Abstract
NOD2 is a cytosolic pathogen recognition receptor that regulates susceptibility to a variety of infections and chronic diseases. Burkholderia pseudomallei, a facultative intracellular bacterium, causes the tropical infection melioidosis. We hypothesized that NOD2 may participate in host defense in melioidosis. We performed a series of in vitro assays and in vivo experiments, and analyzed the association of human genetic variation with infection to delineate the contribution of NOD2 to the host response to B. pseudomallei. We found that transfection with NOD2 mediated NF-κB activation induced by B. pseudomallei stimulation of HEK293 cells. After low dose inoculation with aerosolized B. pseudomallei, Nod2-deficient mice showed impaired clinical responses and permitted greater bacterial replication in the lung and dissemination to the spleen compared to wild type mice. IL-6 and KC levels were higher in the lungs of Nod2-deficient mice. In a cohort of 1,562 Thai subjects, a common genetic polymorphism in the NOD2 region, rs7194886, was associated with melioidosis and this effect was most pronounced in women. rs7194886 was not associated with differences in cytokine production induced by whole blood stimulation with the NOD2 ligand, MDP, or B. pseudomallei. These findings are the first to characterize the role of NOD2 in host defense in mammalian melioidosis.
Keywords: Burkholderia pseudomallei, melioidosis, NOD2, innate immunity, genetic variation, animal model, pneumonia, sepsis
Introduction
Melioidosis is a severe infection caused by the soil saprophyte Burkholderia pseudomallei. Endemic in parts of southeast Asia and northern Australia, the disease is often characterized by severe sepsis, indicative of a dysregulated host immune response (1). Pneumonia is a common manifestation of disease, either secondarily due to hematogenous spread, or due to primary inhalation of bacteria (1).
Toll-like receptors (TLRs), membrane-associated pathogen recognition receptors (PRRs), have already been implicated in governing the host response in murine and human melioidosis (2–5). TLR2, a sensor of lipopeptides, is deleterious in mice infected with B. pseudomallei via the intranasal route, an effect that is most apparent several days following infection (4). TLR4 recognizes LPS and contributes to bacterial containment in the first 24 hours of murine respiratory infection with B. pseudomallei or with the related organism B. thailandensis, but has no effect on mortality (2, 4, 6). However, human genetic polymorphisms in TLR4 are associated with susceptibility to melioidosis (3). In patients with melioidosis, a nonsense polymorphism in TLR5, a flagellin sensor, is associated with survival (5). A complementary set of PRRs - the nucleotide binding oligomerization domain (NOD)-like receptors (NLRs) - exists in the cytosol. These proteins contain an N-terminal caspase and recruitment domain (CARD), a central nucleotide binding oligomerization domain, and a C-terminal leucine rich repeat. NLRs can be broadly divided into 1) those (such as NLRC4) that contribute to the assembly of the inflammasome, a molecular platform that permits caspase-1 activation, and 2) the non-inflammasome NLRs, such as NOD1 and NOD2 (7). NOD2 is expressed in monocytes, macrophages, dendritic cells, intestinal Paneth cells, and lung epithelial cells (8, 9). Upon ligation of bacterial cell wall component muramyl dipeptide (MDP) by NOD2, CARD proteins RIP2/RICK and CARD9 are recruited and signaling occurs via MAPK and NF-κB pathways (10). Because of the cytosolic location of NLRs, they may function synergistically with TLRs to promote a pro-inflammatory state (11).
B. pseudomallei is a facultative intracellular pathogen that readily escapes from endosomes into the cytosol (12), highlighting the potential importance of NLRs and related cytosolic signaling pathways in host defense in melioidosis. In mice, the type III secretion system of B. pseudomallei is detected by NLRC4 and resistance to murine melioidosis requires both NLRC4 and caspase-1 (13–15). B. pseudomallei up-regulates NOD2 expression in a mouse macrophage cell line (16) but otherwise the function of NOD2 in melioidosis is largely unknown. Given the established function of specific TLRs in melioidosis and potential for synergistic effects of cytosolic receptors, we hypothesized that NOD2 may play a role in modulating host defense in melioidosis. To test this hypothesis, we examined the role of NOD2 in regulating innate immune responses to B. pseudomallei in vitro and in a murine model of respiratory melioidosis, and we tested the association of human genetic NOD2 polymorphisms with disease in a cohort of Thai subjects.
Materials and Methods
Bacteria
B. pseudomallei 1026b (17) was used for all experiments. Heat-killing was accomplished by growing bacteria for 6 h shaking at 180 rpm at 37°C in Luria–Bertani (LB) broth (2). Bacteria were washed twice in sterile PBS and resuspended in PBS before being heat-killed for 45–60 minutes at 65°C. Bacterial concentration and confirmation of successful killing was determined by quantitative culture. For in vivo studies, B. pseudomallei 1026b was grown in LB broth shaking in air at 37°C, washed twice, resuspended in PBS containing 20% glycerol, and frozen at −80°C. Immediately before each aerosol infection experiment, the freezer stock was thawed and diluted in PBS to the desired concentration as previously described (18).
Transfections
HEK293 cells were seeded at 50,000 cells/well in a 96 well flat-bottomed tissue culture plate to reach at least 90% confluency at the time of transfection. After two days, cells were simultaneously transfected and stimulated. Cells were transfected using FuGENE HD (Roche, Mannheim, Germany) at a ratio of 6 µL FuGENE HD per 2 µg DNA according to the manufacturer’s instructions. Vector DNA transfected consisted of 10 ng/well NF-κB-ELAM firefly luciferase, 1 ng/well control pRL-TK Renilla luciferase (Promega, Madison, WI) and 0.1 ng/well either empty vector or huNOD2-WT-pEF6 (19). After transfected DNA was applied to all wells, cells were immediately stimulated in triplicate with media, control NOD2 ligand MDP (Invivogen, San Diego, CA), or heat-killed bacteria. Cells were incubated overnight. The following day, cells were washed three times with D-PBS and lysed with 20 µL/well Passive Lysis Buffer included with Dual-Luciferase Reporter Assay System (Promega, Madison, WI). Activation of NF-κB was determined in 10 µL/well cell lysate using the Dual-Luciferase Reporter Assay System and a Veritas Microplate Luminometer (Turner BioSystems/Promega, Sunnyvale, CA).
Mouse model of melioidosis
Animals
Specific pathogen-free C57BL/6 mice were obtained from the Jackson Laboratory (Bar Harbor, ME). Nod2−/− mice backcrossed eight generations onto a C57BL/6 background are previously described (19). Mice were housed in isolator cages with ad lib access to chow and water, and were monitored one to two times daily. All animal experiments were approved by the University of Washington Institutional Animal Care and Use Committee.
Infection
Mice were exposed to aerosolized bacteria in a 24 port cylindrical nose-only exposure chamber (In-Tox Products, Moriarty, NM) (18). Aerosols were generated by a MiniHeart Hi-Flo nebulizer (Westmed, Tucson, AZ) driven at 8 L/min with 7 L/min simultaneous dilution air for 10 minutes followed by 5 minutes washout period. Pressure and airflow were controlled by an AeroMP aerosol management platform (Biaera Technologies, Frederick, MD). Bacterial deposition in each experiment was determined from quantitative culture of lung tissue from four mice sacrificed immediately after infection. Animals were examined daily for illness or death and their clinical condition recorded. When indicated, abdominal surface temperatures were measured using a Ranger MX4P digital infrared thermometer (Raytek, Santa Cruz, CA, USA). Ill animals with temperatures <21.5°C or a combination of ruffled fur, eye crusting, hunched posture and lack of resistance to handling were deemed terminal and euthanized. Spontaneous death was not required as an endpoint.
Bacterial quantification
Forty eight hours after infection mice were sacrificed. The left lung and spleen each were homogenized in 1 mL sterile PBS. One hundred microliters each of homogenate and 10-fold serial dilutions were plated in duplicate on LB agar or Ashdown’s medium. Colonies were counted after 2–4 days of incubation at 37°C or up to a week incubating at room temperature.
Lung histology and quantitative morphometry
The right lung was fixed in 4% paraformaldehyde as previously described (18). Lung tissue was embedded in paraffin, sectioned and stained with hematoxylin and eosin; sections were examined by a veterinary pathologist who was blinded to group assignment. The number of focal inflammatory lesions, area of each lesion, and total tissue area in one representative section from each mouse were determined using Nikon NIS-Elements software.
Cytokine measurements
Left lung homogenates in PBS were diluted 1:1 in lysis buffer containing 2× protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany), incubated on ice for 30 min, and then centrifuged at 1500×g. Supernatants were collected and stored at −80°C until assayed for cytokines. Whole blood was centrifuged, serum removed and stored at −80°C until assayed. IFN-γ, IL-10, IL-12p70, IL-1β, IL-6, KC, and TNF-α were measured in lung homogenates and serum using a 7-plex electrochemiluminescence detection assay (Meso Scale Discovery Gaithersburg, MD, USA) and read on the MSD Sector Imager 2400.
Human subjects
Clinical cohort
Human genetic analyses were performed on 614 inpatients with culture-proven melioidosis admitted to Sappasithiprasong Hospital, Ubon Ratchathani, Thailand from July 1999 through December 2005, and 950 ambulatory control subjects recruited at the hospital through 2010. Subsets of this cohort have been described previously (3, 5). DNA was extracted from blood using a Nucleon BACC3 kit (GE Healthcare, Buckinghamshire, UK). Consent for enrollment into clinical studies of melioidosis was obtained from subjects or their representatives at the time of recruitment.
Ev-vivo blood stimulation
Three hundred Thai subjects donating blood at the blood donation center at Sappasithiprasong Hospital, Ubon Ratchathani, Thailand were recruited for a blood sample as previously described [Chantratita, submitted; West, JI, 2013]. Subjects were included if they indicated they were between the ages of 18 and 60 years and did not report any history of immunodeficiency or inflammatory conditions, chronic diseases, pregnancy in the past six months, anti-inflammatory medication use in the past week, antibiotic use in the past month, vaccination in the past six months, heavy exercise or alcohol consumption in the past 24 hours, or smoking in the past month. Those who met enrolment criteria gave written informed consent to participate and provided a post-donation blood sample in citrate tubes. A complete blood count with differential was determined for each subject in the hospital laboratory. A batch of 96 well immuno-assay plates were generated by adding 20 µl of innate immune ligands and heat killed bacteria in appropriate concentrations to each well. Plates were frozen at −80°C until the day of use when they were thawed to 37°C. 380 µl of fresh whole blood anticoagulated with citrate from each subject was mixed 1:1 with RPMI media and added to each well. For this study, the stimulants analyzed were MDP 10 µg/ml (Invivogen) and heat-killed B. pseudomallei 1026b 2.5 × 106 CFU/ml in the 127 female subjects. Plates were placed on a shaker at 37°C under 5% CO2 for 6 hours before being spun down and the plasma supernatant removed and frozen at −80°C. Cytokine concentrations were subsequently assayed using R&D Systems reagents on a Luminex multiplex bead system. For this study, the analysis was restricted to IL-6, TNF-α, IL-1β, and IL-10. Due to out-of-range values in the multiplex assay, IL-1β concentrations for B. pseudomallei were determined by ELISA (BD Biosciences). DNA was extracted from whole blood using the QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany).
Human subjects
The University of Washington Human Subjects Division Institutional Review Board; the Ethical Review Committee for Research in Human Subjects, Ministry of Public Health, Thailand; the Ethical Review Committee for Research in Human Subjects, Sappasithiprasong Hospital, Ubon Ratchathani, Thailand; and the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand approved the studies.
Polymorphism selection and genotyping
Human single nucleotide polymorphisms in the NOD2 gene region that are associated with Crohn’s disease or leprosy were identified from the published literature. The frequency of these variants in Asian populations was determined using the NCBI dbSNP database and the Genome Variation Server (http://gvs.gs.washington.edu/GVS/). Seven common variants were selected for assay design and analysis. Genotyping was performed using ABI TaqMan assays on an ABI Prism 7900 (Carlsbad, CA).
Statistics
Comparisons of two groups of data expected to follow a normal distribution were made using Student’s t test. CFUs were log10 transformed before analysis. Counts of murine signs of illness were compared using a test of proportions. Survival analyses were performed with the log rank test. For multivariate analysis of genetic associations, logistic regression was performed adjusting for age, gender, and diabetes. Effect modification was determined by testing the incorporation of an interaction variable into the regression model, using the likelihood ratio test. Cytokine concentrations from the ex-vivo blood stimulation study were normalized to monocyte count and log10 transformed before analysis by linear regression using an additive genetic model, adjusting for age and batch. Statistics were performed with GraphPad Prism 4.0 (San Diego, CA) or Stata 11.2 (College Station, TX). A two sided p value of ≤ 0.05 was considered significant.
Results
B. pseudomallei activates innate immunity in a NOD2-dependent manner
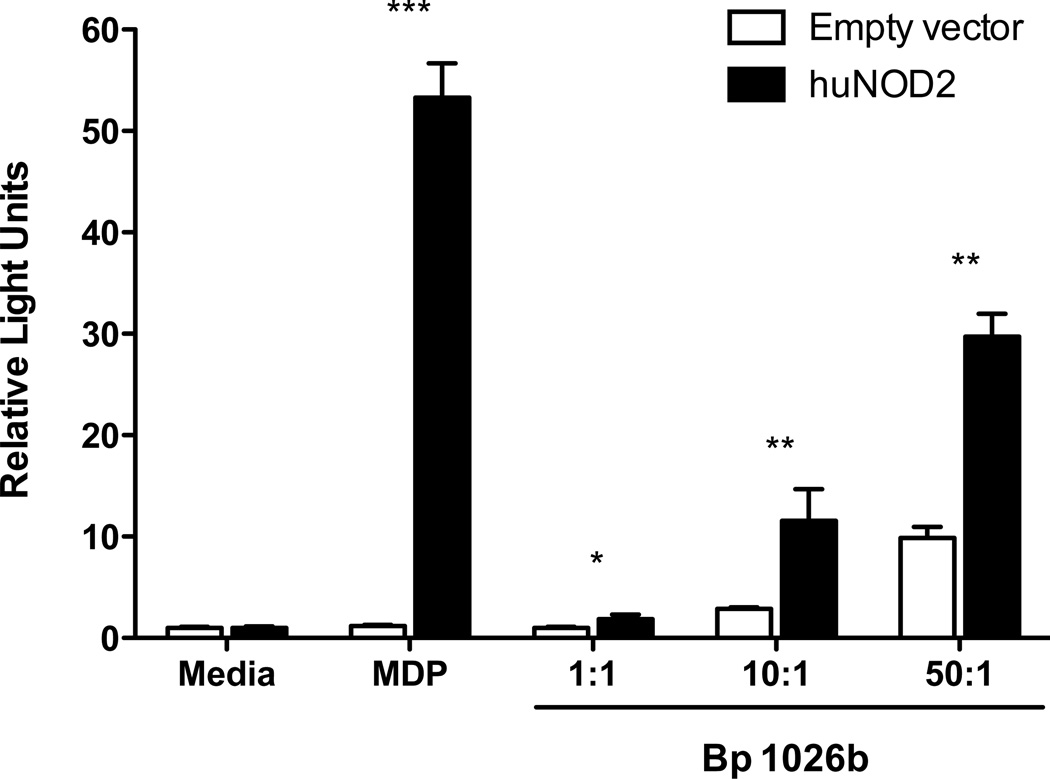
We first sought to determine whether NOD2 mediates innate immune activation by B. pseudomallei in vitro. We transfected HEK293 cells with human NOD2 constructs (19) and stimulated the cells with heat-killed B. pseudomallei at several concentrations (Fig. 1). We measured NF-κB activation by co-transfecting an NF-κB-ELAM-luciferase construct and control thymidine kinase Renilla luciferase construct, and quantifying relative light units. In NOD2-transfected cells we observed a significant increase in NF-κB activation induced by B. pseudomallei, indicating recognition of B. pseudomallei motifs by NOD2.
Figure 1. NOD2 mediates activation of innate immunity by B. pseudomallei.
HEK293 cells transfected with huNOD2 or empty vector and NF-κB-ELAM firefly luciferase and control thymidine kinase Renilla luciferase were stimulated overnight with media, MDP, or heat-killed B. pseudomallei 1026b at various bacteria:cell ratios. Cells were then lysed and NF-κB activation determined by relative light emission (firefly/Renilla luciferases). Data displayed are means ± SD of triplicate conditions normalized to empty vector media values. The data shown are from one of six representative and independent experiments. *, p≤0.05; **, p≤0.01; ***, p≤0.001.
Nod2-deficiency impairs the clinical response in low-dose murine respiratory melioidosis
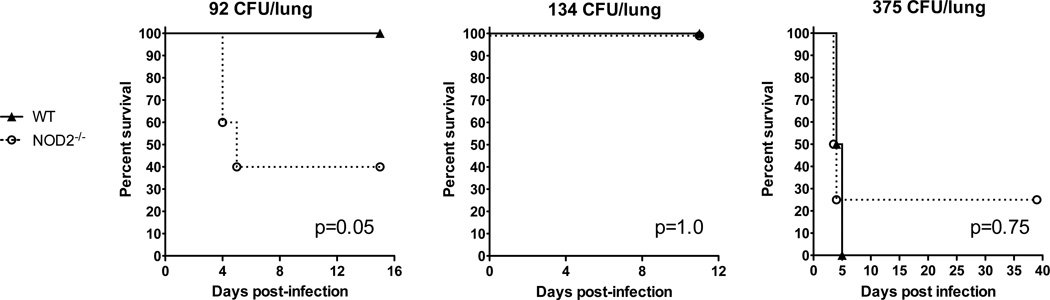
We then examined the effect of Nod2-deficiency in a murine model of respiratory melioidosis. We chose this model because inhalation is a route of infection in human melioidosis and because pneumonia is one of the most common manifestations of severe disease. We have developed a reproducible model of murine pneumonia by aerosolizing B. pseudomallei to mice (18). In this model, we have found that the median lethal deposition dose to C57BL/6 mice is 334 CFU/lung. However, there is a narrow range for lethality as all C57BL/6 mice infected with 292 CFU/lung survive whereas 100% of mice infected with 375 CFU/lung die. We conducted several experiments to test whether Nod2-deficiency altered survival from melioidosis. We deposited 92 CFU B. pseudomallei per lung by aerosol to wild type C57BL/6 and Nod2-deficient mice, and monitored survival. At this dose, all wild type mice survived but only two of five Nod2-deficient mice survived (p=0.05 by the log rank test) (Fig. 2).
Figure 2. Effect of Nod2 on survival in murine respiratory melioidosis.
Wild type and Nod2-deficient mice were infected with 92, 134, or 375 CFU/lung aerosolized B. pseudomallei 1026b and monitored at least daily for survival n = 5 per group, 8 per group, and 4 per group, respectively. Each experiment was performed independently. The p values were determined by the log rank test.
In a second experiment at a marginally higher dose we deposited 134 CFU/lung to wild type and Nod2-deficient mice (Fig. 2). All eight mice in each group survived for duration of the experiment (p=1.0). Although this experiment did not replicate the survival advantage of Nod2 seen previously, there were clear differences in clinical condition between mouse strains (Table I). Following infection, the wild type mice remained clinically well for the entire 11 day clinical monitoring period. In contrast, Nod2-deficient mice showed overt signs of illness manifesting primarily as coat scruffiness and hunching beginning at day one following infection. By day two, all eight Nod2-deficient mice were scruffy and hunched whereas no wild type mouse showed any clinical signs of infection. This difference persisted until the end of the first week when the Nod2-deficient mice began to show clinical improvement.
Table I.
Murine clinical responses in respiratory melioidosis
| Number of mice displaying clinical signs following infection with 134 CFU B. pseudomallei/lunga |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Condition | Mouse strain | Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | Day 11 |
| Impaired mobility | WT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NOD2−/− | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Coat scruffiness | WT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NOD2−/− | 4* | 8*** | 4* | 8*** | 8*** | 4* | 4* | 4* | |
| Hunching | WT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NOD2−/− | 4* | 8*** | 8*** | 8*** | 8*** | 8*** | 4* | 0 | |
| Eye crusting | WT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NOD2−/− | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
Total N=8 per group for all days
p≤0.05;
p≤0.001 for difference between mouse strains for each condition
In a third experiment targeting an even higher inoculation, we deposited 375 CFU/lung to wild type and Nod2-deficient mice (Fig. 2). Infection was lethal to all the wild type and three of four Nod2-deficient mice within five days, without any clear separation of survival curves (p=0.75). Collectively, these experiments suggested that NOD2 did not clearly augment survival in murine melioidosis, especially at higher doses. For infection with lower doses of B. pseudomallei, however, NOD2 imparted detectable clinical benefit. To maximize our sensitivity to detect pertinent phenotypes, we therefore targeted lower inoculating doses in subsequent experiments.
Nod2-deficiency permits greater bacterial replication and dissemination in sub-lethal murine respiratory melioidosis
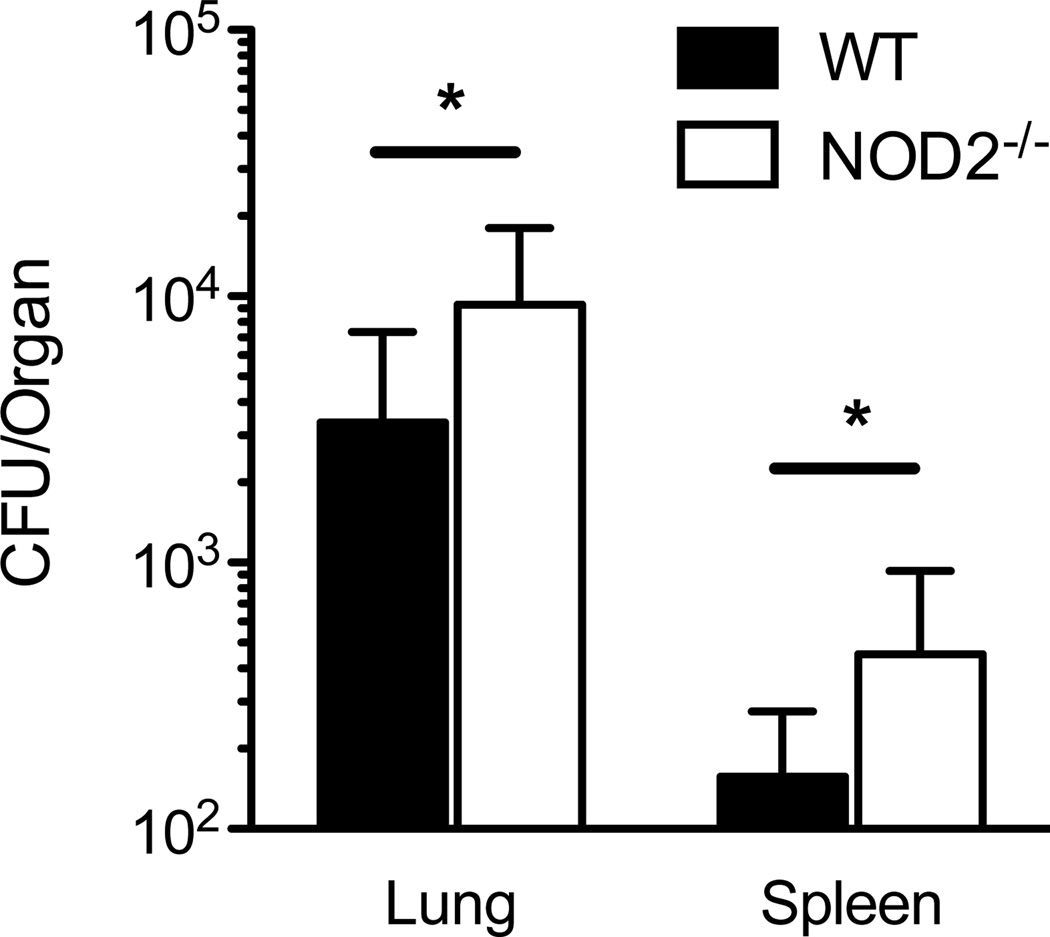
To test whether NOD2 facilitates bacterial containment in murine respiratory melioidosis, we elected to aerosolize a low deposition dose of B. pseudomallei to wild type and Nod2-deficient mice and quantify bacterial burdens in the lung and spleen. In wild type C57BL/6 mice, we have observed that a deposition dose of 56 CFU/lung results in significant replication (up to 104 CFU) in the lung and dissemination to the liver and spleen by 24 hours following infection (18). There is continued increase in bacterial burdens and associated inflammation from 24 to 48 hours following sub-lethal infection that diminishes by 96 hours (18). To identify maximal NOD2-dependent differences after low dose infection, we therefore deposited 57 CFU B. pseudomallei in the lungs of wild type and Nod2-deficient mice and quantified bacterial burdens in the lung and spleen 48 hours after infection (Fig. 3). There was pulmonary replication and dissemination to the spleen in both groups of mice but significantly more in Nod2-deficient mice compared to wild type mice (a difference in mean CFU of 5.9 × 103 in the lungs and 300 in the spleens). These data indicated a role for NOD2 in containment of B. pseudomallei after low dose airborne infection.
Figure 3. Nod2-deficiency permits greater bacterial replication and dissemination in murine respiratory melioidosis.
Wild type and Nod2-deficient mice were infected with 57 CFU/lung aerosolized B. pseudomallei 1026b. Lungs and spleens were harvested and quantitatively cultured 48 hours after infection. Data displayed are means ± SD and represent one experiment. *, p≤0.05. N= 8 per group.
Lung IL-6 and KC are higher in respiratory melioidosis in Nod2-deficient mice
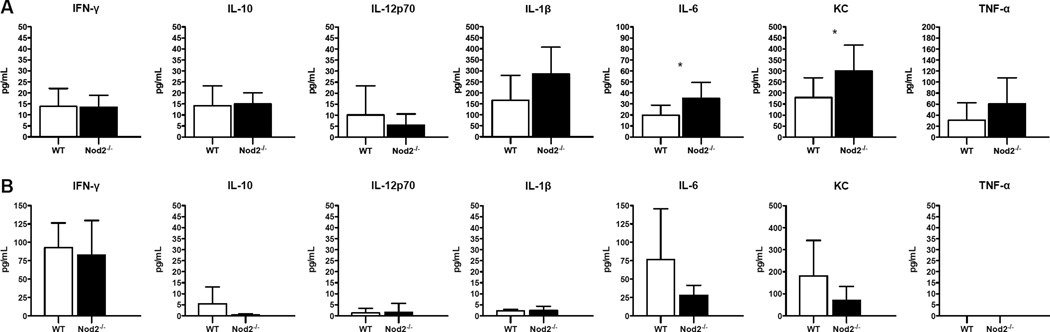
To determine whether the mechanism of impaired bacterial containment in the absence of NOD2 may be due to inadequate cytokine production, we measured levels of a panel of cytokines and chemokines in murine lung homogenate and serum 48 hours after aerosol infection with 57 CFU B. pseudomallei (Fig. 4). After this low dose-infection, overall levels of mediators were low, but we detected greater release of IL-6 and KC in the lungs of Nod2-deficient mice. There were no differences in the levels of serum inflammatory mediators. Thus, the impaired clearance of B. pseudomallei in Nod2-deficient mice was not associated with a blunted cytokine response at 48 hours.
Figure 4. Lung IL-6 and KC levels are higher in Nod2-deficient mice with respiratory melioidosis.
Cytokines and chemokines were measured in lung homogenate (A) and serum (B) from wild type and Nod2-deficient mice 48 hours after aerosol infection with 57 CFU/lung B. pseudomallei 1026b. N= 8 per group for lung homogenate and N=7 per group for serum. Data displayed are means ± SD and represent one experiment. *, p≤0.05.
Nod2-deficiency does not alter histologic changes in the lung during melioidosis
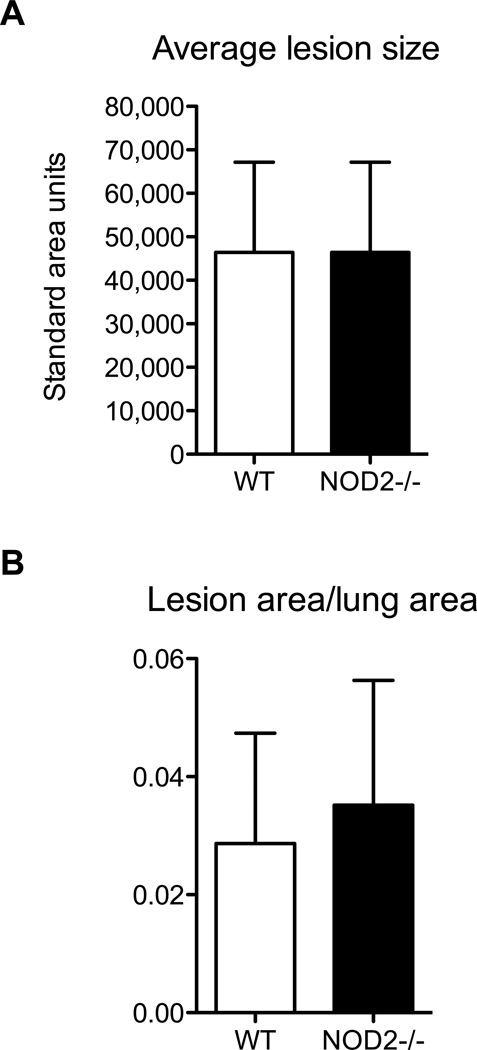
To further determine how NOD2 might regulate pulmonary inflammation in vivo, we compared lung sections from wild type or Nod2-deficient mice 48 hours after infection with 57 CFU/lung B. pseudomallei. Qualitatively the scattered foci of infection observed in the lung parenchyma of wild type mice were no different in Nod2-deficient mice and were similar to our previous report (18). We quantified the average size of the inflammatory foci and determined the ratio of total foci area to total lung area (Fig. 5). We did not detect any differences in these measures between wild type and Nod2-deficient mice, demonstrating that the greater bacterial burdens and pulmonary cytokine release in the absence of NOD2 was not associated with any increase in histologically apparent inflammation.
Figure 5. Nod2-deficiency does not alter pulmonary histopathology in respiratory melioidosis.
Lungs were fixed 48 hours after aerosol infection of wild type and Nod2-deficient mice with 57 CFU/lung B. pseudomallei 1026b. Hematoxylin and eosin staining was performed. The size of focal infiltrates (A) and the ratio of total infiltrate to lung area (B) were quantified for each group of mice. N= 8 per group. Data displayed are means ± SD and represent one experiment. p>0.05 for comparisons between wild type and Nod2-deficient values.
A common NOD2 region genetic variant is associated with melioidosis
Our in vitro and murine studies supported a modest role for NOD2 as a contributor to the host response to B. pseudomallei infection under experimental conditions. However, we sought additional evidence for a role for NOD2 in human melioidosis. Human genetic variation in innate immune genes has been associated with infection in numerous studies and offers insight into the importance of specific elements of human host defense (20). We hypothesized that genetic variation in NOD2 may be associated with melioidosis in a cohort of Thai subjects (3, 5). To test this, we examined the published literature for NOD2 region single nucleotide polymorphisms that had been previously associated with disease. Although non-synonymous NOD2 variants are associated with Crohn’s disease (21), none are common in Asian populations (minor allele frequency ≤1%). Other NOD2 region single nucleotide polymorphisms have been associated with leprosy or its clinical outcomes in Nepalese or Chinese populations (22–24). We chose seven candidate polymorphisms from these populations to genotype in our Thai cohort of 612 culture confirmed melioidosis cases and 950 ambulatory controls.
Five assays were successful (rs2287195, rs9302752, rs7194886, rs751271, and rs1077861) with a call rate of at least 97.5%. In the control subjects, linkage disequilibrium, a measure of non-random allelic association of different linked polymorphisms (r2), ranged from 0.07 – 0.97 for these variants. We confirmed the lack of deviation from Hardy Weinberg equilibrium for each variant in the controls and analyzed the genotype frequencies in cases and controls using an additive genetic model adjusted for age, gender, and diabetes (Table II). We did not apply a conservative Bonferroni correction given the observed linkage disequilibrium. We found that carriage of the minor T allele at rs7194886 was associated with melioidosis (OR 1.32, 95% CI 1.03–1.70, p=0.029). rs7194886 is located about 5.8 kilobases upstream of NOD2 and has a minor allele frequency of 0.08 in control subjects. The magnitude of effect was stronger in a recessive model (OR 3.60, 95% CI 1.07–12.16, p=0.039). Unexpectedly, we observed a substantial effect of gender on the association (likelihood ratio p=0.01; Supplemental Table I). For females, the odds ratio for melioidosis in carriers of the variant was 12.56, 95% CI: 1.53–102.97, whereas for males the odds ratio was 0.41, 95% CI: 0.04–4.03. These data, although limited to an association, imply that NOD2 may mediate susceptibility to human melioidosis.
Table II.
Association of NOD2 variants with melioidosis
| SNP | Genotype | Control | Case | HWE | Genotype associationb | |
|---|---|---|---|---|---|---|
| p valuea | OR (95%CI) | p value | ||||
| rs2287195 | TT | 642 | 382 | 0.30 | 1.11 (0.91–1.34) | 0.30 |
| TC | 281 | 178 | ||||
| CC | 24 | 19 | ||||
| rs9302752 | TT | 646 | 392 | 0.27 | 1.13 (0.93–1.37) | 0.23 |
| TC | 279 | 178 | ||||
| CC | 23 | 21 | ||||
| rs7194886 | CC | 793 | 488 | 0.26 | 1.32 (1.03–1.70) | 0.029 |
| CT | 151 | 108 | ||||
| TT | 4 | 8 | ||||
| rs751271 | TT | 684 | 426 | 0.61 | 1.11 (0.91–1.37) | 0.31 |
| TG | 244 | 168 | ||||
| GG | 19 | 13 | ||||
| rs1077861 | AA | 715 | 443 | 0.21 | 1.13 (0.90–1.41) | 0.28 |
| AT | 218 | 151 | ||||
| TT | 11 | 8 | ||||
HWE, Hardy Weinberg equilibrium
Additive genetic model adjusting for age, gender, and diabetes
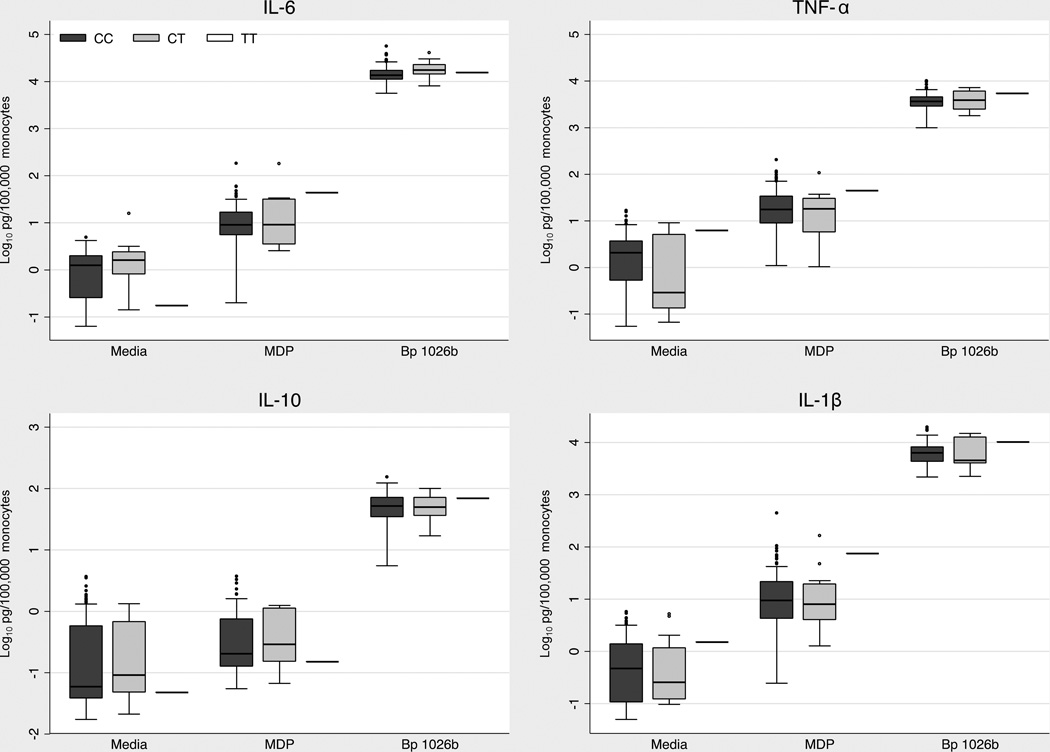
We next sought to determine whether rs7194886 has a demonstrable functional effect on the inflammatory response to B. pseudomallei. We stimulated whole blood from 127 healthy Thai women with the NOD2 agonist MDP or heat killed B. pseudomallei. We measured IL-6, TNF-α, IL-10, and IL-1β responses in plasma (Fig. 6). Cytokine responses to 10 µg/ml of MDP were several orders of magnitude lower than responses induced by B. pseudomallei, but were nonetheless readily detectable. We found no differences in cytokine concentrations based on rs7194886 genotype. Thus in this assay we could not ascribe a functional effect to this polymorphism.
Figure 6. Whole blood cytokine responses to NOD2 agonists by rs7194886 genotype.
Monocyte-normalized plasma IL-6, TNF-α, IL-1β, and IL-10 levels induced by stimulation of whole blood from 127 healthy female subjects at 37°C for six hours with medium alone, MDP 10 µg/ml (Invivogen) and heat-killed B. pseudomallei 1026b 2.5 × 106 CFU/ml. Boxes show the median and interquartile range; whiskers show upper and lower adjacent values. N= 111 (CC), 15 (CT), 1(TT). p>0.05 for all comparisons between genotypes.
Discussion
We show in this work that NOD2 activation by B. pseudomallei induces activation of the innate immune transcription factor NF-κB, that the absence of NOD2 does not alter survival but has a modest effect on clinical response, bacterial containment, and cytokine release in the murine lung after low-dose infection with B. pseudomallei in vivo, and that human NOD2 region genetic variation is associated with susceptibility to melioidosis. This is the first study of the role of NOD2 in B. pseudomallei infection in vivo and in humans, advancing our understanding of mammalian host defense to this facultative intracellular pathogen.
A growing body of literature points to the importance of NOD2 in facilitating the host response to experimental bacterial infections yet the role of this receptor is infection-specific (23, 25–29). For example, in murine pneumonia caused by E. coli or C. pneumoniae, NOD2 contributes to bacterial containment in the lung (25, 26). However, in murine pneumonia due to S. aureus or L. pneumophila, NOD2 is redundant for this purpose (19, 28, 30). Our data indicate that, while there is no clear NOD2-dependent effect on murine survival after lethal or sub-lethal B. pseudomallei infection, NOD2 facilitates pulmonary containment of B. pseudomallei after low dose infection. In the absence of NOD2, our results show that bacterial replication and dissemination is enhanced and are accompanied by clinical signs of illness and increased production of low levels of IL-6 and KC in the lung. Interestingly, this cytokine profile differs from E. coli pneumonia, in which Nod2−/− cytokine responses are blunted despite augmented bacterial counts in the lung (25). This difference may be explained by B. pseudomallei’s behavior as a facultative intracellular pathogen. Recently, Pudla et al showed that NOD2 knockdown impairs expression of the negative cytokine signaling regulator SOCS3 in B. pseudomallei-infected macrophages (16). Together, these data imply that the mechanism by which NOD2 contributes to containment in murine melioidosis is not due to lack of cytokine production. Recent work indicates that NOD2 may control intracellular pathogens by facilitating autophagy (31), a recognized suppressor of B. pseudomallei survival in mammalian cells (32). This mechanism should be investigated in future studies.
Human genetic variation in NOD2 is well described and NOD2 variation is associated with inflammatory disorders such as Crohn’s disease (21), Blau syndrome (33), sarcoidosis (34), asthma and chronic obstructive pulmonary disease (35, 36). rs7194886 is an intergenic variant upstream of NOD2 that is associated with leprosy in a genome wide association study in Chinese subjects and with type I reversal reactions and erythema nodosum leprosum in a candidate gene analysis in Nepalese subjects (22, 23). rs7194886 is not associated with leprosy in Vietnamese subjects (24); however, the variant is associated with tuberculosis in Chinese individuals (37). Thus, there is increasing evidence of its importance in mycobacterial infection in Asian populations. As such, its association with susceptibility to melioidosis – another intracellular infection that shares a number of clinical and pathophysiologic similarities with tuberculosis (38) – is quite plausible. Interestingly, the 1000 Genomes Project shows that the minor allele frequency of the variant is 0.45 in Europeans, 0.27 in Americans, 0.24 in Africans, but only 0.09 in East Asians (39). This suggests that there may be some selective pressure against the variant that is particularly pronounced in Asian populations. The effect of rs7194886 on NOD2 function is presently unknown; rs7194886 scores low as a regulatory variant in the RegulomeDB (40) but may lie within a haplotype with regulatory function. While our ex-vivo blood stimulation assay did not reveal genotype-dependent differences in cytokine responses induced by MDP or by killed B. pseudomallei, putative differences in NOD2-dependent autophagy would not be detected in this assay. Thus, while intriguing, our association of rs7194886 with melioidosis does not indicate causation. Our ongoing investigations are focused both on elucidating the architecture of variation in the NOD2 gene region in Asians and on identifying functional effects that may underlie the association of rs7194886 with disease.
Our observation of a gender-specific effect of a NOD2 region genetic variant is of particular interest. To date, few studies have examined the role of gender in governing the association of host genetic makeup with infectious disease. Several authors have reported effects of gender in genetic studies of lipids, obesity or cardiovascular disease (41–44). It is possible that an unmeasured confounder may account for the gender-dependent genetic association in the case-control cohort. To our knowledge, gender has not been previously implicated in NOD2-dependent signaling, although gender-related differences in innate immune activation are well described (45–47). Our findings prompt further evaluation of the role of gender in modulating innate immune function.
An important consideration is the possibility of population admixture confounding our genetic association results. In previous analyses of a large subset of this cohort, however, we have not identified significant population stratification (3). Validation of genetic associations in independent populations is also desirable, although to our knowledge there are few other cohorts of melioidosis patients of sufficient size to permit this.
Together, our data comprise the most comprehensive investigation to date on NOD2 as a contributor to host defense in melioidosis and add to the expanding literature implicating cytosolic pathogen recognition receptors in the host response to intracellular infection.
Supplementary Material
Acknowledgements
Tony Han and Loren Kinman assisted with animal experiments. Premjit Amornchai, Aunchalee Thanwisai, and Malinee Oyuchua extracted DNA. Chris Goss provided statistical advice.
Funding
Funding was provided by National Institutes of Health (UL1TR000423, K08HL094759, U54AI057141, K08AI080952); the Puget Sound Partners for Global Health; the Parker B. Francis Foundation; and a Wellcome Trust Career Development Award in Public Health and Tropical Medicine 087769/Z/08/Z to NC. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.Wiersinga WJ, Currie BJ, Peacock SJ. Melioidosis. The New England journal of medicine. 2012;367:1035–1044. doi: 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- 2.West TE, Ernst RK, Jansson-Hutson MJ, Skerrett SJ. Activation of Toll-like receptors by Burkholderia pseudomallei. BMC immunology. 2008;9:46. doi: 10.1186/1471-2172-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.West TE, Chierakul W, Chantratita N, Limmathurotsakul D, Wuthiekanun V, Emond MJ, Hawn TR, Peacock SJ, Skerrett SJ. Toll-like receptor 4 region genetic variants are associated with susceptibility to melioidosis. Genes and immunity. 2012;13:38–46. doi: 10.1038/gene.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiersinga WJ, Wieland CW, Dessing MC, Chantratita N, Cheng AC, Limmathurotsakul D, Chierakul W, Leendertse M, Florquin S, de Vos AF, White N, Dondorp AM, Day NP, Peacock SJ, van der Poll T. Toll-Like Receptor 2 Impairs Host Defense in Gram-Negative Sepsis Caused by Burkholderia pseudomallei (Melioidosis) PLoS Med. 2007;4:e248. doi: 10.1371/journal.pmed.0040248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.West TE, Chantratita N, Chierakul W, Limmathurotsakul D, Wuthiekanun V, Myers ND, Emond MJ, Wurfel MM, Hawn TR, Peacock SJ, Skerrett SJ. Impaired TLR5 Functionality Is Associated with Survival in Melioidosis. Journal of immunology. 2013;190:3373–3379. doi: 10.4049/jimmunol.1202974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West TE, Hawn TR, Skerrett SJ. Toll-like receptor signaling in airborne Burkholderia thailandensis infection. Infection and immunity. 2009;77:5612–5622. doi: 10.1128/IAI.00618-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franchi L, Park JH, Shaw MH, Marina-Garcia N, Chen G, Kim YG, Nunez G. Intracellular NOD-like receptors in innate immunity, infection and disease. Cellular microbiology. 2008;10:1–8. doi: 10.1111/j.1462-5822.2007.01059.x. [DOI] [PubMed] [Google Scholar]
- 8.Shaw MH, Reimer T, Kim YG, Nunez G. NOD-like receptors (NLRs): bona fide intracellular microbial sensors. Current opinion in immunology. 2008;20:377–382. doi: 10.1016/j.coi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inohara Chamaillard, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annual review of biochemistry. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. International immunology. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourhis LL, Werts C. Role of Nods in bacterial infection. Microbes and infection / Institut Pasteur. 2007;9:629–636. doi: 10.1016/j.micinf.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Harley VS, Dance DA, Tovey G, McCrossan MV, Drasar BS. An ultrastructural study of the phagocytosis of Burkholderia pseudomallei. Microbios. 1998;94:35–45. [PubMed] [Google Scholar]
- 13.Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breitbach K, Sun GW, Kohler J, Eske K, Wongprompitak P, Tan G, Liu Y, Gan YH, Steinmetz I. Caspase-1 mediates resistance in murine melioidosis. Infection and immunity. 2009;77:1589–1595. doi: 10.1128/IAI.01257-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ceballos-Olvera I, Sahoo M, Miller MA, Del Barrio L, Re F. Inflammasome-dependent pyroptosis and IL-18 protect against Burkholderia pseudomallei lung infection while IL-1beta is deleterious. PLoS pathogens. 2011;7:e1002452. doi: 10.1371/journal.ppat.1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pudla M, Kananurak A, Limposuwan K, Sirisinha S, Utaisincharoen P. Nucleotide-binding oligomerization domain-containing protein 2 regulates suppressor of cytokine signaling 3 expression in Burkholderia pseudomallei-infected mouse macrophage cell line RAW 264.7. Innate immunity. 2011;17:532–540. doi: 10.1177/1753425910385484. [DOI] [PubMed] [Google Scholar]
- 17.West TE, Myers ND, Limmathurotsakul D, Liggitt HD, Chantratita N, Peacock SJ, Skerrett SJ. Pathogenicity of high-dose enteral inoculation of Burkholderia pseudomallei to mice. The American journal of tropical medicine and hygiene. 2010;83:1066–1069. doi: 10.4269/ajtmh.2010.10-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West TE, Myers ND, Liggitt HD, Skerrett SJ. Murine pulmonary infection and inflammation induced by inhalation of Burkholderia pseudomallei. International journal of experimental pathology. 2012;93:421–428. doi: 10.1111/j.1365-2613.2012.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berrington WR, Iyer R, Wells RD, Smith KD, Skerrett SJ, Hawn TR. NOD1 and NOD2 regulation of pulmonary innate immunity to Legionella pneumophila. European journal of immunology. 2010 doi: 10.1002/eji.201040518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vannberg FO, Chapman SJ, Hill AV. Human genetic susceptibility to intracellular pathogens. Immunological reviews. 2011;240:105–116. doi: 10.1111/j.1600-065X.2010.00996.x. [DOI] [PubMed] [Google Scholar]
- 21.Abraham C, Cho JH. Functional consequences of NOD2 (CARD15) mutations. Inflammatory bowel diseases. 2006;12:641–650. doi: 10.1097/01.MIB.0000225332.83861.5f. [DOI] [PubMed] [Google Scholar]
- 22.Zhang FR, Huang W, Chen SM, Sun LD, Liu H, Li Y, Cui Y, Yan XX, Yang HT, Yang RD, Chu TS, Zhang C, Zhang L, Han JW, Yu GQ, Quan C, Yu YX, Zhang Z, Shi BQ, Zhang LH, Cheng H, Wang CY, Lin Y, Zheng HF, Fu XA, Zuo XB, Wang Q, Long H, Sun YP, Cheng YL, Tian HQ, Zhou FS, Liu HX, Lu WS, He SM, Du WL, Shen M, Jin QY, Wang Y, Low HQ, Erwin T, Yang NH, Li JY, Zhao X, Jiao YL, Mao LG, Yin G, Jiang ZX, Wang XD, Yu JP, Hu ZH, Gong CH, Liu YQ, Liu RY, Wang DM, Wei D, Liu JX, Cao WK, Cao HZ, Li YP, Yan WG, Wei SY, Wang KJ, Hibberd ML, Yang S, Zhang XJ, Liu JJ. Genomewide association study of leprosy. The New England journal of medicine. 2009;361:2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 23.Berrington WR, Macdonald M, Khadge S, Sapkota BR, Janer M, Hagge DA, Kaplan G, Hawn TR. Common polymorphisms in the NOD2 gene region are associated with leprosy and its reactive states. The Journal of infectious diseases. 2010;201:1422–1435. doi: 10.1086/651559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant AV, Alter A, Huong NT, Orlova M, Van Thuc N, Ba NN, Thai VH, Abel L, Schurr E, Alcais A. Crohn's disease susceptibility genes are associated with leprosy in the Vietnamese population. The Journal of infectious diseases. 2012;206:1763–1767. doi: 10.1093/infdis/jis588. [DOI] [PubMed] [Google Scholar]
- 25.Theivanthiran B, Batra S, Balamayooran G, Cai S, Kobayashi K, Flavell RA, Jeyaseelan S. NOD2 signaling contributes to host defense in the lungs against Escherichia coli infection. Infection and immunity. 2012;80:2558–2569. doi: 10.1128/IAI.06230-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Shimada K, Chen S, Dempsey PW, Sorrentino R, Alsabeh R, Slepenkin AV, Peterson E, Doherty TM, Underhill D, Crother TR, Arditi M. The NOD/RIP2 pathway is essential for host defenses against Chlamydophila pneumoniae lung infection. PLoS pathogens. 2009;5:e1000379. doi: 10.1371/journal.ppat.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Opitz B, Puschel A, Schmeck B, Hocke AC, Rosseau S, Hammerschmidt S, Schumann RR, Suttorp N, Hippenstiel S. Nucleotide-binding oligomerization domain proteins are innate immune receptors for internalized Streptococcus pneumoniae. The Journal of biological chemistry. 2004;279:36426–36432. doi: 10.1074/jbc.M403861200. [DOI] [PubMed] [Google Scholar]
- 28.Kapetanovic R, Nahori MA, Balloy V, Fitting C, Philpott DJ, Cavaillon JM, Adib-Conquy M. Contribution of phagocytosis and intracellular sensing for cytokine production by Staphylococcus aureus-activated macrophages. Infection and immunity. 2007;75:830–837. doi: 10.1128/IAI.01199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 30.Frutuoso MS, Hori JI, Pereira MS, Junior DS, Sonego F, Kobayashi KS, Flavell RA, Cunha FQ, Zamboni DS. The pattern recognition receptors Nod1 and Nod2 account for neutrophil recruitment to the lungs of mice infected with Legionella pneumophila. Microbes and infection / Institut Pasteur. 2010;12:819–827. doi: 10.1016/j.micinf.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nunez G, Girardin SE, Philpott DJ. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nature immunology. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 32.Cullinane M, Gong L, Li X, Lazar-Adler N, Tra T, Wolvetang E, Prescott M, Boyce JD, Devenish RJ, Adler B. Stimulation of autophagy suppresses the intracellular survival of Burkholderia pseudomallei in mammalian cell lines. Autophagy. 2008;4:744–753. doi: 10.4161/auto.6246. [DOI] [PubMed] [Google Scholar]
- 33.Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, Hugot JP. CARD15 mutations in Blau syndrome. Nature genetics. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 34.Kanazawa N, Okafuji I, Kambe N, Nishikomori R, Nakata-Hizume M, Nagai S, Fuji A, Yuasa T, Manki A, Sakurai Y, Nakajima M, Kobayashi H, Fujiwara I, Tsutsumi H, Utani A, Nishigori C, Heike T, Nakahata T, Miyachi Y. Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-kappaB activation: common genetic etiology with Blau syndrome. Blood. 2005;105:1195–1197. doi: 10.1182/blood-2004-07-2972. [DOI] [PubMed] [Google Scholar]
- 35.Weidinger S, Klopp N, Rummler L, Wagenpfeil S, Baurecht HJ, Gauger A, Darsow U, Jakob T, Novak N, Schafer T, Heinrich J, Behrendt H, Wichmann HE, Ring J, Illig T. Association of CARD15 polymorphisms with atopy-related traits in a population-based cohort of Caucasian adults. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2005;35:866–872. doi: 10.1111/j.1365-2222.2005.02269.x. [DOI] [PubMed] [Google Scholar]
- 36.Kinose D, Ogawa E, Hirota T, Ito I, Kudo M, Haruna A, Marumo S, Hoshino Y, Muro S, Hirai T, Sakai H, Date H, Tamari M, Mishima M. A NOD2 gene polymorphism is associated with the prevalence and severity of chronic obstructive pulmonary disease in a Japanese population. Respirology. 2012;17:164–171. doi: 10.1111/j.1440-1843.2011.02069.x. [DOI] [PubMed] [Google Scholar]
- 37.Pan H, Dai Y, Tang S, Wang J. Polymorphisms of NOD2 and the risk of tuberculosis: a validation study in the Chinese population. International journal of immunogenetics. 2012;39:233–240. doi: 10.1111/j.1744-313X.2011.01079.x. [DOI] [PubMed] [Google Scholar]
- 38.Koh GC, Schreiber MF, Bautista R, Maude RR, Dunachie S, Limmathurotsakul D, Day NP, Dougan G, Peacock SJ. Host responses to melioidosis and tuberculosis are both dominated by interferon-mediated signaling. PloS one. 2013;8:e54961. doi: 10.1371/journal.pone.0054961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome research. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durlach A, Clavel C, Girard-Globa A, Durlach V. Sex-dependent association of a genetic polymorphism of cholesteryl ester transfer protein with high-density lipoprotein cholesterol and macrovascular pathology in type II diabetic patients. The Journal of clinical endocrinology and metabolism. 1999;84:3656–3659. doi: 10.1210/jcem.84.10.6064. [DOI] [PubMed] [Google Scholar]
- 42.Senti M, Fernandez-Fernandez JM, Tomas M, Vazquez E, Elosua R, Marrugat J, Valverde MA. Protective effect of the KCNMB1 E65K genetic polymorphism against diastolic hypertension in aging women and its relevance to cardiovascular risk. Circulation research. 2005;97:1360–1365. doi: 10.1161/01.RES.0000196557.93717.95. [DOI] [PubMed] [Google Scholar]
- 43.Corella D, Guillen M, Portoles O, Sorli JV, Alonso V, Folch J, Saiz C. Gender specific associations of the Trp64Arg mutation in the beta3-adrenergic receptor gene with obesity-related phenotypes in a Mediterranean population: interaction with a common lipoprotein lipase gene variation. Journal of internal medicine. 2001;250:348–360. doi: 10.1046/j.1365-2796.2001.00894.x. [DOI] [PubMed] [Google Scholar]
- 44.Ordovas JM. Gender, a significant factor in the cross talk between genes, environment, and health. Gender medicine. 2007;4(Suppl B):S111–S122. doi: 10.1016/s1550-8579(07)80052-0. [DOI] [PubMed] [Google Scholar]
- 45.Lefevre N, Corazza F, Duchateau J, Desir J, Casimir G. Sex differences in inflammatory cytokines and CD99 expression following in vitro lipopolysaccharide stimulation. Shock. 2012;38:37–42. doi: 10.1097/SHK.0b013e3182571e46. [DOI] [PubMed] [Google Scholar]
- 46.Imahara SD, Jelacic S, Junker CE, O'Keefe GE. The influence of gender on human innate immunity. Surgery. 2005;138:275–282. doi: 10.1016/j.surg.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 47.Pennell LM, Galligan CL, Fish EN. Sex affects immunity. Journal of autoimmunity. 2012;38:J282–J291. doi: 10.1016/j.jaut.2011.11.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.