator methionine (40) (fig. S4).
Completing the in vivo analysis of N-terminal actin processing, we also detected N-terminal peptides corresponding to posttranslational acetylation of β and γ actin at 2Glu and 2Asp (fig. S4). Furthermore, we identified 23 proteins that occurred in both the free and the acetylated N-termini forms (table S13), including examples of all of the acetylation types described earlier, thereby supporting incomplete fidelity for acetylation (41, 42) shown here for the first time in a mammalian tissue.
Finally, we analyzed the sequence specificity of N-terminal pyroQ formation (fig. S5) derived from 181 peptides (table S14). Overall, our in vivo tissue analyses corroborate previous in vitro studies (20, 22, 23, 34, 35) showing that cotranslational N-terminal acetylation of proteins at, or immediately past, the initiator methionine is sequence-dependent, and that this sequence differs from that of posttranslational acetylation, thereby indicating the activity of different N-terminal acetyltransferases for these distinct modifications. These sequence motifs agree well with predictions from TopFIND, an integrated knowledge base for protein termini (39, 43), and thus directly corroborate these findings in a mammalian tissue.
Changes to the N-terminome in inflammation
To determine quantitative changes in the skin N-terminome upon treatment with TPA, we ranked 1690 quantifiable N-terminal peptides by their log2(wt-TPA/wt-ctrl) ratios and calculated a cutoff for TPA-dependent regulation based on experimental variation, which was derived from the ratio distribution of natural N-termini (Fig. 2E) (44). Among 127 high-ratio (WT-TPA/WT-ctrl) N-terminal peptides from 57 proteins (TPA only), 18 were natural protein N-termini, indicating new local synthesis or import to the tissue from the inflammatory exudate. In support of this, many were acute phase proteins from the serum (for example, A2M, HP, SAA1, SAA2, and SAA4), were produced in epidermal cells in response to TPA (for example MMP3, fig. S6), or were released from resident or infiltrating inflammatory cells [such as, IL-1β receptor-like 1 (IL1RL1), CXCL2, CXCL5/LIX, CXCL7, and MPO]. Several proteins characteristic of the inflammatory response were also cleaved in TPA-treated skin, including the pro-inflammatory proteins S100A8 (Fig. 3A) and S100A9 (Fig. 3B and fig. S7).
Fig. 3.
Examples of proteolytic processing events identified during inflammation and epidermal differentiation. (A and B) Proteolytic processing of S100A8 and S100A9 during inflammation generates neo-N-termini with reporter ions in the wild-type (114) and knockout (117) channels with equal ratios vs. normal skin. This indicates the higher abundance and increased cleavage of both proteins during inflammation in mice of both genotypes. The acetylated original mature N-terminus of S100A9 was also detected (fig. S7). Western blotting analysis was used for confirmation. (C) Known cleavages related to the inflammatory stimulus. (D) Proteolytic turnover during epidermal differentiation. The peptide in parentheses was identified but with a probability of correct identification below the cutoff for high-confidence peptide identifications. Domains in LEKTI are numbered according to sequence homology with the human protein.
Examples of different biological processes that were identified by iTRAQ-TAILS in vivo included release of bradykinin from kininogen (Fig. 3C), and removal of an N-terminal dipeptide in CXCL5 at 1AP↓ SS by dipeptidyl peptidase 4 (DPPIV) (Fig. 3C), which inactivates the chemokine (20). Proteolytic maturation by profilaggrin endoproteinase 1 (45), N-terminal cyclization of filaggrin, and the breakdown of corneodesmosin, all of which are crucial steps in epidermal differentiation and the desquamation of corneocytes from the epidermis, respectively (31, 46), were also identified (Fig. 3D and table S15). For corneodesmosin, a major component of corneodesmosomes, N-terminal aminopeptidase activity was evident at the N-terminus. A neo-N-terminus at position 378, a likely product of kallikrein 7 activity, was also identified in vivo, matching previous in vitro data (47). Finally, the kallikrein inhibitor lympho-epithelial kazal-type-related inhibitor was identified by neo-N-termini of 3 cleavage products of furin that each are inhibitors for distinct kallikreins (48).
Changes in protease and inhibitor expression in inflammation
To determine the inflammatory protease and inhibitor repertoire [also known as the protease degradome (14)] present in skin and that was responsible for the proteolytic fingerprint, we performed CLIP-CHIP protease and inhibitor microarray analysis. In wild-type mice, the genes encoding 234 proteases and inhibitors were statistically significantly altered in expression during inflammation (Fig. 4A and table S16), including a marked increase in the expression of genes encoding specific MMPs (Mmp3, Mmp8, Mmp13), cathepsins (CatB, CatS), kallikrein 6 and their cognate inhibitors, Timp1 and Stefin A1/A2/A3, indicating tight regulation of the increased protease activity. The genes encoding several serpin inhibitors were decreased in expression, enabling the higher activity of their protease targets in coagulation and inflammation (49). ProMMP3 was substantially increased at the mRNA and protein levels during inflammation (Fig. 4A and fig. S6). Similarly, cathepsin B showed an increase in abundance at both the mRNA (∼2-fold) and protein levels (∼2.5-fold) (Fig. 1D and Fig. 4A). Even though Mmp2 mRNA amounts were unaltered in wild-type mice upon treatment with TPA, MMP2 protein was present in an increased amount as the activated 62 kD form (Fig. 1C) (13). The mRNA abundance for the closely related Mmp9 was slightly lower in TPA-treated skin than in normal skin (Fig. 4A), but as a major neutronphil protease, MMP9 protein was strongly increased in abundance by TPA compared to vehicle treatment in both the wild-type and knockout mouse skin samples, consistent with the inflammatory cell influx (Fig. 1B). Unlike MMP2, MMP9 was present only in the proteolytically inactive zymogen form (Fig. 1C). Other proteases might also have divergent abundances at the mRNA and protein levels. Thus, although the abundances of proteases follow discrete profiles during the course of inflammation that one point in time cannot capture, at the 48-hour stage of the inflammatory response, some MMPs were increased in abundance at the mRNA level whereas others were increased at the levels of protein abundance and activation, as shown for MMP2, or at the protein level alone, as for MMP9.
Fig. 4.
Changes in protease and inhibitor transcript abundances during inflammation and upon loss of MMP2. (A) Intensity log-ratio (M) vs. mean log intensity (A) plot of differentially regulated protease-related transcripts (proteases, inhibitors, inactive homologs) in inflamed vs. normal wild-type skin (table S16). Total RNA from TPA-treated and control skin from wild-type mice was analyzed by the CLIP-CHIP microarray. Selected proteases and inhibitors are labeled. Values are the averages of two replicates; the green and red dotted lines show the median and mean A-value of the dataset, respectively. A-values give relative intensity information that characterizes the sensitivity of the cDNA microarray analysis. (B) Comparison of changes in protease web–related gene expression induced by TPA in the skin of wild-type and Mmp2-/- mice as assessed by CLIP-CHIP analysis (table S17). Log2-ratios for genes that are ≥ two-fold (dotted lines) increased or decreased in expression in TPA-treated wild-type skin (WT) are plotted against log2-ratios for the corresponding genes in skin from MMP2 knockout mice (KO). Only genes with expression intensities (A-value) above the median were included. WT-, not regulated in wild-type; KO-, not regulated in Mmp2-/-.
Altered protease and inhibitor expression in MMP2-deficient mouse
Genes encoding 87 proteases and inhibitors showed statistically significantly altered expression changes when comparing the inflammatory response in inflamed skin from MMP2-deficient mice to that of wild-type mice, with 34 alterations in normal skin (Fig. 4B and table S17). Mmp11, Mmp28, cathepsin L (CatL), and kallikrein 9 (KLK 9) were greater than two-fold increased in expression upon exposure to TPA in the MMP2-knockout mice, but were unaffected in wild-type skin, whereas the gene encoding cathepsin J was induced only in the wild-type mice. Furthermore, the lack of MMP2 influenced the TPA-dependent expression of several inhibitors of the serpin and cystatin families, such as serpin 2, serpin 2b, and cystatin 6.
Reduced exudation of acute phase proteins into the skins of Mmp2-/- mice
To refine the analysis of the general proteolytic response in TPA-induced skin inflammation, we used multiplex iTRAQ-TAILS to investigate the role of MMP2 by analyzing Mmp2-/- mice (Fig. 1A and fig. S1). Although there were no differences between wild-type mice and MMP2-deficient mice in the inflammatory cell infiltrate or epidermal hyperproliferation, as revealed by histological analysis (Fig. 1B), there was a general reduction in the extent of exudation of acute phase proteins, including complement factors and general plasma proteins, such as murinoglobulin and serotransferrin, which were all statistically significantly reduced in the inflamed skin of MMP2-deficient animals compared to that of wild-type mice. We compared the proteins that were quantified in ≥ 3 iTRAQ-preTAILS samples (Fig. 5A and table S18), and we validated the reduction in SAA1 and haptoglobin abundance by Western blotting analysis and subsequent densitometry (Fig. 5B and fig. S8).SAA1 was completely absent from non-inflamed skin of both genotypes and was only present in wild-type skin upon inflammation. Haptoglobin was also lower in uninflamed skin from Mmp2-/- compared to wild-type mice but increased statistically significantly stronger in inflamed skin from wild-type than from knockout animals. Notably, haptoglobin protein amounts in the plasma were similar before and during the inflammation of wild-type and knockout mice, and the abundance of this protein showed no difference in inflamed livers from both genotypes (Fig. 5C and fig. S9). Hence, the observed local changes at the site of inflammation were not a result of differences between mouse genotypes in systemic levels of acute phase proteins. Thus, in the Mmp2-/- mouse, there was an unexpected greatly diminished increase in acute phase and serum proteins in the skin upon inflammation.
Fig. 5.
Decrease in the extent of protein exudation in inflamed Mmp2-/- skin and in MMP2-dependent proteolysis in vivo. (A) Differential abundances of proteins in TPA-treated skin from wild-type and Mmp2-/- mice. 976 proteins were quantified in at least 3 of 5 experiments and were analyzed for their statistically significant differential abundance between TPA-treated wild-type and Mmp2-/- skin (two-fold, P ≤ 0.05, Limma moderated t-statistic, Benjamini-Hochberg multiple testing correction). (B) Western blotting analysis of acute phase proteins in TPA-treated and control skin from wild-type and Mmp2-/- mice. All skin samples analyzed by iTRAQ-TAILS were also analyzed by Western blotting for the acute phase proteins SAA1 and haptoglobin with specific antibodies. (C) Western blotting analysis of haptoglobin in plasma from MMP2-deficient animals (n = 3 mice, two of which are shown) and wild-type controls (n = 3 mice, two of which are shown) before and after treatment with a model of mustard oil– induced ear skin inflammation. (D) Quantitative categorization of N-termini in inflamed skin from Mmp2-/- and wild-type mice based on natural N-termini. The distribution of quantitative ratios for natural N-termini in TPA-treated wild-type and Mmp2-/- skin was used to define cutoffs for statistically significant changes in the abundances of N-terminal peptides in inflamed wild-type vs. knockout skin. (E) Protein assignment to N-termini categorized by position in the mature protein and differential abundance in inflamed skin from wild-type and Mmp2-/- mice. The 4-way Venn diagram indicates how many proteins were identified by which categories of N-terminal peptides. N-termini in overlapping categories correspond to different N-terminal peptides from the same protein. Hence, four proteins were identified by their natural N-terminus, which were not altered in abundance, and by the increased abundance of neo-N-termini (only one per protein). (F) Assignment of N-termini to C1 inhibitor (C1 Inh) and apolipoprotein C1 (APOC1). C1 Inhibitor was identified by its natural N terminus with equal abundance in both genotypes and by a single neo-N-terminus with a high log2(wt-TPA/ko-TPA) ratio (1.70) at position 471, which indicated MMP2-dependent cleavage at the reactive bond. The high-ratio N-terminus assigned to APOC1 indicated an MMP2-dependent cleavage by dipeptidyl peptidase 4 (DPPIV) and which is known to cut at sites with this sequence.
Direct inactivation of C1 inhibitor by MMP2 and control of vascular permeability
We hypothesized that in Mmp2-/- mice there was less of an increase in vascular permeability in inflammation compared to that in wild-type mice, and hence there was less exudation of serum and acute phase proteins into the tissue. To identify potential substrate mediators, we followed the same approach as was performed for the analysis of the general proteolytic response to inflammation (Fig. 2E), but we focused on abundance ratios of N-terminal peptides in TPA-treated wild-type versus TPA-treated MMP2-deficient skin (Fig. 5D). We determined a log2(wt-TPA/ko-TPA)-ratio of 0.90 as the cutoff for N-termini that were present in statistically significantly greater amounts in the wild-type mice than in the MMP2 knockout mice. Notably, two serpins (C1 inhibitor and A1AT4) and two apolipoproteins (APOA1 and APOC1) were cleaved only in the wild-type mice and were identified as MMP2-dependent substrates by two criteria. First, these proteins were found by a single high ratio free neo-N-terminal peptide (starting position ≥ 3) (Fig. 5E), indicating a MMP2-dependent cleavage event. Second, their natural N-termini were equally present in inflamed skin of both genotypes (that is, they had a ratio centered on 1.0 for their natural N-termini). This means that the high ratio of the cleaved neo-N-termini did not arise from increased amounts of the protein by new synthesis (and retaining the same turnover rate) or from tissue import from serum exudate as a cleaved protein.
The analysis strategy was supported for APOA1 and APOC1 in the iTRAQ-preTAILS analysis when ratios for multiple peptides per protein were averaged (Fig. 5A). However, lacking N-terminal information, such as from shotgun analyses, meant that these iTRAQ-preTAILS data were not definitive and required the power of specific N-termini–enrichment to functionally dissect proteolytic modifications in vivo. Interestingly, the neo-N-terminus of APOC1 identified by iTRAQ-TAILS matched exactly with a DPPIV cleavage site and so indicated an MMP2-dependent event that was indirect (Fig. 5F). Consistent with a general positive influence of MMP2 on DPPIV, the neutrophil chemoattractant CXCL5 was cleaved and inactivated by DPPIV (20), as shown by a very high log2(wt-TPA/ko-TPA) ratio of 4.98 for the characteristic neo-N-terminus starting after the 1AP↓SS cleavage (Fig. 3C).
C1 inhibitor is the major inhibitor of plasma kallikrein that releases the important vasodilator bradykinin from kininogen (Fig. 6A). We detected C1 inhibitor in equivalent amounts in the skin of mice of both genotypes by its natural mature N-terminus with a log2(WT-TPA/KO-TPA)-ratio of 0.41 (being below the cutoff this ratio means it was not significantly altered in abundance), and by a neo-N-terminus that was present in a much higher amount in the presence of MMP2 [log2(WT-TPA/KO-TPA)-ratio = 1.70] than in its absence (Fig. 5F). The sequence of the neo-N-terminal peptide matched with a cleavage event at the reactive bond of C1 inhibitor that renders it inactive towards its target serine proteases. Incubating C1 inhibitor with active MMP2 generated two bands as analyzed by SDS-PAGE (Fig. 6B). The first band of 4 kD resulted from release of the C-terminus of the inhibitor after cleavage at the reactive bond (Fig. 6A), and the second band of 95 kD corresponded to the heavily glycosylated full-length C1 inhibitor being shifted to a lower mass after proteolytic release of the C-terminal 4 kD fragment.
Fig. 6.
Control of vascular permeability by MMP2-dependent regulation of bradykinin release. (A) Extended Ingenuity pathway analysis demonstrating the control of bradykinin release from kininogen by MMP2. Proteins identified by N-terminal peptides are in red. A higher log2-ratio for the plasma kallikrein (KLKB1)-generated neo-N-terminus of kininogen-1 (KNG1a) than for the natural N-terminus of plasma kallikrein indicates stimulation of the protease by MMP2 activity. (B) Direct cleavage of C1 inhibitor (C1 Inh) by MMP2. SDS-PAGE and silver staining demonstrated the release of the expected 4-kD fragment from human recombinant C1 inhibitor by active MMP2 (lower gel), and the corresponding reduction in the molecular mass of the remaining N-terminal part of the protein (upper gel) only in the absence of the MMP2 inhibitor Marimastat (Marim.). (C) Inactivation of C1 inhibitor activity on plasma kallikrein by MMP2. Generation of bradykinin (BDK, m/z = 1060) from plasma kallikrein was monitored by MALDI-TOF in relation to a spike in the control peptide (m/z = 1296) in the presence and absence of the C1 inhibitor that had either been pre-incubated with MMP2 or buffer control. Representative spectra recorded after 30 min of incubation are shown. (D) Quantification of the relative amount of bradykinin release from five replicated MALDI-TOF measurements after 60 min of incubation. Error bars represent the SD. (E) Time-course of bradykinin generation as recorded by MALDI-TOF. Values represent the averages of two independent experiments. (F and G) Representative images of wild-type (WT) and Mmp2-/- mice subjected to the Miles assay for vascular permeability and showing the fold-increase in Evans blue dye release from mustard oil–treated vs. vehicle-treated ears (n = 3 mice; *P < 0.05 by Student's t test).
By identifying the neo-N-terminal peptide of the kallikrein-cleaved light chain of kininogen in wild-type mouse skin (Fig. 3C), we could estimate the extent of bradykinin release. Notably, this was more than four-fold higher in inflamed skin from wild-type mice than in inflamed skin from Mmp2-/- mice (Fig. 6A). Under the same conditions, the natural N-terminus of mature plasma kallikrein was only two-fold more abundant in wild-type samples than in MMP2 knockout samples, suggesting positive MMP2-dependent regulation at the level of kallikrein activity. Indeed, only full-length C1 inhibitor, but not the protein cleaved by MMP2, inhibited the kallikrein-mediated excision of bradykinin from kininogen, as was revealed by MALDI-TOF mass spectrometry analysis of cleavage assays (Fig. 6C to E). To confirm that MMP2 modified vascular permeability during inflammation, we monitored Evans blue dye exudation in the Miles assay. Vascular permeability in inflamed ear skin tissue was substantially reduced in Mmp2-/- mice compared to that in wild-type controls (Fig. 6F, G). Hence, this second model of inflammation validated a previously uncharacterized in vivo function of MMP2, namely, the proteolytic inactivation of C1 inhibitor leading to bradykinin release and thus an increase in vascular permeability.
Activation of the complement cascade by MMP2 through inactivation of C1 inhibitor
In addition to regulating plasma kallikrein activity, C1 inhibitor strictly controls the complement cascade by inhibiting the C1r and C1s serine proteases of the C1 complex and MASP in the lectin pathway (9, 50). Therefore, the inactivation of C1 inhibitor by MMP2 should also enhance complement activation. We tested this hypothesis by mining our iTRAQ-TAILS dataset, which enabled the detailed quantitative and positional analysis of complement pathway activation in vivo by the identification of 21 N-terminal and neo-N-terminal peptides of 7 complement proteins (Fig. 7A). Complement C4 was identified by the N-terminus of its unprocessed α-chain and in its activated form by the N-terminal peptide of C4b (Fig. 7B). The latter fragment is generated either by activated complement C1 in the classical pathway or by mannan-binding lectin-associated serine protease 2 (MASP2) during lectin-induced complement activation, which in turn requires its binding protein mannose-binding protein A (MBL) (5). We identified both MASP2 and MBL by their mature protein N-termini after signal peptide removal. Being shifted by one amino acid for MBL (see UniProtKB/Swiss-Prot), this again illustrates how terminomics can correctly annotate databases (correct position confirmed by SignalP 3.0 prediction (51), fig. S10). The activity of C4b was confirmed by the neo-N-terminal peptide corresponding to cleavage of the C3 α-chain by C3-convertase (C4bC2a) resulting in the formation of C3b (Fig. 7C). We also used iTRAQ-TAILS to detect the N terminus of the unprocessed C3 α-chain. Complement factor I (IF), which was identified by its light-chain N-terminal peptide, inactivates C3, a cleavage event that was identified from the N-termini of the C3 degradation fragments C3dg and C3c. Thus, by identifying the N-termini of complement proteins in vivo, we globally identified activation and inactivation events of complement proteins, thus discovering previously uncharacterized cleavage sites and validating in vivo a long history of in vitro analyses with purified proteins.
Fig. 7.
N-terminal analysis of the complement network and its control by MMP2. (A) Extended Ingenuity pathway analysis. A part of the classical and lectin complement pathway is shown with proteins that have N-terminal peptides identified by iTRAQ-TAILS shown in red. High log2-ratios indicate higher abundance in wild-type mice than in MMP2-deficient mice. (B) Detection of the N termini of complement C4 and their validation by Western blotting analysis. Fragments generated by known cleavages and, where available, their log2-ratios are indicated. (C) Detection of the N-termini of complement C3, as described for (B). Peptides in parentheses were identified but with a probability of correct identification below the cutoff for high-confidence peptide identifications. (D) Inhibition of complement activity by C1 inhibitor (C1 Inh) as assessed in a hemolytic assay with red blood cells. Pre-incubation of C1 inhibitor with active MMP2 abolishes its inhibitory activity on complement activation. Data from three experiments performed in duplicate and with two different inhibitor preparations. Data are means ± SE. n = number of measurements For each incubation type and with the following concentrations of C1 Inh, the n numbers indicate the number of measurements: 2.73 μM (n = 4), 3.64 μM (n = 4), 4.55 μM (n = 6), 6.36 μM (n = 6), 7.27 μM (n = 2). **P ≤ 0.01; ***P ≤ 0.0001 by Student's t test.
Consistent with the MMP2-dependent inactivation of C1 inhibitor, we found in skin from MMP2-knockout animals the diminished induction of C4b, C3b and their inactivation products C4d′, C3dg, and C3c [shown by high log2(wt-TPA/ko-TPA)-ratios of their N termini] (Fig. 7B, C and table S8). We validated these findings by Western blotting analysis of the inflamed skin (Fig. 7B and C). Indeed, C1 inhibitor that had been cleaved by incubation with active MMP2 no longer inhibited hemolysis in a complement hemolytic assay (Fig. 7D). The amounts of N-terminal peptides for MASP2 and MBL were also increased in inflamed skin from wild-type mice compared to that in inflamed skin from MMP2-deficient mice (Fig. 7A), augmenting the effect in vivo. Hence, in addition to vascular permeability control by MMP2, we validated that in wild-type mice, MMP2 exerts control on complement activation in vivo by indirectly increasing C1 and MASP activity.
Discussion
We present the first system-wide analysis of the murine skin proteome and its perturbation during inflammation. Applying multiplex iTRAQ-TAILS N-terminomics enabled us to concomitantly dissect global proteolytic events and MMP2-dependent processing in vivo. From the differences in substrate cleavages that occurred upon genetic loss of MMP2, it was evident that MMP2 had effects in addition to its ability to remodel the extracellular matrix, which traditionally has been interpreted as the main function of MMPs (52). In identifying and quantifying global changes in the inflammatory response at the levels of proteins and protease substrates, we found that MMP2 regulated multiple proteolytic pathways that are integral to the inflammatory process. In particular, cleavage at the reactive bond of the C1 inhibitor by MMP2 decreased the amount of intact functional C1 inhibitor in vivo, and thereby controlled vessel permeability and complement activation during inflammation. Our proteomics analyses showed that inflammation in the skin of wild-type mice resulted in an increased exudate of serum proteins, but that in the absence of MMP2, the increase in the exudate of acute phase, complement, and serum proteins was lost because of the impaired increase in vascular permeability. Mechanistically, this was associated with reduced bradykinin release in the skin of MMP2 knockout mice, as quantified by the iTRAQ-TAILS analyses. The in vivo proteomic evidence was supported by in vitro MALDI-TOF analyses showing that cleavage of the C1 inhibitor at the reactive bond by MMP2 rendered C1 inhibitor unable to block kallikrein-mediated cleavage of the high molecular weight kininogen, which led to increased bradykinin release. In addition, we co firmed this MMP2-related defect in vascular permeability in a second inflammation model by showing the substantially reduced exudation of Evans Blue dye in inflamed skin on the ears of mustard oil–treated Mmp2-/- mice compared to that in wild-type mice.
Through cleavage of VEGF-inhibitory binding proteins, such as CCN1 and HARP, MMP2 mobilizes VEGF (53), which also potently enhances endothelial permeability during inflammation (54). Bradykinin has a similar vascular effect to that of VEGF and is also an inducer of angiogenesis (55). Hence, the reduced bradykinin production in Mmp2-/- tissue correlates with the reduced angiogenesis previously observed in MMP2-deficient animals (24). Thus, by adding to the roles of MMP2 in regulating chemokine activity by precise cleavage (11, 56), we have shown that MMP2 has additional higher-order roles in inflammation.
C1 inhibitor also controls the complement system whereby intact C1 inhibitor reduces the extent of complement activation (9). Concomitant with cleavage of C1 inhibitor, the in vivo TAILS analyses showed multiple cleavages of the major components of both the C3 convertase (C4bC2a) and C5 convertase (C4bC2aC3b) complexes—complement C4 and C3—which were substantially reduced in abundance in the inflamed skin of MMP2 knockout mice compared to that of wild-type mice. Thus, with markedly increased amounts of the complement proteins MASP2 and MBL in the inflamed skin of wild-type mice compared to Mmp2-/- mice, the extent of complement activation was reduced in the knockout animals. The mechanistic link between these two observations was confirmed in vitro: Whereas a reduction in sheep erythrocyte hemolysis by complement activation occurred after the addition of intact C1 inhibitor, complement activation and hemolysis were unchanged in the presence of the MMP2-cleaved form of the C1 inhibitor. Thus, our studies reveal the critical importance of previously uncharacterized upstream regulation of the C1 inhibitor by MMP2-dependent processing as a key control point of multiple inflammatory pathways. Given that much of the complement cascade was originally described from experiments with purified proteins, it comes as no surprise that previously uncharacterized cleavage sites were found when we studied the complement pathway in vivo, a context in which all of the proteins and their binding partners are present at normal concentrations.
By proteome simplification through N-terminal peptide enrichment, iTRAQ-TAILS expands the dynamic range of peptide analysis by proteomics, thus leading to the identification of many rare proteins that were not observed through the iTRAQ-preTAILS analyses. It is surprising that ∼44% of the normal skin proteins identified commenced at sites lying distal to the expected protein start sites. This was despite stringent measures to inhibit all proteases at the tissue-harvesting stage and afterwards, so revealing an unexpectedly high homeostatic rate of proteolysis in skin to generate new stable protein forms. During the inflammatory response, the rate of proteolysis increased, thus truncating the N-termini of ∼60% of the proteins identified, many of which had altered activities as a result. Such a qualitative increase in proteolytic processing was a result of the increased abundance of mRNAs for proteases during skin inflammation that we identified. However, given the alterations in the abundances of mRNAs for 87 proteases and inhibitors that occurred after loss of a single protease (MMP2), it is difficult to ascribe actions to a single protease in the in vivo milieu in which hundreds of proteases and inhibitors operate in an interconnected protease web (6, 57). Indeed, such marked perturbations in inflammation on an Mmp2-/- background emphasize this important caveat in interpreting protease knockout mice in models of disease.
Contributing to the increase in the amounts of neo-N-termini are proteins released from infiltrating immune cells as well as from the inflammatory exudate of blood plasma, which also contains an increased proportion of neo-N-terminal peptides (58). Exploiting the negative selection strategy of iTRAQ-TAILS, we used quantification of the natural N-terminus as an approach to discriminate between MMP2-dependent, direct effects and MMP2-dependent, indirect effects. Thereby, by multiplex quantitative analysis we determined the following. First, we discriminated the TPA-regulated cleavage fragments that are mechanistically important during inflammation from the cleavage fragments that were generated in similar amounts in both normal and inflamed skin, and thus were less likely to play key roles in inflammation. Second, we identified those proteins with a mature original N-terminal peptide ratio (wild-type/Mmp2-/-) centered on 1.0 that were not derived from differential new synthesis or were imported in the inflammatory cell infiltrate or exudate from the blood in mice of both genotypes. Third, in combination with the high ratio and unique neo-N-terminal peptides of these proteins, we discerned effects of MMP2 on proteolytic processing versus those on changes in protein abundances in the skin (Fig. 5E). This again emphasizes the need for quantitative terminomics approaches, such as TAILS, to analyze proteolysis in vivo. Many proteins are potentially direct substrates of MMP2, but without identification of mature N-termini, these cannot be assigned with certainty and will require validation. In addition to identifying MMP2 substrates, iTRAQ-TAILS enabled the identification of intact proteins and diverse proteolytic events in skin inflammation, such as the trimming of N-termini by aminopeptidases, activation of enzymes and chemokines, precursor protein processing, and release of cryptic growth factors from extracellular matrix proteins. Collectively, these processes form a proteolytic signature of the in vivo global activity of those proteases active at 48 hours after treatment with TPA.
Hence, we devised and implemented a new approach to dissect proteolytic events in complicated tissue responses in vivo. By layering positional information on top of quantitative proteomics, we characterized and quantified signaling proteolytic events in a mammalian tissue in an unbiased and system-wide manner, and thereby identified previously uncharacterized substrates and functions of MMP2. As revealed by proteomics in vivo, and as confirmed by functional assays in vitro, MMP2 proteolytically inactivates the serine protease C1 inhibitor by a single and high-precision cleavage within its functional domain, and thereby regulates downstream processes of vascular permeability and complement activation during inflammation. These results demonstrate the usefulness and power of multiplexed quantitative TAILS analyses as a mechanistically transparent hypothesis-generating approach, and emphasize the need for terminomics to complement other analyses to understand complex processes in vivo. Overall, the present degradomics-based study enabled us to dissect and reconstruct the proteolytic events during complicated inflammatory tissue responses and it demonstrated how lack of a single protease perturbed the interconnected signaling pathways by simultaneous modulation of its components at the level of gene expression, protein abundance, and activity.
Materials and Methods
Mouse skin inflammation
Mmp2 knockout and wild-type mice on an FVB background (provided by Dr. Lynn Matrisian and Dr. Barbara Fingleton, Vanderbilt University, Nashville, TN) were maintained as homozygous lines as approved by the UBC Animal Care Committee. 12-O-tetradecanoylphorbol-13-acetate (TPA, Sigma) was used to induce skin inflammation (59). Eight mice (4 knockout and 4 wild-type females, 3 months old) were anesthetized by inhalation of isoflurane and the fur on their backs was shaved. Each of 4 animals (2 wild-types and 2 knockouts) received either 100 nanomoles of TPA in 200 μl of acetone or acetone alone. Animals were euthanized after 48 hours with carbon dioxide followed by cervical dislocation. The treated back skin was excised, immediately frozen in liquid nitrogen and stored at -80°C. Alternatively, to establish the proteomic background of wild-type and MMP2-deficient mouse skin without inflammation, 2 animals (1 wild-type and 1 knockout) were sacrificed and the shaved back skin was collected without previous application of TPA or acetone.
Skin proteome preparation
To enrich for secreted extracellular proteins, we used detergent-free conditions to preserve cell and nuclear membranes as much as possible. Samples were quickly transferred from dry ice into a vial with a minimal volume of ice-cold buffer [100 mM hepes (pH 8.0), 10 mM EDTA, with Complete EDTA-free protease inhibitor cocktail, Roche, 1 tablet/10 ml]. The tissues were disrupted by a 1-min pulse with an Ultra-Turrax T25 homogenizer (Janke & Kunkel) and placed on ice. The resulting suspensions were sedimented by centrifugation at 17000 g for 10 min at 4°C. The secreted-protein-rich supernatant from the middle portion of the tube was carefully aspirated, being cautious to disturb neither the hair and cell debris at the bottom of the tube nor the lipid layer at the top. Amino acids and metabolites were removed by protein precipitation (with 16.6% TCA for 30 min on ice) and the pelleted proteins were resuspended in a minimal volume of 50 mM sodium hydroxide, which was then adjusted to pH 8.0 with water and a stock solution of 1 M hepes (pH 8.0). Protein concentrations were measured by Bradford assay (Bio-Rad) and samples were assessed by silver staining after SDS-PAGE.
TAILS workflow and mass spectrometry analysis
Skin proteomes of TPA-treated and control mice were analyzed by TAILS with 4plex iTRAQ (Applied Biosystems) (22) as previously described (21). The experimental and labeling scheme is shown in fig. S1, with 8 animals being separately analyzed in five different four plex analyses. Three technical replicates were performed in which iTRAQ labels were swapped. For each four-plex analysis, protein was quantified for iTRAQ-TAILS (250 μg for experiments 1 and 2, and, to increase peptide numbers, 500 μg for experiments 3 to 5). In comparing non-treated wild-type and knockout animals by 2plex-CLIP-TRAQ-TAILS, 500 μg/label was used. iTRAQ-TAILS enriches for protein N-terminal peptides that are naturally blocked or are N-terminal blocked and labeled by iTRAQ. Unlike conventional tryptic peptide iTRAQ labeling, in iTRAQ-TAILS, the performed before trypsinization at the whole protein level. In blocking and labeling the N-terminal peptides of proteins and stable cleavage products, this process differentiates these from the internal tryptic and C-terminal peptides of the protein now bearing a free N-terminal α-amino group. This group is reactive to aldehyde groups on HPG-ALD polymer (Flintbox Innovation Network: http://www.flintbox.com/public/project/1948/) under reductive conditions, thus enabling the unbound blocked and labeled N-terminal peptides to be recovered by ultrafiltration for mass spectrometry analysis (“iTRAQ-TAILS” samples). For all samples a shotgun proteomics analysis was also performed on an aliquot before N-terminal peptide enrichment (“iTRAQ-preTAILS”). Samples were pre-fractionated offline on an Agilent Technologies 1200 series HPLC with a PolySULFOETHYL A 100 mm × 4.6 mm, 5 μm, 300 A column (Poly LC Inc.) (22). The resulting 10 to 13 peptide fractions were analyzed by tandem MS/MS after inline nano-LC separation (C18 150 mm × 100 μm column at a flow rate of 100-200 nL min-1). MS data were collected with a QStar XL Hybrid ESI mass spectrometer (Applied Biosystems; MDS-Sciex) equipped with Analyst QS software version1.1 (ABI/MDS SCIEX) using settings previously described (22).
MS2 peptide assignment, secondary validation, and iTRAQ quantification
MS2 scans were searched against a mouse International Protein Index (IPI) protein database (v.3.24) (52,414 protein entries) by Mascot version 2.2 software (Matrix Science) or X! Tandem 2007.07.01 release as described previously (22), with the following parameters: Semi-ArgC cleavage specificity with up to two missed cleavages, cysteine carbamidomethylation, and peptide lysine iTRAQ were set as fixed modifications; N-terminal iTRAQ, N-terminal acetylation, and methionine oxidation were set as variable modifications. To identify peptides with N-terminal pyroglutamate, spectra were searched with Mascot allowing pyro-Glu (N-term Q) as a variable modification. Peptide tolerance and MS/MS tolerance were both set at 0.4 Daltons, and the scoring scheme used was ESI-QUAD-TOF. Search results were evaluated on the Trans Proteomic Pipeline (TPP v4.2, rev 0, Build 200811181145) with PeptideProphet, iprophet, and ProteinProphet for peptide and protein identification, and Libra for quantification of iTRAQ reporter ion intensities. For iTRAQ-preTAILS analyses and for calculation of isoform assignment scores (IAS) (23) Mascot searches were performed and protein lists compiled with PeptideProphet and ProteinProphet with a probability of 0.9 resulting in low error rates (0.7 to 0.9%). For iTRAQ- and CLIP-TRAQ-TAILS analyses, Mascot and X! Tandem searches were combined by iProphet (probability ≥0.95) after PeptideProphet analyses. This cutoff corresponds to an actual error rate of 0.6 to 0.7% as shown by sensitivity-error rate analysis in iProphet (see the relevant supplementary tables) confirming a highly stringent <1% error rate.
Statistical data analysis and peptide annotation
For quantification of proteins in iTRAQ-preTAILS analyses, iTRAQ-labeled, and thus quantifiable, peptides for each protein were extracted from probability-filtered ProteinProphet output lists and quantified by summing up iTRAQ reporter ion intensities that had a minimum value of 20 in at least one channel from each assigned spectrum with an in-house Perl script. For accurate ratio averaging for peptides of a protein and identified from multiple spectra of individual peptides, reporter ion values from each quantified peptide assigned to a protein were again summed and normalized to the sum of all channels to give the final intensity weighted relative quantification value for the protein (23). Before statistical analysis, the values in all four channels were subjected to quantile normalization to correct for intra-experimental variation. Proteins with statistically significant differential abundance were determined by building a subset of proteins with normalized quantification values in at least 3 of 5 experiments, applying moderated t-statistics (Bioconductor – Limma) with Benjamini-Hochberg multiple testing correction, and extracting entries with a change in abundance between conditions of at least two-fold and a corrected P value of ≤0.05. Finally, lists were sorted and heatmaps generated with mean centered quantification values. To quantify N-terminal peptides in iTRAQ-TAILS analyses, raw iTRAQ reporter ion intensities in iProphet output lists were quantile normalized in all channels for each experiment after extracting only N-terminal labeled or naturally blocked semitryptic peptides and applying a minimum intensity threshold of 15 for iTRAQ (22) and 30 for CLIP-TRAQ (23) experiments, respectively. Then, the union of peptides from all experiments was formed and intensity weighted abundance ratios were calculated from all spectra per peptide with models described previously for iTRAQ (22) and CLIP-TRAQ (23) experiments. The quantification confidence factor (QCF) for each ratio was calculated to discriminate higher from lower confidence peptide ratios (23). Standard deviations are included in the QCF calculated from experimentally derived ion intensity–dependent standard deviations and normalized to a maximum of 10 to enable intra- and inter-experimental comparisons to be made. Thus, a QCF of 0 is an average quantification confidence, whereas QCF values > 0 and < 0 correspond to higher or lower confidence, respectively. N-terminal peptides were assigned to proteins and annotated for their position, and isoform assignment scores (IAS) were calculated by in-house Perl scripts with compiled protein lists from iTRAQ-preTAILS and iTRAQ-TAILS analyses (tables S1 and S19), respectively, as described previously (22, 23).The experimental variation was derived from the ratio distribution of natural N-termini (44). A normal distribution was fitted to the data after removal of outliers by including only values between the 10th to 90th percentiles. This had a mean near 0 and so validated our normalization and single-peptide quantification strategies. The resulting standard deviation was used to calculate P values with help of the Gaussian error function and defined a log2(wt-TPA/wt-ctrl) ratio of >1.19 and <-1.19 (P<0.05) and a log2(wt-TPA/ko-TPA) ratio of >0.90 and <-0.90 (P < 0.05) as cutoffs for peptides that were statistically significantly increased or decreased in abundance by TPA in wild-type skin or were significantly more or less abundant in TPA-treated skin from wild-type mice than in TPA-treated skin from Mmp2-/-mice, respectively. The cutoff of a log2-ratio of 3.32 for singleton N-terminal peptides that are present only in one condition was experimentally defined and statistically validated previously (22).For statistical analyses and the generation of heatmaps, a local installation of the CARMAweb 1.4 frontend to Bioconductor (60) was used. Venn diagrams were generated with“Venny” (61) or “Overlapper” (http://faculty.ucr.edu/∼tgirke/Documents/R_BioCond/My_R_Scripts/overLapper.R) in R (v2.8.1, http://www.r-project.org). For GO enrichment analyses, we used the BiNGO plug-in to Cytoscape (62), and for pathway analyses of protein lists, we used the IPA system from Ingenuity Systems.
CLIP-CHIP microarray analysis
Total RNA from individual mouse skin samples was isolated and purified with TRIzol (Invitrogen, Inc.). Reverse transcription and amplification were performed with a Message Amp II aRNA amplification kit (Ambion) as described previously (26). RNA Cy3 and Cy5 labeling was performed with a ULS aRNA fluorescent labeling kit (Kreatech Diagnostics). All hybridizations were performed in duplicate in a common reference approach with universal mouse RNA (Stratagene) as a standard in all samples. Microarray slides were scanned on a 428 Array Scanner (MWG), images were analyzed with ImaGene 6.1 (Biodiscovery), and fold changes were calculated and visualized with CARMAweb 1.4 (63) and Prism 4.0 (GraphPad Software). To evaluate the effect of MMP2 on TPA-induced changes in the mRNA abundances of proteases and related gene products, we calculated log2(ko-TPA/ko-ctrl) ratios from the CLIP-CHIP analysis and compared them to log2 (wt-TPA/wt-ctrl) ratios from the same experiment applying a two-fold cutoff.
Western blotting analysis and gelatin zymography
Equal amounts of protein from tissue lysates were precipitated by acetone, resolved by SDS-PAGE, transferred to Immobilon-FL PVDF membranes (Millipore), and visualized on a Li-Cor Odyssey with primary polyclonal antibodies and the corresponding Alexa Fluor-conjugated secondary antibodies (Molecular Probes). Goat anti-murine complement C3 (Cooper Biomedical), goat anti-human complement C4 (QuidelQ), rabbit anti-actin (Sigma), and antibodies against murine SAA1 (goat), haptoglobin (sheep), S100A8 (goat), and S100A9 (goat) were obtained from R&D systems. Densitometric analysis was performed with ImageJ and statistically evaluated using Prism 4.0 (GraphPad Software). The gelatinolytic activity in skin lysates (5 μg total protein/lane) and of pro- and active forms of recombinant human MMP2 and MMP9 as standards was monitored by gelatin zymography (10% SDS-PAGE gel with 1 mg/ml gelatin).
Semi-quantitative bradykinin release assay
Plasma kallikrein (0.2 μM) was pre-incubated in a total volume of 5 μl for 2hours at 37°C alone or in a 1:10 molar ratio with C1 inhibitor that had been untreated or was cleaved by MMP2. 1.0 μl of precincubated plasma kallikrein was then added to 9 μl of 2.0 μM single chain high molecular weight kininogen in 20 mM hepes (pH 8.0), 150 mM NaCl, 5 mM CaCl2, and with 0.2 μM des-Arg1 bradykinin and 0.25 μM angiotensin I as internal reference masses (Sequazyme, Applied Biosystems). After 0, 30, 60, or 90 min, 0.5 μl of the reaction were spotted in duplicate on a MALDI plate and mixed with an equal volume of alpha-cyano-4-hydroxycinnamic acid (5 mg/ml in 70% acetonitrile and 0.1% trifluoroacetic acid (TFA)). Samples were desalted on-plate by two washes with 1 μl of ice-cold 0.1% TFA before analysis. For end-point measurements, reactions were performed in quintuplicate in volumes of 5 μl and stopped after 60 min by adding 1.0 μl of 1% TFA. The generation of bradykinin was monitored on a MALDI-TOF mass spectrometer (Applied Biosystems, Voyager-DE STR). The des-Arg1 bradykinin signal (m/z 904.47) was used for normalization and to avoid saturation of the detector by the signals from bradykinin (m/z 1060.59) or angiotensin I (m/z 1296.69). 500 to 800 spectra were collected and averaged for every sample. Baseline reduction and noise removal were performed with Data Explore version 4.6 software. Spectra were exported as ASCII files, and the amount of bradykinin release was quantified relative to the angiotensin I signal in Microsoft Excel.
Miles assay for vascular permeability
12-week-old male Mmp2-/- and wild-type control mice were anesthetized with 2% isoflurane/98% oxygen, and injected into the tail vein with Evans blue reagent (30 mg/kg) in medical grade saline. One minute after injection, the right ear was treated with 30 μl of 5% (v/v) phenyl isothiocyanate (Sigma) in mineral oil (Sigma) delivered drop wise to the ventral and dorsal surfaces. The left ear was treated with mineral oil alone. After 30 min, the mice were euthanized and the ears removed and weighed. Ears were then extracted in 1 ml of formamide (Sigma) at 60°C for 48 hours. The absorbance readings (at 600 nm) for each extract were compared to those of Evans blue standards.
Complement activation assay
The classical pathway hemolytic assay was performed as described previously (64) with antibody-sensitized sheep red blood cells (Complement Technologies) and human serum (CBR). Serpin inhibition assays were performed with 0.003x plasma dilution and increasing amounts of full-length or MMP2-digested C1 inhibitor (Complement Technologies). The results were normalized to the corresponding controls, that is, buffer alone or MMP2-containing buffer, and expressed as the percentage of hemolysis relative to that in a hemolysis reaction that did not contain C1 inhibitor.
Supplementary Material
Fig. S1. iTRAQ labeling scheme for TAILS experiments.
Fig. S2. Skin inflammatory proteome (iTRAQ-preTAILS).
Fig. S3. Distribution of acetylated and non-acetylated N termini with preferred residues.
Fig. S4. Dynamics of actin acetylation revealed by iTRAQ-TAILS in vivo.
Fig. S5. Consensus sequence for cyclized N-Termini after pyroglutamate formation.
Fig. S6. Original mature N terminus of stromelysin-1 (MMP3) identified by iTRAQ-TAILS.
Fig. S7. Original mature N terminus of S100A9 identified by iTRAQ-TAILS.
Fig. S8. Densitometric analysis of Western blotting analyses shown in Fig. 5B and C.
Fig. S9. Western blot analysis of haptoglobin abundance in livers of Mmp2-/- mice and wild-type controls upon mustard oil induced skin inflammation.
Fig. S10. Prediction of signal peptide removal for mannose-binding protein A (MBL1).
Tables S1 to S19. Spectra files.
Acknowledgments
We thank the following members of the University of British Columbia Centre for Blood Research: W. Chen for mass spectrometer operation, J. Kizhakkedathu for kindly providing the HPG-ALD polymer, D. Devine for antibodies against complement C3 and C4 proteins, and E. Conway and M. Krisinger for advice on the complement activation assay. In addition, we thank U. Hassiepen, Novartis, Basel for providing kallikrein and high molecular weight kininogen and advice, and S. Olson (University of Illinois at Chicago) for providing C1 inhibitor.
Funding: U.a.d.K was supported by a German Research Foundation (DFG) research fellowship and A .P. was supported by the UBC Centre for Blood Research Strategic Training Program in Transfusion Science. C.M.O. holds a Canada Research Chair in Metalloproteinase Proteomics and Systems Biology. This work was supported by a grant from the CIHR as well as with an Infrastructure Grant from Michael Smith Foundation for Health Research. US NIH grants 5R01CA084360 and 5R01CA157781 to B.F.
Footnotes
Author contributions: U.a.d.K. and A.P. participated in project design, performed TPA treatment of mice, and interpreted the data; U.a.d.K. did the bioinformatics analysis and drafted the manuscript; A.P. performed TAILS analyses, zymograms and Western blots, protease and complement assays, and revised the manuscript. U.E. performed the MALDI-TOF bradykinin release assays; B.F. performed Miles assays; C.M.O. participated in project design, bioinformatics strategy and was responsible for project supervision, data interpretation, and manuscript writing and provided grant support.
Competing interests: The authors declare that they have no competing interests
Data and materials availability: Mass spectrometry raw data (mzXML files) may be downloaded from the ProteomeCommons.org Tranche system (https://trancheproject.org) with the hashes indicated in the supplementary tables.
References
- 1.Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol. 2004;4:617–629. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 2.Doucet A, Butler GS, Rodriguez D, Prudova A, Overall CM. Metadegradomics: toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Mol Cell Proteomics. 2008;7:1925–1951. doi: 10.1074/mcp.R800012-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Lange PF, Overall CM. Protein TAILS: when termini tell tales of proteolysis and function. Curr Opin Chem Biol. 2012 doi: 10.1016/j.cbpa.2012.11.025. http://dx.doi.org/10.1016/j.cbpa.2012.1011.1025. [DOI] [PubMed]
- 4.Riddel JP, Jr, Aouizerat BE, Miaskowski C, Lillicrap DP. Theories of blood coagulation. J Pediatr Oncol Nurs. 2007;24:123–131. doi: 10.1177/1043454206298693. [DOI] [PubMed] [Google Scholar]
- 5.Ehrnthaller C, Ignatius A, Gebhard F, Huber-Lang M. New insights of an old defense system: structure, function, and clinical relevance of the complement system. Mol Med. 2011;17:317–329. doi: 10.2119/molmed.2010.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overall CM, Kleifeld O. Tumour microenvironment - opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 7.Puente XS, Sanchez LM, Overall CM, Lopez-Otin C. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan AP, Ghebrehiwet B. The plasma bradykinin-forming pathways and its interrelationships with complement. Mol Immunol. 2010;47:2161–2169. doi: 10.1016/j.molimm.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Davis AE, 3rd, Lu F, Mejia P. C1 inhibitor, a multi-functional serine protease inhibitor. Thromb Haemost. 2010;104:886–893. doi: 10.1160/TH10-01-0073. [DOI] [PubMed] [Google Scholar]
- 10.Cox JH, Overall CM. In: The Cancer Degradome: Proteases and Cancer Biology. Edwards D, Høyer-Hansen G, Blasi F, Sloane BF, editors. Springer New York: 2008. pp. 519–539. [Google Scholar]
- 11.McQuibban GA, Gong JH, Tam EM, McCulloch CA, Clark-Lewis I, Overall CM. Inflammation dampened by gelatinase A cleavage of monocyte chemoattractant protein-3. Science. 2000;289:1202–1206. doi: 10.1126/science.289.5482.1202. [DOI] [PubMed] [Google Scholar]
- 12.Corry DB, Rishi K, Kanellis J, Kiss A, Song Lz LZ, Xu J, Feng L, Werb Z, Kheradmand F. Decreased allergic lung inflammatory cell egression and increased susceptibilityto asphyxiation in MMP2-deficiency. Nat Immunol. 2002;3:347–353. doi: 10.1038/ni773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overall CM, Wrana JL, Sodek J. Transcriptional and post-transcriptional regulation of 72-kDa gelatinase/type IV collagenase by transforming growth factor-beta 1 in human fibroblasts. Comparisons with collagenase and tissue inhibitor of matrix metalloproteinase gene expression. J Biol Chem. 1991;266:14064–14071. [PubMed] [Google Scholar]
- 14.Lopez-Otin C, Overall CM. Protease degradomics: a new challenge for proteomics. Nat Rev Mol Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 15.Tam EM, Morrison CJ, Wu YI, Stack MS, Overall CM. Membrane protease proteomics: Isotope-coded affinity tag MS identification of undescribed MT1-matrix metalloproteinase substrates. Proc Natl Acad Sci U S A. 2004;101:6917–6922. doi: 10.1073/pnas.0305862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean RA, Overall CM. Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol Cell Proteomics. 2007;6:611–623. doi: 10.1074/mcp.M600341-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Butler GS, Dean RA, Tam EM, Overall CM. Pharmacoproteomics of a metalloproteinase hydroxamate inhibitor in breast cancer cells: dynamics of membrane type 1 matrix metalloproteinase-mediated membrane protein shedding. Mol Cell Biol. 2008;28:4896–4914. doi: 10.1128/MCB.01775-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.auf dem Keller U, Schilling O. Proteomic techniques and activity-based probes for the system-wide study of proteolysis. Biochimie. 2010;92:1705–1714. doi: 10.1016/j.biochi.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 19.Impens F, Colaert N, Helsens K, Plasman K, Van Damme P, Vandekerckhove J, Gevaert K. MS-driven protease substrate degradomics. Proteomics. 2010;10:1284–1296. doi: 10.1002/pmic.200900418. [DOI] [PubMed] [Google Scholar]
- 20.Kleifeld O, Doucet A, auf dem Keller U, Prudova A, Schilling O, Kainthan RK, Starr AE, Foster LJ, Kizhakkedathu JN, Overall CM. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat Biotechnol. 2010;28:281–288. doi: 10.1038/nbt.1611. [DOI] [PubMed] [Google Scholar]
- 21.Kleifeld O, Doucet A, Prudova A, Auf dem Keller U, Gioia M, Kizhakkedathu JN, Overall CM. Identifying and quantifying proteolytic events and the natural N terminome by terminal amine isotopic labeling of substrates. Nat Protoc. 2011;6:1578–1611. doi: 10.1038/nprot.2011.382. [DOI] [PubMed] [Google Scholar]
- 22.Prudova A, auf dem Keller U, Butler GS, Overall CM. Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics. Mol Cell Proteomics. 2010;9:894–911. doi: 10.1074/mcp.M000050-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.auf dem Keller U, Prudova A, Gioia M, Butler GS, Overall CM. A statistics-based platform for quantitative N-terminome analysis and identification of protease cleavage products. Mol Cell Proteomics. 2010;9:912–927. doi: 10.1074/mcp.M000032-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh T, Tanioka M, Yoshida H, Yoshioka T, Nishimoto H, Itohara S. Reduced angiogenesis and tumor progression in gelatinase A-deficient mice. Cancer Res. 1998;58:1048–1051. [PubMed] [Google Scholar]
- 25.Mueller MM. Inflammation in epithelial skin tumours: old stories and new ideas. Eur J Cancer. 2006;42:735–744. doi: 10.1016/j.ejca.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Kappelhoff R, auf dem Keller U, Overall CM. Analysis of the degradome with the CLIP-CHIP microarray. Methods Mol Biol. 2010;622:175–193. doi: 10.1007/978-1-60327-299-5_10. [DOI] [PubMed] [Google Scholar]
- 27.Alford JG, Stanley PL, Todderud G, Tramposch KM. Temporal infiltration of leukocyte subsets into mouse skin inflamed with phorbol ester. Agents Actions. 1992;37:260–267. doi: 10.1007/BF02028118. [DOI] [PubMed] [Google Scholar]
- 28.Furstenberger G, Richter H, Argyris TS, Marks F. Effects of the phorbol ester 4-O-methyl-12-O-tetradecanoylphorbol-13-acetate on mouse skin in vivo: evidence for its uselessness as a negative control compound in studies on the biological effects of phorbol ester tumor promoters. Cancer Res. 1982;42:342–348. [PubMed] [Google Scholar]
- 29.Zeeuwen PL. Epidermal differentiation: the role of proteases and their inhibitors. Eur J Cell Biol. 2004;83:761–773. doi: 10.1078/0171-9335-00388. [DOI] [PubMed] [Google Scholar]
- 30.Park GT, Lim SE, Jang SI, Morasso MI. Suprabasin, a novel epidermal differentiation marker and potential cornified envelope precursor. J Biol Chem. 2002;277:45195–45202. doi: 10.1074/jbc.M205380200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGrath JA, Uitto J. The filaggrin story: novel insights into skin-barrier function and disease. Trends Mol Med. 2008;14:20–27. doi: 10.1016/j.molmed.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi T, Liang SB, Matsuyoshi N, Zhou S, Miyachi Y, Sonobe H, Ohtsuki Y. Loss of T-cadherin (CDH13, H-cadherin) expression in cutaneous squamous cell carcinoma. Lab Invest. 2002;82:1023–1029. doi: 10.1097/01.lab.0000025391.35798.f1. [DOI] [PubMed] [Google Scholar]
- 33.Hansson M, Olsson I, Nauseef WM. Biosynthesis, processing, and sorting of human myeloperoxidase. Arch Biochem Biophys. 2006;445:214–224. doi: 10.1016/j.abb.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Helbig AO, Gauci S, Raijmakers R, van Breukelen B, Slijper M, Mohammed S, Heck AJ. Profiling of N-acetylated protein termini provides in-depth insights into the N-terminal nature of the proteome. Mol Cell Proteomics. 2010;9:928–939. doi: 10.1074/mcp.M900463-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Damme P, Arnesen T, Gevaert K. Protein alpha-N-acetylation studied by N-terminomics. FEBS J. 2011;278:3822–3834. doi: 10.1111/j.1742-4658.2011.08230.x. [DOI] [PubMed] [Google Scholar]
- 36.Martinez A, Traverso JA, Valot B, Ferro M, Espagne C, Ephritikhine G, Zivy M, Giglione C, Meinnel T. Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics. 2008;8:2809–2831. doi: 10.1002/pmic.200701191. [DOI] [PubMed] [Google Scholar]
- 37.Goetze S, Qeli E, Mosimann C, Staes A, Gerrits B, Roschitzki B, Mohanty S, Niederer EM, Laczko E, Timmerman E, Lange V, Hafen E, Aebersold R, Vandekerckhove J, Basler K, Ahrens CH, Gevaert K, Brunner E. Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLoS Biol. 2009;7:e1000236. doi: 10.1371/journal.pbio.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y, Lee Y, Kim B, Shin Y, Nam S, Kim P, Kim N, Chung WH, Kim J, Lee S. ECgene: an alternative splicing database update. Nucleic Acids Res. 2007;35:D99–103. doi: 10.1093/nar/gkl992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lange PF, Overall CM. TopFIND a knowledgebase linking protein termini with function. Nat Methods. 2011;8:703–704. doi: 10.1038/nmeth.1669. [DOI] [PubMed] [Google Scholar]
- 40.Polevoda B, Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J Mol Biol. 2003;325:595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- 41.Arnesen T, Van Damme P, Polevoda B, Helsens K, Evjenth R, Colaert N, Varhaug JE, Vandekerckhove J, Lillehaug JR, Sherman F, Gevaert K. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci U S A. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Damme P, Hole K, Pimenta-Marques A, Helsens K, Vandekerckhove J, Martinho RG, Gevaert K, Arnesen T. NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation. PLoS Genet. 2011;7:e1002169. doi: 10.1371/journal.pgen.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lange PF, Huesgen PF, Overall CM. TopFIND 2.0—Linking protein termini with proteolytic processing and modifications altering protein function. Nucleic Acids Res. 2012;40:D351–361. doi: 10.1093/nar/gkr1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gioia M, Foster LJ, Overall CM. Cell-based identification of natural substrates and cleavage sites for extracellular proteases by SILAC proteomics. Methods Mol Biol. 2009;539:131–153. doi: 10.1007/978-1-60327-003-8_8. [DOI] [PubMed] [Google Scholar]
- 45.Resing KA, Thulin C, Whiting K, al-Alawi N, Mostad S. Characterization of profilaggrin endoproteinase 1. A regulated cytoplasmic endoproteinase of epidermis. J Biol Chem. 1995;270:28193–28198. doi: 10.1074/jbc.270.47.28193. [DOI] [PubMed] [Google Scholar]
- 46.Meyer-Hoffert U. Reddish, scaly, and itchy: how proteases and their inhibitors contribute to inflammatory skin diseases. Arch Immunol Ther Exp (Warsz) 2009;57:345–354. doi: 10.1007/s00005-009-0045-6. [DOI] [PubMed] [Google Scholar]
- 47.Caubet C, Jonca N, Brattsand M, Guerrin M, Bernard D, Schmidt R, Egelrud T, Simon M, Serre G. Degradation of corneodesmosome proteins by two serine proteases of the kallikrein family, SCTE/KLK5/hK5 and SCCE/KLK7/hK7. J Invest Dermatol. 2004;122:1235–1244. doi: 10.1111/j.0022-202X.2004.22512.x. [DOI] [PubMed] [Google Scholar]
- 48.Deraison C, Bonnart C, Lopez F, Besson C, Robinson R, Jayakumar A, Wagberg F, Brattsand M, Hachem JP, Leonardsson G, Hovnanian A. LEKTI fragments specifically inhibit KLK5, KLK7, and KLK14 and control desquamation through a pH-dependent interaction. Mol Biol Cell. 2007;18:3607–3619. doi: 10.1091/mbc.E07-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heutinck KM, ten Berge IJ, Hack CE, Hamann J, Rowshani AT. Serine proteases of the human immune system in health and disease. Mol Immunol. 2010;47:1943–1955. doi: 10.1016/j.molimm.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 50.Ambrus G, Gal P, Kojima M, Szilagyi K, Balczer J, Antal J, Graf L, Laich A, Moffatt BE, Schwaeble W, Sim RB, Zavodszky P. Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on recombinant catalytic fragments. J Immunol. 2003;170:1374–1382. doi: 10.4049/jimmunol.170.3.1374. [DOI] [PubMed] [Google Scholar]
- 51.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 52.Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: a tail of a frog that became a prince. Nat Rev Mol Cell Biol. 2002;3:207–214. doi: 10.1038/nrm763. [DOI] [PubMed] [Google Scholar]
- 53.Dean RA, Butler GS, Hamma-Kourbali Y, Delbe J, Brigstock DR, Courty J, Overall CM. Identification of candidate angiogenic inhibitors processed by matrix metalloproteinase 2 (MMP-2) in cell-based proteomic screens: disruption of vascular endothelial growth factor (VEGF)/heparin affin regulatory peptide (pleiotrophin) and VEGF/Connective tissue growth factor angiogenic inhibitory complexes by MMP-2 proteolysis. Mol Cell Biol. 2007;27:8454–8465. doi: 10.1128/MCB.00821-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mehta D, Malik AB. Signaling mechanisms regulating endothelial permeability. Physiol Rev. 2006;86:279–367. doi: 10.1152/physrev.00012.2005. [DOI] [PubMed] [Google Scholar]
- 55.Parenti A, Morbidelli L, Ledda F, Granger HJ, Ziche M. The bradykinin/B1 receptor promotes angiogenesis by up-regulation of endogenous FGF-2 in endothelium via the nitric oxide synthase pathway. FASEB J. 2001;15:1487–1489. [PubMed] [Google Scholar]
- 56.McQuibban GA, Gong JH, Wong JP, Wallace JL, Clark-Lewis I, Overall CM. Matrix metalloproteinase processing of monocyte chemoattractant proteins generates CC chemokine receptor antagonists with anti-inflammatory properties in vivo. Blood. 2002;100:1160–1167. [PubMed] [Google Scholar]
- 57.Kruger A. Functional genetic mouse models: promising tools for investigation of the proteolytic internet. Biol Chem. 2009;390:91–97. doi: 10.1515/BC.2009.015. [DOI] [PubMed] [Google Scholar]
- 58.Wildes D, Wells JA. Sampling the N-terminal proteome of human blood. Proc Natl Acad Sci U S A. 2010;107:4561–4566. doi: 10.1073/pnas.0914495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Durchdewald M, Beyer TA, Johnson DA, Johnson JA, Werner S, auf dem Keller U. Electrophilic chemicals but not UV irradiation or reactive oxygen species activate Nrf2 in keratinocytes in vitro and in vivo. J Invest Dermatol. 2007;127:646–653. doi: 10.1038/sj.jid.5700585. [DOI] [PubMed] [Google Scholar]
- 60.Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z. CARMAweb: comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 2006;34:W498–503. doi: 10.1093/nar/gkl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007 http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- 62.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 63.Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, et al. CARMAweb: comprehensive R-and bioconductor-based web service for microarray data analysis. Nucleic Acids Research. 2006 doi: 10.1093/nar/gkl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morgan BP. Methods Mol Biol. Humana Press; Totowa, N.J.: 2000. Complement methods and protocols; p. ix.p. 268. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. iTRAQ labeling scheme for TAILS experiments.
Fig. S2. Skin inflammatory proteome (iTRAQ-preTAILS).
Fig. S3. Distribution of acetylated and non-acetylated N termini with preferred residues.
Fig. S4. Dynamics of actin acetylation revealed by iTRAQ-TAILS in vivo.
Fig. S5. Consensus sequence for cyclized N-Termini after pyroglutamate formation.
Fig. S6. Original mature N terminus of stromelysin-1 (MMP3) identified by iTRAQ-TAILS.
Fig. S7. Original mature N terminus of S100A9 identified by iTRAQ-TAILS.
Fig. S8. Densitometric analysis of Western blotting analyses shown in Fig. 5B and C.
Fig. S9. Western blot analysis of haptoglobin abundance in livers of Mmp2-/- mice and wild-type controls upon mustard oil induced skin inflammation.
Fig. S10. Prediction of signal peptide removal for mannose-binding protein A (MBL1).
Tables S1 to S19. Spectra files.