Abstract
Epidemiological and genetic data support the notion that schizophrenia and bipolar disorder share genetic risk factors. In our previous genome-wide association (GWA) study, meta-analysis and follow-up (totaling as many as 18,206 cases and 42,536 controls), we identified four loci showing genome-wide significant association with schizophrenia. Here we consider a mixed schizophrenia and bipolar disorder (psychosis) phenotype (addition of 7,469 bipolar disorder cases, 1,535 schizophrenia cases, 333 other psychosis cases, 808 unaffected family members and 46,160 controls). Combined analysis reveals a novel variant at 16p11.2 showing genome-wide significant association (rs4583255[T], OR = 1.08, P = 6.6 × 10−11). The new variant is located within a 593 kb region that substantially increases risk of psychosis when duplicated. In line with the association of the duplication with reduced body mass index (BMI), rs4583255[T] is also associated with lower BMI (P = 0.0039 in the public GIANT consortium dataset; P = 0.00047 in 22,651 additional Icelanders).
Keywords: schizophrenia, bipolar disorder, association, 16p11.2, cross-disorder
Introduction
Two structural variants, a balanced t(1;11) translocation interrupting the DISC1 gene and a microdeletion at 22q11.2, were the first genetic polymorphisms to show compelling evidence of association with schizophrenia1, 2. More recently, additional microdeletions and microduplications conferring risk of schizophrenia and, in some cases, bipolar disorder have been uncovered3-10. These copy number variants (CNVs) confer high to moderate relative risk, however, because they typically change copy number of multiple genes, and may also affect regulation of genes at their margins, they do not generally implicate individual genes.
Common single nucleotide polymorphisms (SNPs) are currently, in addition to structural variants, convincing risk factors for schizophrenia and bipolar disorder, with alleles at more than 20 loci reported to show genome-wide significant association with at least one of the disorders11-29. None of these low-risk variants are located inside structural polymorphisms previously shown to be susceptibility factors for schizophrenia or bipolar disorder. Nevertheless, first principles and data from other disorders predict the existence of common variants conferring risk through the same genes as rare structural alleles30. The identification of common risk variants within CNV regions may aid in uncovering the causal gene or genes of a CNV, or help to elucidate other aspects of a CNV’s association with disease.
Two loci have been reported to harbor common alleles showing genome-wide significant association with both schizophrenia and bipolar disorder13, 16, 23, 24. In addition, several common variants initially displaying genome-wide significant association with one of the disorders have been shown, in subsequent studies, to confer risk of the other31, 32. Investigations considering schizophrenia and bipolar disorder as a single phenotype also support shared risk alleles16, 19, 22, and an overlapping polygenetic component has been described by several studies21, 28. These genetic data are consistent with current epidemiological investigations, which predict shared genetic risk factors for schizophrenia and bipolar disorder33.
Previously, we carried out a schizophrenia GWA study, SGENE-plus, followed by meta-analysis of the top 1500 results with data from the International Schizophrenia Consortium (ISC) and the Molecular Genetics of Schizophrenia (MGS) group15. Loci having P values less than 1 × 10−4 (covered by 39 SNPs located in 33 genomic regions) were followed up in a data set of up to 10,260 schizophrenia cases and 23,500 controls14. In this work , we broaden our phenotype of interest to psychosis (schizophrenia, bipolar disorder and related psychoses), examining the same group of follow-up SNPs in a data set augmented by 7,469 bipolar disorder cases, 1,535 schizophrenia cases, 333 other psychosis cases, 808 unaffected family members and 46,160 controls.
Materials and methods
Samples
The genome-wide typed (“SGENE-plus”; 2,663 cases and 13,498 controls) and meta-analysis (“SGENE-plus+ISC+MGS”) samples (in total, 7,946 cases and 19,036 controls) used here were identical to those used in our previous schizophrenia GWA study and meta-analysis15 . The primary psychosis follow-up samples employed consisted of follow-up samples from our previous GWA follow-up study (9,246 schizophrenia cases and 22,356 controls)14, plus an additional 9,337 psychosis cases (1,535 schizophrenia, 7,469 bipolar disorder, 333 related psychoses) and 46,968 controls/unaffected family members. The primary follow-up samples were genotyped or imputed for all follow-up markers. The secondary follow-up samples consisted of 1,014 cases and 1,144 controls from the Göttingen Research Association for Schizophrenia (GRAS)34, 35 study. These samples, which also had been used for secondary follow-up in our previous GWA follow-up study14, were genotyped for SNPs that were genome-wide significant in the combined meta-analysis and primary follow-up samples. Table 1 summarizes the schizophrenia and psychosis datasets used in previous and current work, and Supplementary Table 1 includes details on the individual study groups. The autism samples (3,672 cases, 16,103 controls, 4,206 family members) derived from AGP, AGRE and nine European study groups (Supplementary Table 2). Further information on ascertainment and diagnosis for the psychosis and autism samples is provided in the Supplementary Material.
Table 1. Relevant datasets.
|
N
|
||||||
|---|---|---|---|---|---|---|
| Dataset | case phenotype |
markers examined |
cases | controls + family members |
initial use | overlap with other sets |
| SGENE-plus GWAS | SZ | 314,868 | 2,663 | 13,498 | Stefansson15 | no |
| SGENE-plus+ISC+MGS | SZ | 1,500 | 7,946 | 19,036 | Stefansson15 | includes SGENE-plus GWAS |
| primary schizoprenia follow-up |
SZ | 39 | 9,246 | 22,356 | Steinberg14 | no |
| primary psychosis follow-up |
SZ, BP, rel | 39 | 18,583 | 69,324 | this work | includes primary schizophrenia follow-up |
| secondary follow-up | SZ | 8; 11 | 1,014 | 1,144 | Steinberg14 | no |
Genotyping and association analysis
Genotyping was carried out using Illumina and Affymetrix genome-wide arrays,Centaurus assays (Nanogen), Taqman assays, the Sequenom MassArray iPLEX genotyping system and the Roche LightCycler480 system (Supplementary Tables 1 and 2). Quality control and imputation were performed, by study group, as described in the Supplementary Methods. Case-control or family-based association analyses were carried out for each study group. For the case-control analyses, population stratification was controlled for using genomic control or principal components. Summary statistics from the various study groups were combined as described previously15. BMI measurements were adjusted for age and sex, and inverse standard normal transformed. Analysis was carried out by regressing the adjusted, transformed data on rs4583255[T] count.
Expression Analysis
For the three brain data sets36-38, expression levels were inverse normal transformed and regressed on the number of rs4583255-T alleles with gender, age at death, post-mortem interval, brain source, expression experiment batch, pH (Colantuoni et al36 only), sample expression level based on the total number of transcripts detected (Webster et al38 only) and Alzheimer’s disease patient status (Webster et al38 only) as covariates. To incorporate data from different brain regions (Gibbs et al37) or different probes (KCTD13 in Colantuoni et al36) derived from the same individual, a mixed-effects model with individual as a random effect was used. Results from the three data sets were combined using inverse-variance weighted meta-analysis. The Dutch whole blood data set included control samples from two studies39, 40. Analysis was performed using linear regression in Plink41 taking age and gender as covariates. The Icelandic blood data set has been described previously42, and analysis was carried out as detailed in that work42.
Results
We assembled a psychosis (schizophrenia, bipolar disorder and related psychoses) primary follow-up dataset made up of 36 study groups containing a total of 18,583 cases, 68,516 controls and 808 unaffected family members (Supplementary Table 1). In each study group, allelic association analysis was carried out for 39 SNPs from 33 genomic regions (these SNPs covered P values less than 1 × 10−4 in the SGENE-plus+ISC+MGS meta-analysis at r2 = 0.3). Results from the various study groups were combined using inverse-variance weighted meta-analysis.
At 31 of the 33 loci, ORs in the psychosis follow-up group were in the same direction as in the discovery data set (SGENE-plus+ISC+MGS) (Supplementary Table 3). A similar pattern had been observed in the schizophrenia follow-up set—ORs were in the same direction at 30 of the 33 loci14. These results indicate that the set of variants chosen for follow-up was enriched for risk alleles (P = 7.0 × 10−7 for schizophrenia, and P = 6.5 × 10−8 for psychosis).
Next, we performed a joint analysis of the discovery and psychosis follow-up sets. To account for testing two phenotypes (schizophrenia and psychosis), the genome-wide significance threshold was set at P < (5 × 10−8)/2, or 2.5 × 10−8. Five SNPS, residing at three loci, exceeded this threshold (Supplementary Table 3). Two of the loci—the MHC region and 11q21.2 near NRGN—had been genome-wide significant in the previous schizophrenia analysis; a third locus, in TAOK2 at 16p11.2, was novel (Supplementary Table 3). Following the addition of data from a further 1,014 schizophrenia cases and 1,144 controls, the variant at the novel locus, rs4583255[T], was associated with psychosis with increased significance (OR = 1.08, P = 6.6 × 10−11, Table 1). rs4583255[T]’s association with psychosis fit the multiplicative model (P = 0.42), and there was no evidence of OR heterogeneity (P = 0.71, I2 = 0, Supplementary Table 4).
In examination of the follow-up samples by diagnosis, the novel variant, rs4583255[T], showed significant association with both schizophrenia and bipolar disorder (P = 0.0011 and 0.00026), with OR of 1.06 and 1.08, respectively (independent controls were used for the two analyses; see Supplementary Table 5). We also investigated association with bipolar disorder for variants that had shown genome-wide significant association with schizophrenia in our previous study14. Following correction for eight tests, rs12807809[T], near NRGN, was significantly associated with bipolar disorder (P = 0.0023) with an OR identical to that of the schizophrenia follow-up samples (OR = 1.09). The remaining schizophrenia susceptibility variants did not show nominally-significant association with bipolar disorder—yet OR confidence intervals for the two disorders overlapped for at least some variants at all loci (Supplementary Table 5).
Intriguingly, the newly-identified SNP is located in a nearly 600 kb region that confers risk of schizophrenia and bipolar disorder when duplicated5, 6, 28. Copy number gain of the region also is associated with autism6, 43-45, reduced head circumference46, 47, and low BMI47. We obtained large data sets to examine association of rs4583255[T] with both autism and BMI. Based on 3,672 cases, 16,103 controls and 4,206 unaffected family members from the Autism Genetic Resource Exchange (AGRE), the Autism Genome Project (AGP) and nine European study groups (Supplementary Table 2), we found no evidence of association with autism spectrum disorder (ASD), strict autism or multiplex ASD (ASD, OR = 1.00, P = 0.98; strict autism, OR = 1.02, P = 0.66; multiplex ASD, OR = 1.07, P = 0.22; Supplementary Table 6), although power to detect association at the OR found in the follow-up psychosis samples was modest (at a 0.05 significance level, power was about 57% for ASD, 42% for strict autism, and 23% for multiplex ASD). In contrast, we found significant association of rs4583255[T] with low BMI in the published GIANT consortium GWAS dataset of 123,865 individuals48 (P = 0.0039) and in 22,651 Icelanders who were not included in the GIANT study (P = 0.00047).
Recently, a study examining the effect of altered expression of 16p11.2 CNV region genes on zebrafish head size identified KCTD13 as the major driver of head size change, with MAPK3 and MVP named as possible modifiers49. These results motivated us to examine association of rs4583255[T] with expression of KCTD13, MAPK3, and MVP in human brain. Using data from three publicly-available data sets with at least 50 European-ancestry adult brains each (total N = 565)36-38, we found that rs4583255[T] was significantly associated with expression of MAPK3 (effect = 0.12 s.d., P = 0.011), but not significantly associated with expression of KCTD13 or MVP (Supplementary Table 7). We also investigated association of rs4583255[T] with gene expression in blood using data sets from Iceland (N=972)42 and the Netherlands (N = 437)39, 40. Consistent with the brain results, rs4583255[T] was significantly associated with higher expression of MAPK3 (for Iceland , P = 9.4 × 10−15; for the Netherlands, P = 0.014 for probe 3870601,and P = 0.042 for probe 234040), but not significantly associated with expression of KCTD13 or MVP.
Discussion
In this study, we uncovered a novel variant at 16p11.2, rs4583255[T], showing genome-wide significant association with psychosis (OR = 1.08, P = 6.6 × 10−11). In follow-up samples, ORs were similar for schizophrenia and bipolar disorder (OR = 1.06 and 1.08, respectively), and association was significant for both (P = 0.0011 and P = 0.00026, respectively). Thus, rs4583255[T] is a compelling example of a genetic variant that confers risk across traditional diagnostic boundaries.
Among the variants that showed genome-wide significant association with schizophrenia in our previous study14,only rs12807809[T] showed significant association with bipolar disorder in the current work. Nevertheless, OR confidence intervals for schizophrenia and bipolar disorder overlapped for most risk alleles. Very large data sets will be necessary to establish conclusively where these variants fall on the spectrum of conferring risk of one disorder, exclusively, to conferring equal risk of either.
To our knowledge, this is the first case in which a common risk allele showing genome-wide significant association with psychosis has turned out to be located within a CNV that had been previously associated with psychosis. Both copy number gain and loss of the 16p11.2 region are associated with multiple phenotypes. Duplication is associated with psychosis5, 6, 28, both copy number gain and loss are associated with autism and developmental delay6, 43-45, and duplication and deletion lead to reduction and enlargement, respectively, of head circumference and BMI46, 47.
In this work, we found that rs4583255[T] also confers risk of reduced BMI (P = 0.0039 in GIANT, P = 0.00047 in additional Icelanders). This result supports the suggestion, made previously47, that the duplication’s effects on psychosis and BMI have a single origin, presumably in the brain. We did not find evidence of association of rs4583255[T] with autism, although we were somewhat underpowered to detect an effect of the same size as in psychosis, especially for sub-phenotypes.
We found that rs4583255[T] was associated with increased expression in adult brain and blood of MAPK3, one of the 16p11.2 genes identified as involved in causing head circumference changes in zebrafish49. Caution is required in interpretation of this result, however, as the significance in brain is marginal, and, furthermore, gene expression in the pre-adult brain may be most relevant for the development of psychosis. Data from only extremely small numbers of European-ancestry brains at pre-adult stages were available; thus, investigation of the association of rs4583255[T] with gene expression at these stages was precluded.
In conclusion, in this work, we broadened our phenotype of interest to psychosis, identifying a new common risk allele, rs4583255[T], with similar ORs for schizophrenia and bipolar disorder. The novel variant is located within a duplication previously associated with psychosis, and, in line with the duplication’s effects, also confers risk of low BMI. In the future, knowledge of this common variant association may prove useful to studies aimed at further understanding the mechanism through which the duplication exerts its effects on neurodevelopmental and anthropomorphic phenotypes.
Supplementary Material
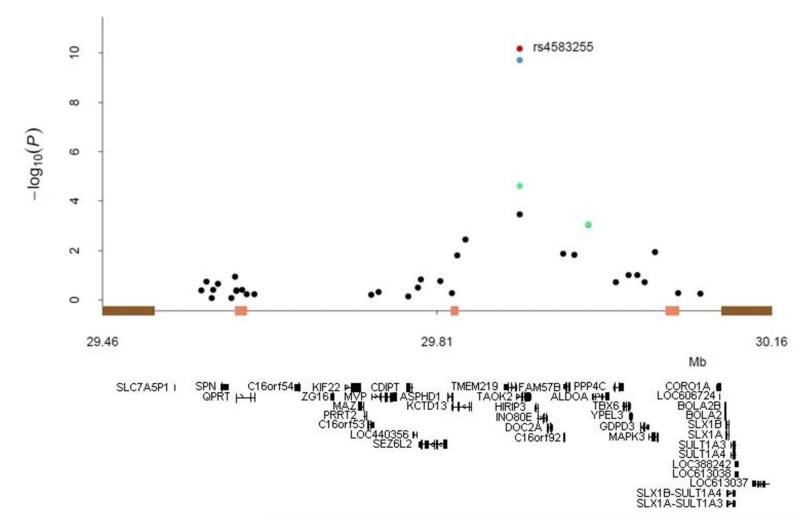
Figure 1.
Association results and structure of the 16p11.2 region. Bars on the x-axis indicate segmental duplications (brown) and recombination hotspots (pink). Association results are illustrated for SGENE-plus (black), SGENE-plus+MGS+ISC (green), SGENE-plus+MGS+ISC plus the primary psychosis follow-up (blue), and SGENE-plus+MGS+ISC plus the primary psychosis and secondary schizophrenia follow-up (red). RefSeq genes in the region are shown below the plot.
Table 2. Genome-wide association of rs4583255[T] with psychosis.
|
N
|
|||||
|---|---|---|---|---|---|
| study group | cases | controls | family members |
OR (95% CI) | P value |
| SGENE-plus+ISC+MGS (SZ) | 7,946 | 19,036 | 0 | 1.10 (1.05, 1.15) | 2.5 × 10−5 |
| primary psychosis follow-up (SZ,BP,rel) | 18,583 | 68,516 | 808 | 1.07 (1.04, 1.10) | 9.2 × 10−7 |
| secondary follow-up (SZ) | 1,014 | 1,144 | 0 | 1.10 (0.97, 1.24) | 0.14 |
| combined | 27,543 | 88,696 | 808 | 1.08 (1.05, 1.10) | 6.6 × 10−11 |
SZ, schizophrenia; BP, bipolar disorder; rel, related psychoses; OR, odds ratio; CI, confidence interval
Acknowledgements
We would like to thank the subjects, their families and the recruitment centre staff. We would also like to acknowledge the help of Maria Dolores Moltó (Genetics Department, Valencia University, CIBERSAM), Eduardo Paz and Ramón Ramos-Ríos (Complexo Hospitalario de Santiago), and the contribution of Fundación Botín.
This study makes use of seven external, publicly-available datasets. First, it makes use of data generated by the Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) project whose principal investigators were Jeffrey A. Lieberman, M.D., T. Scott Stroup, M.D., M.P.H., and Joseph P. McEvoy, M.D.. The CATIE trial was funded by a grant from the National Institute of Mental Health (N01 MH900001) along with MH074027 (PI PF Sullivan). Genotyping was funded by Eli Lilly and Company. Second, the GAIN/BiGs datasets used in this work were obtained from the database of Genotypes and Phenotypes (dbGaP) found at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000017.v3.p1. Third, the study uses samples genotyped using the Ilumina 550K platform by the Pritzker Consortium, supported by the Pritzker Neuropsychiatric Disorders Research Fund L.L.C. The Pritzker Consortium includes scientists at the University of Michigan (H. Akil and S. J. Watson, Site Directors, and Michael Boehnke, lead on bipolar genotyping effort); Stanford University (Rick Myers and Alan Schatzberg, Site Directors); the University of California at Davis (Ted Jones, Site Director); the University of California at Irvine (William Bunney, Site Director); and the Weill Medical College of Cornell University (Jack Barchas, Site Director). Fourth, the work uses data from the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD) project, led by Gary Sachs, M.D., and coordinated by Massachusetts General Hospital in Boston, MA (NIMH grant number was 2N01MH080001-001). Fifth, this study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113 and 085475. Sixth, we gratefully acknowledge the resources provided by the Autism Genetic Resource Exchange (AGRE) Consortium* and the participating AGRE families. The Autism Genetic Resource Exchange is a program of Autism Speaks and is supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to Clara M. Lajonchere (PI). Seventh, the Autism Genome Project (AGP) data sets used for the analysis described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number, phs000267.v1.p1. Submission of the data to dbGaP was provided by Dr. Bernie Devlin on behalf of the Autism Genome Project (AGP). Collection and submission of the data to dbGaP were supported by a grant from the Medical Research Council (G0601030) and the Wellcome Trust (075491/Z/04), Anthony P. Monaco, P.I., University of Oxford.
This work was also supported by the European Union [grant numbers LSHM-CT-2006-037761 (Project SGENE), PIAP-GA-2008-218251 (Project PsychGene), HEALTH-F2-2009-223423 (Project PsychCNVs), HEALTH-F4-2009-242257 (Project ADAMS), IMI-JU-NewMeds]; the National Genome Research Network of the German Federal Ministry of Education and Research (BMBF) [grant numbers 01GS08144 (MooDS-Net), 01GS08147 (NGFNplus)]; the National Institute of Mental Health [R01 MH078075, and N01 MH900001, MH074027 to the Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) project]; the Centre of Excellence for Complex Disease Genetics of the Academy of Finland (grant numbers 213506, 129680); the Biocentrum Helsinki Foundation and Research Program for Molecular Medicine, Faculty of Medicine, University of Helsinki; the Stanley Medical Research Institute; the Danish Council for Strategic Research (grant number 2101-07-0059); H. Lundbeck A/S; the Research Council of Norway (grant number 163070/V50); the Danish Medical Research Council; the South-East Norway Health Authority (grant number 2004-123); the Medical Research Council; Ministerio de Sanidad y Consumo, Spain (grant number PI081522 to J.C.); Xunta de Galicia (grant number 08CSA005208PR to A. Carracedo); the Swedish Research Council; the Wellcome Trust (Wellcome Trust grants 085475/B/08/Z and 085475/Z/08/Z as part of the Wellcome Trust Case Control Consortium 2); the Max Planck Society; Saarland University (grant number T6 03 10 00 - 45 to C.M.F.); the Netherlands Foundation for Brain Research (Hersenstichting) (grant number 2008(1).34 to M. Poot); and Eli Lilly and Company (genotyping for CATIE and part of the TOP sample).
For further acknowledgements, see the Supplementary Material.
Appendix.
Genetic Risk and Outcome in Psychosis (GROUP)
René S. Kahn1, Don H. Linszen2, Jim van Os3, Durk Wiersma4, Richard Bruggeman4, Wiepke Cahn1, Lieuwe de Haan2, Lydia Krabbendam3, & Inez Myin-Germeys3
1Department of Psychiatry, Rudolf Magnus Institute of Neuroscience, University Medical Center Utrecht, Postbus 85060, Utrecht, the Netherlands
2Academic Medical Centre University of Amsterdam, Department of Psychiatry, Amsterdam, NL326 Groot-Amsterdam, the Netherlands
3Maastricht University Medical Centre, South Limburg Mental Health Research and Teaching Network, 6229 HX Maastricht, the Netherlands
4University Medical Center Groningen, Department of Psychiatry, University of Groningen, PO Box 30.001, 9700 RB Groningen, the Netherlands
Wellcome Trust Case Control Consortium 2
Management Committee
Peter Donnelly (Chair)1,2, Ines Barroso (Deputy Chair)3, Jenefer M Blackwell4, 5, Elvira Bramon6 , Matthew A Brown7 , Juan P Casas8 , Aiden Corvin9, Panos Deloukas3, Audrey Duncanson10, Janusz Jankowski11, Hugh S Markus12, Christopher G Mathew13, Colin NA Palmer14, Robert Plomin15, Anna Rautanen1, Stephen J Sawcer16, Richard C Trembath13, Ananth C Viswanathan17, Nicholas W Wood18
Data and Analysis Group
Chris C A Spencer1, Gavin Band1, Céline Bellenguez1, Colin Freeman1, Garrett Hellenthal1, Eleni Giannoulatou1, Matti Pirinen1, Richard Pearson1, Amy Strange1, Zhan Su1, Damjan Vukcevic1, Peter Donnelly1,2
DNA, Genotyping, Data QC and Informatics Group
Cordelia Langford3, Sarah E Hunt3, Sarah Edkins3, Rhian Gwilliam3, Hannah Blackburn3, Suzannah J Bumpstead3, Serge Dronov3, Matthew Gillman3, Emma Gray3, Naomi Hammond3, Alagurevathi Jayakumar3, Owen T McCann3, Jennifer Liddle3, Simon C Potter3, Radhi Ravindrarajah3, Michelle Ricketts3, Matthew Waller3, Paul Weston3, Sara Widaa3, Pamela Whittaker3, Ines Barroso3, Panos Deloukas3.
Publications Committee
Christopher G Mathew (Chair)13, Jenefer M Blackwell4,5, Matthew A Brown7, Aiden Corvin9, Chris C A Spencer1
1 Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, UK; 2 Dept Statistics, University of Oxford, Oxford OX1 3TG, UK; 3 Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, UK; 4 Telethon Institute for Child Health Research, Centre for Child Health Research, University of Western Australia, 100 Roberts Road, Subiaco, Western Australia 6008; 5 Cambridge Institute for Medical Research, University of Cambridge School of Clinical Medicine, Cambridge CB2 0XY, UK; 6 Department of Psychosis Studies, NIHR Biomedical Research Centre for Mental Health at the Institute of Psychiatry, King’s College London and The South London and Maudsley NHS Foundation Trust, Denmark Hill, London SE5 8AF, UK; 7 University of Queensland Diamantina Institute, Brisbane, Queensland, Australia; 8 Dept Epidemiology and Population Health, London School of Hygiene and Tropical Medicine, London WC1E 7HT and Dept Epidemiology and Public Health, University College London WC1E 6BT, UK; 9 Neuropsychiatric Genetics Research Group, Institute of Molecular Medicine, Trinity College Dublin, Dublin 2, Eire; 10 Molecular and Physiological Sciences, The Wellcome Trust, London NW1 2BE; 11 Department of Oncology, Old Road Campus, University of Oxford, Oxford OX3 7DQ, UK , Digestive Diseases Centre, Leicester Royal Infirmary, Leicester LE7 7HH, UK and Centre for Digestive Diseases, Queen Mary University of London, London E1 2AD, UK; 12 Clinical Neurosciences, St George’s University of London, London SW17 0RE; 13 King’s College London Dept Medical and Molecular Genetics, King’s Health Partners, Guy’s Hospital, London SE1 9RT, UK; 14 Biomedical Research Centre, Ninewells Hospital and Medical School, Dundee DD1 9SY, UK; 15 King’s College London Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Denmark Hill, London SE5 8AF, UK; 16 University of Cambridge Dept Clinical Neurosciences, Addenbrooke’s Hospital, Cambridge CB2 0QQ, UK; 17 NIHR Biomedical Research Centre for Ophthalmology, Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology, London EC1V 2PD, UK; 18 Dept Molecular Neuroscience, Institute of Neurology, Queen Square, London WC1N 3BG, UK.
Footnotes
Conflict of interest. The authors declare no conflict of interest.
Supplementary information is available at Molecular Psychiatry’s website
References
- 1.Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ, Millar JK. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13(1):36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- 2.Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci U S A. 1995;92(17):7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ingason A, Rujescu D, Cichon S, Sigurdsson E, Sigmundsson T, Pietilainen OP, et al. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 16(1):17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455(7210):237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 168(3):302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41(11):1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulle JG, Dodd AF, McGrath JA, Wolyniec PS, Mitchell AA, Shetty AC, et al. Microdeletions of 3q29 confer high risk for schizophrenia. Am J Hum Genet. 87(2):229–236. doi: 10.1016/j.ajhg.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rujescu D, Ingason A, Cichon S, Pietilainen OP, Barnes MR, Toulopoulou T, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18(5):988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455(7210):232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vacic V, McCarthy S, Malhotra D, Murray F, Chou HH, Peoples A, et al. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 471(7339):499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue WH, Wang HF, Sun LD, Tang FL, Liu ZH, Zhang HX, et al. Genome-wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat Genet. 43(12):1228–1231. doi: 10.1038/ng.979. [DOI] [PubMed] [Google Scholar]
- 12.Williams HJ, Norton N, Dwyer S, Moskvina V, Nikolov I, Carroll L, et al. Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol Psychiatry. 16(4):429–441. doi: 10.1038/mp.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vassos E, Steinberg S, Cichon S, Breen G, Sigurdsson E, Andreassen OA, et al. Replication Study and Meta-Analysis in European Samples Supports Association of the 3p21.1 Locus with Bipolar Disorder. Biol Psychiatry. doi: 10.1016/j.biopsych.2012.02.040. [DOI] [PubMed] [Google Scholar]
- 14.Steinberg S, de Jong S, Andreassen OA, Werge T, Borglum AD, Mors O, et al. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum Mol Genet. 20(20):4076–4081. doi: 10.1093/hmg/ddr325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460(7256):744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 43(10):977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y, Li Z, Xu Q, Wang T, Li T, Shen J, et al. Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nat Genet. 43(12):1224–1227. doi: 10.1038/ng.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460(7256):753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 43(10):969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rietschel M, Mattheisen M, Degenhardt F, Kahn RS, Linszen DH, Os JV, et al. Association between genetic variation in a region on chromosome 11 and schizophrenia in large samples from Europe. Mol Psychiatry. doi: 10.1038/mp.2011.80. [DOI] [PubMed] [Google Scholar]
- 21.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40(9):1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 23.McMahon FJ, Akula N, Schulze TG, Muglia P, Tozzi F, Detera-Wadleigh SD, et al. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21.1. Nat Genet. 42(2):128–131. doi: 10.1038/ng.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamshere ML, Walters JT, Smith R, Richards AL, Green E, Grozeva D, et al. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry. doi: 10.1038/mp.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40(9):1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cichon S, Muhleisen TW, Degenhardt FA, Mattheisen M, Miro X, Strohmaier J, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 88(3):372–381. doi: 10.1016/j.ajhg.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen DT, Jiang X, Akula N, Shugart YY, Wendland JR, Steele CJ, et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry. doi: 10.1038/mp.2011.157. [DOI] [PubMed] [Google Scholar]
- 28.Bergen SE, O’Dushlaine CT, Ripke S, Lee PH, Ruderfer DM, Akterin S, et al. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol Psychiatry. 17(9):880–886. doi: 10.1038/mp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13(2):197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322(5903):881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muhleisen TW, Mattheisen M, Strohmaier J, Degenhardt F, Priebe L, Schultz CC, et al. Association between schizophrenia and common variation in neurocan (NCAN), a genetic risk factor for bipolar disorder. Schizophr Res. doi: 10.1016/j.schres.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Williams HJ, Craddock N, Russo G, Hamshere ML, Moskvina V, Dwyer S, et al. Most genome-wide significant susceptibility loci for schizophrenia and bipolar disorder reported to date cross-traditional diagnostic boundaries. Hum Mol Genet. 20(2):387–391. doi: 10.1093/hmg/ddq471. [DOI] [PubMed] [Google Scholar]
- 33.Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373(9659):234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papiol S, Begemann M, Rosenberger A, Friedrichs H, Ribbe K, Grube S, et al. A phenotype-based genetic association study reveals the contribution of neuregulin1 gene variants to age of onset and positive symptom severity in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 156B(3):340–345. doi: 10.1002/ajmg.b.31168. [DOI] [PubMed] [Google Scholar]
- 35.Ribbe K, Friedrichs H, Begemann M, Grube S, Papiol S, Kastner A, et al. The cross-sectional GRAS sample: a comprehensive phenotypical data collection of schizophrenic patients. BMC Psychiatry. 10:91. doi: 10.1186/1471-244X-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 478(7370):519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 6(5):e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84(4):445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 42(4):295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saris CG, Horvath S, van Vught PW, van Es MA, Blauw HM, Fuller TF, et al. Weighted gene co-expression network analysis of the peripheral blood from Amyotrophic Lateral Sclerosis patients. BMC Genomics. 2009;10:405. doi: 10.1186/1471-2164-10-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452(7186):423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 43.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358(7):667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 44.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17(4):628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 46.Shinawi M, Liu P, Kang SH, Shen J, Belmont JW, Scott DA, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 47(5):332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacquemont S, Reymond A, Zufferey F, Harewood L, Walters RG, Kutalik Z, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 478(7367):97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Golzio C, Willer J, Talkowski ME, Oh EC, Taniguchi Y, Jacquemont S, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 485(7398):363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.