Abstract
The Cry11Aa protein produced in Bacillus thuringiensis subsp. israelensis, a bacterial strain used worldwide for the control of Aedes aegypti larvae, binds midgut brush border membrane vesicles (BBMV) with an apparent Kd of 29.8 nM. Previously an aminopeptidase N (APN), named AaeAPN2, was identified as a putative Cry11Aa toxin binding protein by pull-down assays using biotinylated Cry11Aa toxin (Chen et al., (2009) Insect Biochem Mol Biol., 39: 688–696). Here we show this protein localizes to the apical membrane of epithelial cells in proximal and distal regions of larval caeca. The AaeAPN2 protein binds Cry11Aa with high affinity, 8.6 nM. The full-length and fragments of AaeAPN2 were cloned and expressed in Escherichia coli. The toxin-binding region was identified and further competitive assays demonstrated that Cry11Aa binding to BBMV was efficiently competed by the full-length AaeAPN2 and the fragments of AaeAPN2b and AaeAPN2e. In bioassays against Ae. aegypti larvae, the presence of full-length and a partial fragment (AaeAPN2b) of AaeAPN2 enhanced Cry11Aa larval mortality. Taken together, we conclude that AaeAPN2 is a binding protein and plays a role in Cry11Aa toxicity.
Keywords: Bacillus thuringiensis, Cry11Aa toxin, aminopeptidase, receptor, binding affinity, midgut, Aedes aegypti
Introduction
Aedes aegypti is the principle vector for dengue and yellow fever diseases, both of which have seen recent reemergence. Control of Aedes mosquitoes during their larval stages has increasingly used Bacillus thuringiensis subsp. israelensis formulations. This bacterium produces inclusions that contain crystalline (Cry4Aa, Cry4Ba, Cry10Aa and Cry11Aa) and cytolytic (Cyt1Aa, Cyt2Ba and Cyt1Ca) proteins, which are produced during the sporulation phase (Berry et al., 2002). Among them, Cry11Aa is one of the most active toxins against Ae. aegypti larvae (Chilcott and Ellar, 1988).
The mechanism of action of Cry toxins has been best studied in lepidopteran insects, where presently four major protein receptors have been identified for Cry1A toxins – cadherins, ABCC transporters, aminopeptidases (APNs) and alkaline phosphatases (ALPs) (for reviews see (Bravo et al., 2005; Pardo-Lopez et al., 2013; Pigott and Ellar, 2007; Soberon et al., 2009)). The activated Cry toxins bind first to the cadherin receptor or ABCC transporter in the microvilli of midgut epithelial cells. Binding to the former is known to trigger toxin oligomerization, and then the toxin oligomers bind to GPI-anchored receptors, APN and/or ALP, leading to membrane insertion and pore formation (Bravo et al., 2005, 2007; Soberon et al., 2009). It is possible a similar process is involved after Cry toxin binding to the ABCC transporter. Membrane insertion and pore formation are thought to lyse the midgut cells ultimately killing larval insect (Soberon et al., 2009). Alternately, in another model, the cadherin alone initiates an intracellular cascade that leads to cell toxicity (Zhang et al., 2006).
Since APNs were identified as Cry1 toxin-binding proteins (Gill et al., 1995; Knight et al., 1995), numerous lepidopteran APNs have been reported to bind Cry1 toxins (Pigott and Ellar, 2007). Cry1Ac toxin interaction with APNs is generally thought to involve glycosylated moieties. For example, Cry1Ac interacts with APNs from M. sexta (Burton et al., 1999; Masson et al., 1995), Heliothis virescens (Gill et al., 1995) and Lymantria dispar (Jenkins et al., 2000) through N-acetyl galactosamine residues (GalNAc). But Bombyx mori APNs are believed to bind toxins in a glycan-independent manner (Atsumi et al., 2005; Yaoi et al., 1997; Yaoi et al., 1999). However, only a few APNs apparently mediate in vivo toxin activity, for example, the silencing of midgut APNs results in reducing Spodoptera litura and H. armigera sensitivity to Cry1C and Cry1Ac, respectively (Rajagopal et al., 2002; Sivakumar et al., 2007). More recent evidence shows that a mutation in APN is associated with Cry1Ac resistance in H. armigera (Zhang et al., 2009) and down-regulation of APN is correlated with cabbage looper resistance to Cry1Ac toxins (Tiewsiri and Wang, 2011). These evidences support a functional role for APN in mediating Cry1 toxicity. In the sequential toxin binding model, cadherin-induced toxin oligomers can bind APN (Bravo et al., 2004).
Recent evidence suggests mosquitocidal toxins also bind a similar set of proteins in the mosquito midgut, and toxin binding APNs have been identified. For example, APNs from Anopheles quadrimaculatus and An. gambiae bind Cry11Ba (Abdullah et al., 2006; Zhang et al., 2008) and APNs from Ae. aegypti bind Cry11Aa (Chen et al., 2009b). Furthermore, all these Cry11A-bound APNs from Ae. aegypti, along with a number of other GPI-anchored proteins are localized in lipid rafts (Bayyareddy et al., 2012). In the case of the 110kDa APN (AaeAPN1) from Ae. aegypti and the 106 kDa APN (AgAPN2) from A. gambiae, partial protein fragments expressed in E. coli bind Cry11Aa and Cry11Ba toxins, respectively (Chen et al., 2009b; Zhang et al., 2008). Moreover, two partial APN fragments show synergistic and inhibitory effects on the Cry11B toxicity in the An. gambiae (Zhang et al., 2010). In addition, recently three predicted glycosylphosphatidylinositol (GPI)-anchored APNs transcript knockdown by RNAi cause Cry4B toxicity decrease in Ae. aegypti (Saengwiman et al., 2011).
Previously we reported another Ae. aegypti APN, named AaeAPN2, also binds Cry11Aa (Chen et al., 2009b). Here we report further characterization of this protein, including its expression and characterization in E. coli, and its immunolocalization in the larval midgut. Cry11Aa toxin binds AaeAPN2 with high affinity, and we also identified a toxin binding region in this APN. Here this cloned APN is referred to as AaeAPN2, which has five amino acids that differ from that of AAEL008155.
Materials and methods
Purification and activation of Cry11Aa toxin
Cry11Aa inclusions were isolated from a recombinant B. thuringiensis strain (Chang et al., 1993), grown in nutrient broth sporulation medium containing 12.5 μg/ml erythromycin at 30°C (Lereclus et al., 1995). Following cell autolysis spores and inclusions were harvested, washed 3X with 1 M NaCl, 10 mM EDTA, pH 8.0 and centrifuged. The resulting pellet was resuspended in 30 ml of the same buffer and purified by NaBr gradients as previously described (Cowles et al., 1995). The purified inclusions were solubilized in 50 mM Na2CO3, pH 10.5 and activated by trypsin (1:20, w/w), and the activated Cry11Aa toxin was purified by ion exchange chromatography (Mono Q, FPLC).
Preparation of membranes from Aedes midguts
Brush border membrane vesicles (BBMV) were prepared from early fourth instar Ae. aegypti larvae midguts as described (Nielsen-Leroux and Charles, 1992). The cell suspension was sonicated, centrifuged at 700g for 10 min, and the supernatant was then centrifuged at 16,000g for 1 h at 4°C. The membrane pellet obtained was used fresh..
Assembly, cloning and analysis of AeaAPN2 gene
Five pairs of primers listed in Table 1 were designed based on AAEL008155 transcript, and used for PCR for isolation of full-length or partial AaeAPN2 cDNA. Total RNA was extracted from early 4th instar larvae midguts using Trizol reagent (Invitrogen). For 5′-rapid amplification of cDNA ends (RACE), a poly(A) tail was added to the 5′ termini of cDNAs transcribed from mRNA by using the gene-specific primer, APN2-C. A 0.2 kb PCR product was amplified from the tailed cDNA by the primers, APN2-5R and adaptor primer. For 3′ RACE, first strand cDNA was synthesized from total RNA with an (dT)17-adaptor primer. Using an adaptor primer and a gene-specific primer, APN2-3F, a 0.7 kb PCR product was obtained by nested PCR. Based on sequences of 5′ RACE and 3′ RACE amplified products, two partial overlapping fragments of ORF were amplified by gene-specific primers of AaeAPN2, APN2-1F / 1R and APN2-2F / 2R, respectively and then were assembled as full-length AaeAPN2 ORF using a unique restriction site, BsiWI, present in both products. In addition, a 815 bp product from the 3′ end of the AaeAPN2 gene was obtained using primers APN1-eF/eR and the protein obtained from this product was used for antibody preparation. All PCR products were cloned into TA cloning vector, pCR2.1 TOPO (Invitrogen) and fully sequenced (Institute of Integrative Genome Biology (IIGB), University of California Riverside).
Table 1.
Primers used in this study
| Primer | Primer sequence (5′-3′) |
|---|---|
| (dT)17-adaptor | 5′-GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT-3′ |
| adaptor | 5′-GACTCGAGTCGACATCGA-3′ |
| APN2-C | 5′-GGCATATTAGCAACCGCATT-3′ |
| APN2-5R | 5′-CGTTCGTTTTCATGGATCG-3′ |
| APN2-3F | 5′-AAAGTGTCGGCCAATACCG-3′ |
| APN2-1F | 5′-GGATCCATGTTGTTGAAGGAAGTGTTTTTGTGCGC-3′ |
| APN2-1R | 5′-CGTACGCAACTGTGTCAAACAGGCCAG-3′ |
| APN2-2F | 5′-CGTACGATAAGTCGGGAAGTGTTCT-3′ |
| APN2-2R | 5′-TCTAGATCACTGTTTGTACGCCGCGAAGAACTC-3′ |
| APN2-eF | 5′-GGATCCAAGTTGACAACAACTATCCCTG-3′ |
| APN2-eR | 5′-AAGCTTACCGAGAAACTCGATAACG-3′ |
Sequence alignments and other sequence analysis were performed using NCBI blast programs and Lasergene (DNAstar). The signal peptide and GPI anchor were predicted by SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) and big PI predictor (http://mendel.imp.ac.at/gpi/gpi_server.html), respectively. The OGPET v1.0 (http://ogpet.utep.edu/OGPET/) and NetNGlyc v1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) programs were used to determine potential O- and N-glycosylation sites, respectively.
Expression of AaeAPN2 in E. coli
The 815 bp PCR product and full-length AaeAPN2 orf, together with four partial overlapping gene fragments cleaved from this full-length AaeAPN2 orf by using appropriate restriction enzymes or obtained by PCR, were cloned into pQE series expression vector (Qiagen). The constructs were transformed into E. coli M15(pREP4) strain, and protein expression induced by addition of 1 mM isopropyl β-D-thiogalactoside (IPTG). Expressed proteins were purified by Ni-NTA resin (Qiagen) under denaturing conditions and resolved in SDS-PAGE.
Alternatively, inclusion bodies were purified from bacterial cultures expressing AaeAPN2 fragments using a B-PER Bacterial Protein extraction reagent following the manufacturer’s instructions (Pierce). The inclusion bodies were dissolved in 0.1M NaOH buffer for 1 h and then dialyzed against 50 mM Na2CO3 (pH 10.5), and protein concentration was measured using the BCA assay (Pierce). Total proteins extracted were analyzed by SDS-PAGE gels and the percentage of protein consisting of AaeAPN2 fragments measured by NIH ImageJ software.
Antibody production and immunolocalization of AaeAPN2 in larval midgut of Ae. aegypti
For antibody production a variable region 29 kDa fragment, AaeAPN2e, was expressed and purified by Ni-NTA resin (Qiagen) under denaturing conditions and resolved in SDS-PAGE. Gels were stained and destained, and the purified protein bands were excised, washed three times and used for antibody development in rabbits.
Fourth instar larvae and guts of Ae. aegypti were fixed overnight in 4% paraformaldehyde (PFA) at 4°C. After fixation, tissues were processed, 8μm thick sections cut and analyzed as described (Kang’ethe et al., 2007). Sections were incubated with protein A-purified rabbit polyclonal anti-AaeAPN2 antibody diluted 1:200 in 1% bovine serum albumin (BSA)/PBST overnight at 40°C, and processed as previously described (Chen et al., 2009b). Images were obtained using a Zeiss Axioplan confocal microscope (LSM 510) located in IIGB, and images were imported in Photoshop for assembly and annotation.
Cry11Aa binding to BBMV
The kinetics of disassociation of Cry11Aa toxin to BBMV was measured using a modified microplate assay. Briefly, in 96-well plates each well was coated with 4 μg BBMV and incubated overnight at 4 °C, and then blocked in blocking buffer (PBST, 0.1% Tween 20 with 5% skim milk) for 1 h at room temperature. For nonspecific binding, increasing amount of biotinylated-Cry11Aa toxin (1–333 nM) was equilibrated with excess unlabeled Cry11Aa protein (10,000 nM) in 100 μl PBST for 1 h at room temperature. The mixtures were then transferred to the plate previously coated with BBMV for 2 h. For total binding, increasing amount of biotinylated-Cry11Aa toxin (1–333 nM) was incubated directly in the plate coated with BBMV as described. The plates were washed with 100 μl PBST, 0.1% Tween 20. Bound biotinylated-Cry11Aa protein was detected by streptavidin-HRP conjugate (1:1500). The HRP activity was revealed with a freshly prepared luminol substrate (Supersignal ELISA pico, Thermo Scientific). An X-ray film was place over the microplate in a darkroom for 1–5 min, and the data quantified by NIH Image J Software and analyzed using Origin (Origin lab). Nonspecific binding was subtracted from total binding to determine specific binding. The concentration corresponding to half maximal response was considered as the dissociation constant (Kd).
Cry11Aa binding to APN2
For the full-length or truncated AaeAPN2 proteins, their dose-dependent binding curves were obtained using a modified ELISA (Perez et al., 2005). In brief, 96-well plates were coated with 0.4 μg Cry11Aa per well and then treated with blocking buffer (PBS, 0.1% Tween 20, 0.5% gelatin) for 1 h at 37°C, washed and then 0–1000 nM AaeAPN2 protein solutions were transferred to the coated plates. After washing, to remove unbound AaeAPN2e, anti-His antibody (1:5000) was added and incubated overnight at 4°C. After additional washes, goat-anti-mouse antibody coupled to alkaline phosphatase (1:2000) was added to wells and incubated for a further 2 h at 37°C. After further washing, alkaline phosphatase activity was revealed with freshly prepared substrate (3 mM nitrophenyl phosphate) and absorbance read at 405 nm with a microplate reader (Molecular Devices). The AaeAPN2a concentration that showed a linear range of binding was used for competitive ELISA below.
Kinetics of Cry11Aa toxin binding to AaeAPN2a was measured by competitive ELISA (Bravo et al., 2004; Dong et al., 2003; Perez et al., 2005). AaeAPN2a, 80 nM, was equilibrated with increasing concentrations of Cry11Aa toxin (0.2 nM to 1000 nM) in 100 μl PBST for 1 h at room temperature. The mixtures were then transferred to plate wells previously coated with Cry11Aa toxin. The detection procedure was then continued as described above. Data were analyzed using Origin (Origin Lab). The concentration corresponding to half maximal absorbance was considered the inhibition constant (Ki) (Dong et al., 2003; Perez et al., 2005).
Competition of APN2 fragments with Cry11Aa binding to BBMV
The ability of AaeAPN2 fragments to compete with toxin binding to BBMV was also measured using biotinylated-Cry11Aa. Briefly, 0.1–1000 nM AaeAPN2 protein fragments were incubated with 15 nM biotinylated-Cry11Aa toxin overnight, and these mixtures were then transferred to the BBMV coated and washed plates described above. After incubation for 2 h, the plates were washed three times with PBS, pH 7.4, and then incubated with streptavidin-HRP conjugate (1:1500) in this PBS buffer. The HRP activities were revealed with a freshly prepared luminal substrate (Supersignal ELISA pico, Thermo scientific) as described above.
Mosquito larvae bioassay
Bioassays were performed with fourth instar of Ae. aegypti larvae (University of California Riverside). Briefly, 20 early fourth instar larvae were placed in water in a 6-oz plastic cup (5 cm cup diameter, Costar, USA) to which an Cry11Aa suspension and APN fragments and PP9 fragment were added to a total volume of 200 ml. The APN and PP9 fragment inclusions were prepared and purified from recombinant E. coli. The Cry11Aa toxin was added at concentration of 150 ng/ml (approximate LC50 of Cry11Aa) while the APN and PP9 fragments were added at concentration of 1000 ng/ml. Mortality was recorded after 24-h incubation at room temperature. All bioassays were examined and repeated at least three times.
Results
Specific binding of Cry11Aa with BBMV
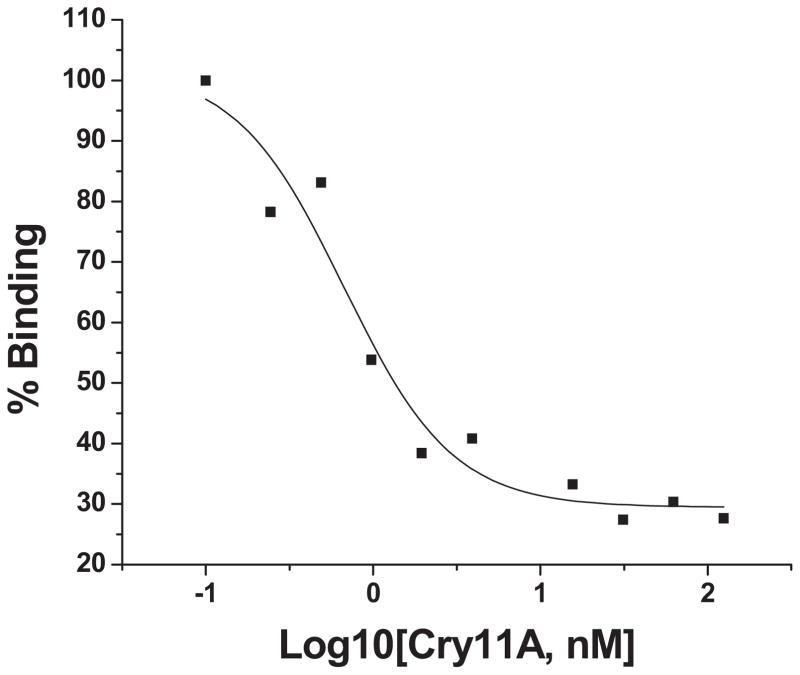
The specific binding of Cry11Aa toxin to freshly prepared Aedes BBMV was determined by using biotinylated Cry11Aa toxin. The density of toxin binding was read with the increasing of biotin labeled toxin for total binding (TT) and in the presence of excess unlabeled Cry11Aa toxin for nonspecific binding (NS) (Fig. 1A). The specific binding was the difference of total and nonspecific binding at each ligand concentration. The Kd for Cry11Aa toxin to BBMV was 29.8 nM (Fig. 1B).
Figure 1. Specificity and affinity of Cry11Aa toxin binding BBMV.
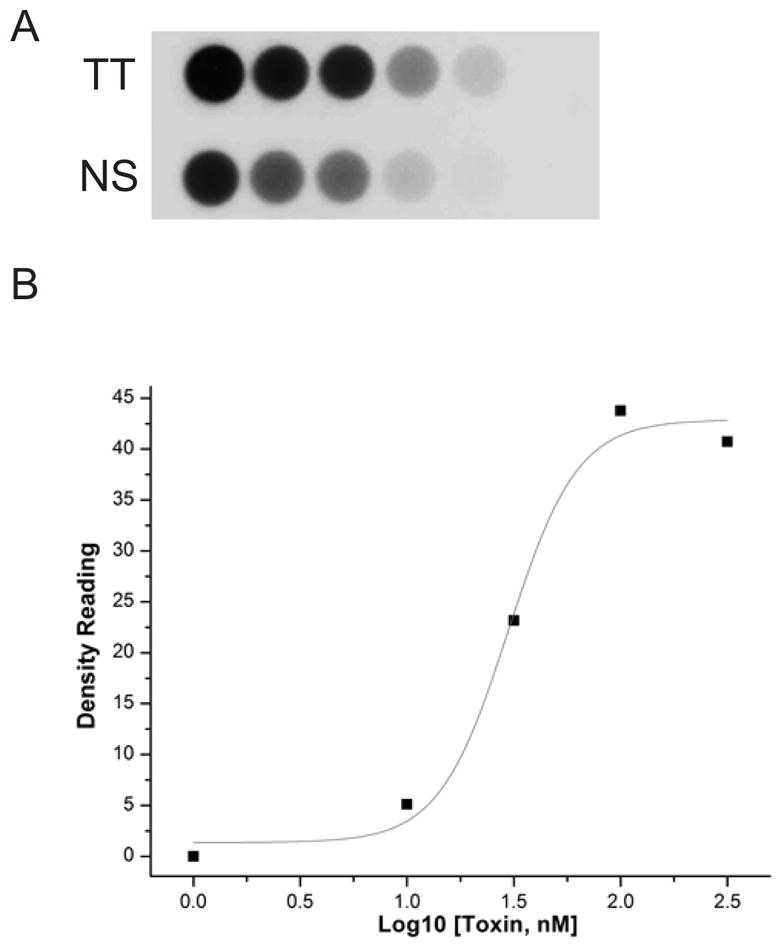
A. Specific binding of Cry11Aa toxin was determined by using biotinylated Cry11Aa toxin binding to Aedes BBMV coated on 96-well plated. Toxin binding was determined with the increasing amounts of biotin-labeled toxin for total binding (TT) and in the presence of excess unlabeled Cry11Aa toxin for nonspecific binding (NS).
B. The specific binding was the difference of total and nonspecific binding at each ligand concentration. The dissociation constant (Kd, 29.8nM) for toxin binding affinity was established using Origin.
Cloning of full-length AaeAPN2
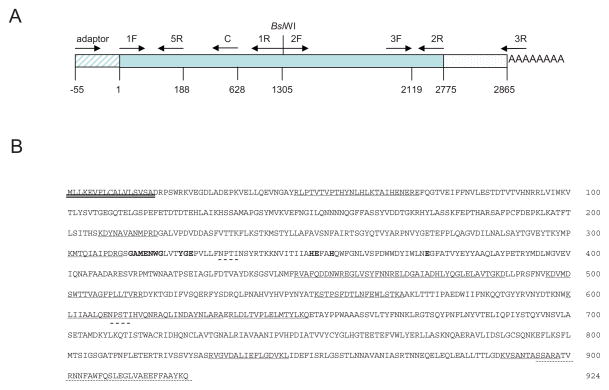
Using pull down assays we previously identified AaeAPN2 as a Cry11Aa toxin binding protein (Chen et al., 2009b). Here the full-length AaeAPN2 cDNA was characterized. To confirm the annotation of the predicted transcript, AAEL008155, 3′- and 5′-RACE were performed using primers deduced from the amino acid fragments detected by mass spectrophotometry. Results from 3′-RACE confirmed the predicted 3′ end sequence in Vectorbase.org was correctly annotated. The 5′ RACE sequences are characteristic of 5′-UTR, ensuring the correct ATG was obtained. This 5′-RACE showed that the 5′ annotation of AAEL008155 in Vectorbase (Aedes aegypti Gene Set 1.3) was incorrect. But we were unable to isolate the full-length ORF in a single PCR using the 5′ and 3′ end sequences obtained. Hence using two partial overlapping fragments, each containing the same unique restriction site BsiWI, were amplified and the full-length AaeAPN2 ORF assembled from these two fragments (Fig. 2A).
Figure 2. Cloning of AaeAPN2 and structural features of the full-length protein.
A. Schematic for isolation of full-length AaeAPN2 cDNA, 5′ RACE product, 3′-RACE product and two partial overlapping fragments. Primer positions are indicated by arrows. B. Twelve peptides identified by mass spectrometry are underlined. The putative N-terminal signal peptide is double-underlined. The GPI-anchor signal peptide is dot-underlined. The predicted N-glycosylation sites are dash-underlined. The consensus gluzincin aminopeptidase motif is in bold and shadow.
The 2775 bp ORF encodes a full-length AaeAPN2 protein of 925 residues, a predicted mass of 104 kDa, with a consensus zinc binding/catalytic site (gluzincin aminopeptidase motif), GAMENWGX3-YRE-X23-HE-X2-H-X18-E (where X is any amino acid) (Hooper, 1994). Twelve peptides that were identified by mass spectrometry (22) are underlined. A 17 amino acid signal peptide sequences in the N-terminus was predicted by SignalP and the C-terminus has a predicted GPI-anchored site with 894Ser, a predicted ω-site. In addition, there are two consensus Asn-Xaa-Ser/Thr sequences, indicating possible N-glycosylation sites in the protein (Fig. 2B).
Cry11Aa binds AaeAPN2 and partial fragments
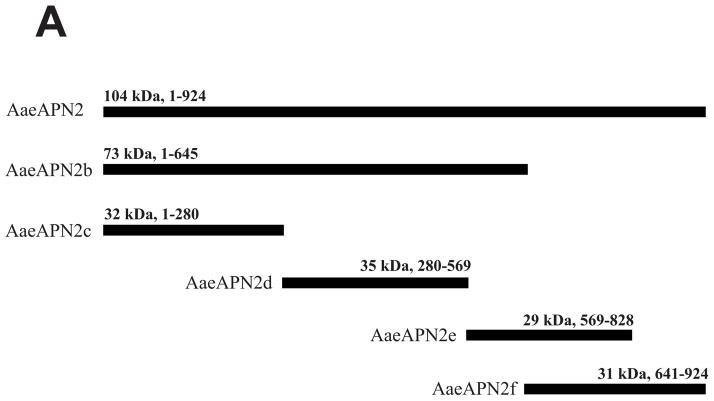
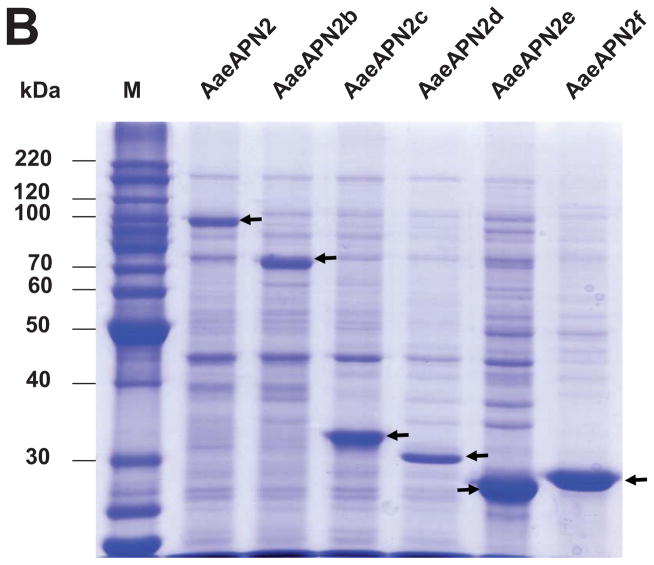
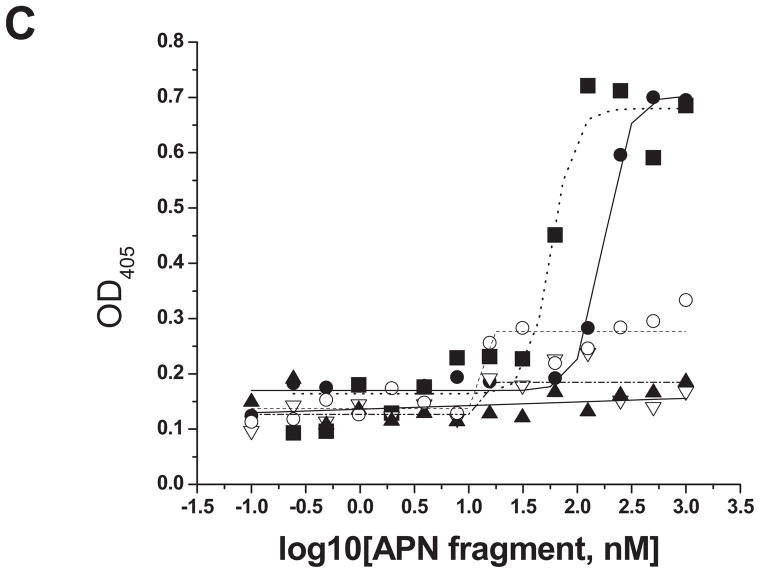
The full-length ORF and five overlapping fragments of AaeAPN2 were cloned and expressed in E. coli. Other than the full-length AaeAPN2 (considered AaeAPN2a), the five partial AaeAPN2 proteins (named AaeAPN2b, c, d, e and f) consist of amino acids 1–645, 1–280, 280–569, 569–828 and 641–924, respectively (Figure 3A). Inclusion bodies of these expressed proteins were extracted and detected by SDS-PAGE (Figure 3B). AaeAPN2, AaeAPN2b and AaeAPN2e proteins bound Cry11Aa, but AaeAPN2c and AaeAPN2d did not bind toxin (Figure 3C). The AaeAPN2e fragment, which also is the least conserved when compared to other APNs, was affinity purified and used as an antigen for antibody development.
Figure 3. Cry11Aa binds AaeAPN2 and its partial fragments.
A. Schematic for isolation of partial AaeAPN2 fragments (b-f). The numbers above each bar refer to the molecular weight of the protein, and amino acid residues are that of the full-length AaeAPN2 protein. A Cry11Aa toxin binding site was mapped to aa 569–641. B. Expression of partial AaeAPN2 proteins. Five overlapping AaeAPN2 cDNA fragments were generated by PCR or by using existing restriction sites. These were then subcloned into pQE series expression vectors and expressed in E. coli M15(pREP4). The inclusion bodies of expressed proteins were obtained and separated by 12% SDS-PAGE gels. Arrows indicate expected size proteins. C. Cry11Aa binding to AaeAPN2 fragments. AaeAPN2a (■), AaeAPN2b (●) and AaeAPN2e (○) show higher binding to immobilized Cry11Aa toxin (0.4 μg) with increased APN protein levels. No binding was observed with AaeAPN2c (▽) and AaeAPN2d (▲).
We then utilized competitive ELISA to measure toxin affinity to the full-length E. coli expressed AaeAPN2, using 8 nM protein, since this binds Cry11Aa in the linear range (Fig. 3C). Increasing Cry11Aa concentrations (0.1–1000nM) displaced the AaeAPN2 protein from binding immobilized Cry11Aa (Figure 4). The apparent constant (Ki) for AaeAPN2a binding to Cry11Aa toxin was 8.6 nM, showing AaeAPN2 binds Cry11Aa toxin with high affinity.
Figure 4. Cry11Aa toxin binds full-length AaeAPN2 proteins with high affinity.
The apparent binding affinity, 8.6 nM, of Cry11Aa toxin to full-length AaeAPN2 protein was determined in a competition assay using 80 nM AaeAPN2a with increasing concentrations of Cry11Aa toxin (0.1–1000 nM). Maximal binding was normalized to the maximal absorbance obtained in the absence of Cry11Aa toxin.
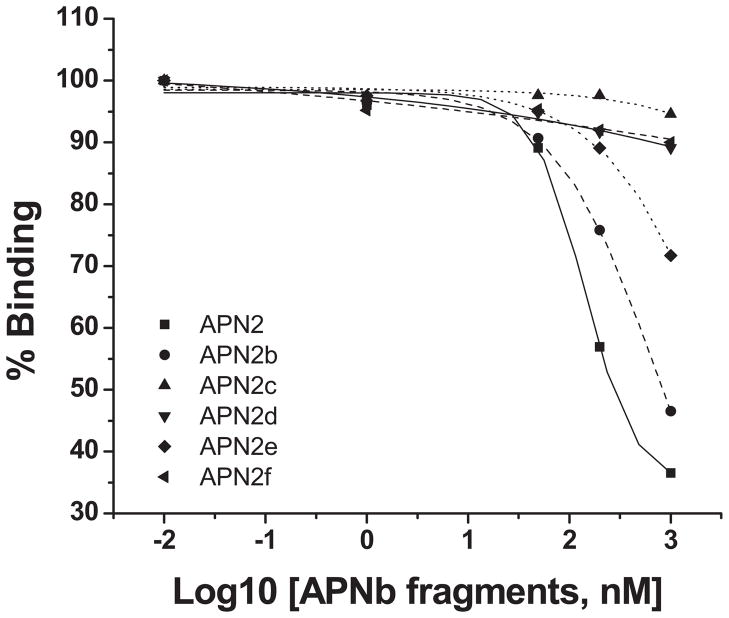
AaeAPN2 fragments that bound Cry11Aa also competed with toxin binding to BBMV
The E. coli expressed AaeAPN2 and the five overlapping fragments (Fig 3A, B) were then analyzed for their ability to affect Cry11Aa binding to BBMV. AaeAPN2 and the partial fragment AaeAPN2b were able to compete readily with Cry11Aa toxin binding to BBMV, while AaeAPN2e competes with toxin binding only at high levels (Fig. 5). Fragments AaeAPN2c, d and f could not compete with Cry11Aa binding. These data together with that in Fig. 3C suggest that part of the Cry11Aa toxin binding domain on AaeAPN2 resides between amino acids 569–641.
Figure 5. AaeAPN2 fragments compete with Cry11Aa toxin binding to Aedes BBMV.
Increasing concentrations of some AaeAPN2 fragments competed with the binding of biotinylated-Cry11Aa toxin to Ae. aegypti BBMV. The full-length AaeAPN2 (■), AaeAPN2b (●) and AaeAPN2e (◆) competed with Cry11Aa binding, with the AaeAPN2f being the least efficient. Whereas the partial fragments, AaeAPN2c (▲), d (▼) and f (◀), did not compete with the binding of Cry11Aa.
Expression of AaeAPN2 in larval midgut of Ae. aegypti
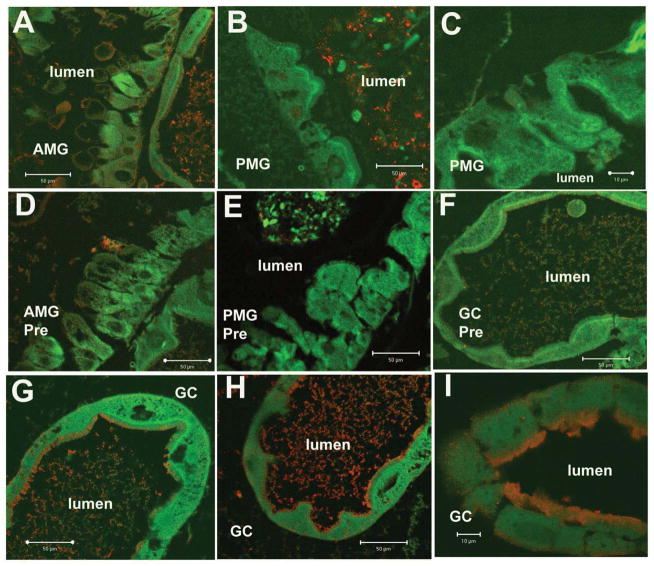
We used the anti-AaeAPN2 antibody (diluted by 1:200) to obtain the localization of in the midgut of fourth instar larvae. Specific immunoreactivity was observed in apical membrane of epithelial cells in proximal and distal regions of gastric caeca (Fig 6., Panels G, H and I). The anterior midgut showed low levels of immunoreactivity (Fig 6., panels A), and no immunoreactivity was observed in the posterior midgut (Fig 6., panels B and C) or the hindgut (data not shown). No specific immunoreactivity was found in tissues probed with preimmune serum using the same dilution as the anti-AaeAPN2 antibody (Fig. 6, panels D, E and F).
Figure 6. AaeAPN2 is expressed in the midgut of larval Ae. aegypti.
Paraffin sections of fourth instar larval midgut were probed with anti-AaeAPN2 antibody. AaeAPN2 was localized in apical membrane of gastric caeca epithelial cells (panels G, H and I, red). Low levels of immunofluorescence were also observed in the anterior midgut (Panel A). No immunofluorescence was observed in the posterior midgut epithelium (panels B, C). No immunoreactivity was detected in control tissues what were incubated with preimmune serum at the same dilutions (1:200) as anti-AaeAPN2 antibody (D, AMG; E, PMG; F, gastric caeca). Scale bars, 100 μm (C, I), 50 μm (A, B, D, F, G, H).
Mosquito bioassay
To test the effect of AaeAPN2 fragments on Cry11Aa toxicity against Aedes larvae, the bioassay was performed using fourth instar larvae and repeated at least three times. The data in table 2 suggest the full-length AaeAPN2 (AaeAPN2a) and the partial fragment, AaeAPN2b, enhanced Cry11Aa toxicity, but other fragments derived from AaeAPN2 did not affect Cry11Aa toxicity.
Table 2.
Effect of APN fragments on Cry11Aa toxicity to Ae. aegypti larvae
| Fed Proteins | %Mortality |
|---|---|
| Cry11Aa | 48.8±2.5 |
| Cry11Aa+APNaa | 76.3±4.8 |
| Cry11Aa+APNb | 67.5±6.5 |
| Cry11Aa+APNc | 50.0±5.0 |
| Cry11Aa+APNd | 51.7±5.8 |
| Cry11Aa+APNe | 51.7±7.6 |
| Cry11Aa+APNf | 46.7±2.9 |
| Cry11Aa+ AeCad (7–11) | 40.0±12.9 |
APN and AeCad fragment inclusions were prepared from recombinant E. coli.
Discussion
The selectivity of B. thuringiensis Cry toxins depends on their ability to bind specific membrane proteins in the midgut of affect insects (Gill et al., 1992). Further, the affinity of these toxins to bind midgut BBMV largely correlates with the toxicity of a particular toxin (Van Rie et al., 1989). The Cry11A toxin binds with relatively good affinity (28.9 nM) to the BBMV from Ae. aegypti larvae. This affinity is quite similar to that in Culex quinquefasciatus, when an affinity of 26 nM was observed (S. M. Dai and S. S. Gill, unpublished results). Not surprisingly, the Cry11Aa toxin is quite toxic to the larvae of both insect species.
These Cry toxins also bind to specific regions of the larval gut. For example, Cry toxins from subsp. israelensis bind to the apical side of distal and proximal caeca and the apical regions of posterior midgut of Ae. aegypti larvae (Charles and de Barjac, 1983; Ravoahangimalala et al., 1993; Zalunin et al., 2002). Recent evidence indicates these midgut regions express proteins that bind Cry11Aa, such as the cadherin and ALP proteins (Chen et al., 2009a; Fernandez et al., 2006). Here we show AaeAPN2 is expressed primarily in the gastric caeca and to a lesser extent in the anterior midgut but not in the posterior midgut (Fig. 6). In contrast, AaeAPN1 was localized to the apical side of posterior midgut epithelial cells, but not in anterior midgut and gastric caeca cells (Chen et al., 2009b). The distribution of these two APNs collectively is consistent with the expression profile of Aedes cadherin and an ALP protein (Chen et al., 2009a; Fernandez et al., 2006)..
However, there are some differences in Cry toxin action in mosquitoes. For instance, Cry11Ba, a toxin related to Cry11Aa, binds an APN with high affinity in two anopheline mosquitoes (Abdullah et al., 2006; Zhang et al., 2008), besides, it also binds to an ALP with affinity of 23.9 nM (Hua et al., 2009). In this study, we observed that AaeAPN2 from Ae. aegypti bound to Cry11Aa toxin with high affinity of 8.6 nM. Since previous work showed AaeAPN1 from the same mosquito species also bound to Cry11Aa toxin with high affinity, it would not be surprising that the Cry11Aa toxin can bind APNs directly without prior binding to cadherin, which is believed to initiate the process of oligomerization of Cry1 toxins in lepidopteran insects. And if these initial observations are further validated, this evidence would suggest that there is some additional complexity within the mechanism of action of mosquito-specific toxins, compared to that of lepidopteran-active toxins. While the site(s) of Cry toxin binding on Aedes mosquito APNs has not been previously established, there has been much greater investigation on Cry1A binding sites in lepidopteran APNs. For example, in eight APN isoforms from B. mori and Plutella xylostella, a highly conserved site, RXXFPXXDEP, has been suggested to be the Cry1A toxin binding region (Nakanishi et al., 2002). However, although three Cry11Ba binding sites on An. gambiae BBMV were pinpointed recently, none of them is located in the conserved region (21, 45). Here we show that this conserved region is apparently not involved in Cry11Aa binding, since AaeAPN2c, which includes this conserved site, does not bind the toxin either (Figs. 3c and 5).
Our data demonstrate that full-length AaeAPN2 and two AaeAPN2 fragments (AaeAPN2b and AaeAPN2e) bind Cry11Aa toxin (Fig. 3c) and also compete with Cry11Aa binding to midgut BBMV isolated from Ae. aegypti mosquito (Fig. 5). In contrast, three other fragments, AaeAPN2c, d, and f that consist of amino acids 1–280, 280–596 and 569–828, respectively, do not bind Cry11Aa, nor do they compete with toxin binding to BBMV. Collectively, the data suggest amino acid 569–641 form part of the Cry11Aa toxin binding region. However, we anticipate this domain is likely part of a larger three-dimensional domain that binds the toxin, since most globular proteins exhibit complicated three-dimensional folding involved in protein-protein interactions. Moreover, it is likely that more than one AaeAPN2 domain is involved in binding Cry11Aa toxin. This, in part, would explain the low binding ability of AaeAPN2e, whereas the larger AaeAPN2b protein, which also include amino acids 569–641, competed more effectively with Cry11Aa binding to BBMV. Not surprisingly full-length AaeAPN2 had the highest Cry11Aa binding and best competed with the toxin binding to Aedes BBMV. Similar to previously reported for AaeAPN1 (Chen et al., 2009b). Here, AaeAPN2, which was expressed in E. coli, are able to bind Cry11Aa by using ELISA technique as well as competes with Cry11Aa toxin binding to BBMV. In bioassays against Ae. aegypti larvae, the presence of full-length and a partial fragment (AaeAPN2b) of AaeAPN2 enhanced Cry11Aa larval mortality but the putative binding region, AaeAPN2e, could not change Cry11Aa toxicity significantly (Table 3). However, for the Cry11Ba receptor, AgAPN2, the comparatively longer partial 70 kDa fragment inhibits Cry11Ba toxicity against An. gambiae during the bioassay, but two truncated fragments derived from 70 kDa fragment show inhibitory and synergistic effects on Cry11Ba toxicity, respectively (45).
AaeAPN2 was identified as a Cry11A-bound protein. Here we show it binds Cry11A toxin with high affinity (Kd=8.6nM).
AaeAPN2 is localized to the apical membrane of epithelial cells in proximal and distal regions of larval caeca.
The toxin-binding regions are narrowed down to two partial AaeAPN2 fragments, AaeAPN2b and AaeAPN2e.
AaeAPN2 and AaeAPN2e can deplete Cry11A binding to BBMV and enhance Cry11A larval mortality against Aedes aegypti.
Acknowledgments
This research was funded in part through grants from the National Institutes of Health,1R01 AI066014 and the University of California Agricultural Experiment Station.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdullah MA, Valaitis AP, Dean DH. Identification of a Bacillus thuringiensis Cry11Ba toxin-binding aminopeptidase from the mosquito, Anopheles quadrimaculatus. BMC Biochem. 2006;7:16. doi: 10.1186/1471-2091-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Mizuno E, Hara H, Nakanishi K, Kitami M, Miura N, Tabunoki H, Watanabe A, Sato R. Location of the Bombyx mori aminopeptidase N type 1 binding site on Bacillus thuringiensis Cry1Aa toxin. Appl Environ Microbiol. 2005;71:3966–3977. doi: 10.1128/AEM.71.7.3966-3977.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayyareddy K, Zhu X, Orlando R, Adang MJ. Proteome analysis of Cry4Ba toxin-interacting Aedes aegypti lipid rafts using geLC-MS/MS. J Proteome Res. 2012;11:5843–5855. doi: 10.1021/pr3006167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C, O’Neil S, Ben-Dov E, Jones AF, Murphy L, Quail MA, Holden MT, Harris D, Zaritsky A, Parkhill J. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp israelensis. Appl Environ Microbiol. 2002;68:5082–5095. doi: 10.1128/AEM.68.10.5082-5095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Gill SS, Soberon M. Bacillus thuringiensis: Mechanisms and Use. In: Gilbert LI, Kostas I, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 6. Elsevier; 2005. pp. 175–205. [Google Scholar]
- Bravo A, Gill SS, Soberon M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon. 2007;49:423–435. doi: 10.1016/j.toxicon.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Gomez I, Conde J, Munoz-Garay C, Sanchez J, Miranda R, Zhuang M, Gill SS, Soberon M. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochim Biophy Acta. 2004;1667:38–46. doi: 10.1016/j.bbamem.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Burton SL, Ellar DJ, Li J, Derbyshire DJ. N-acetylgalactosamine on the putative insect receptor aminopeptidase N is recognised by a site on the domain III lectin-like fold of a Bacillus thuringiensis insecticidal toxin. J Mol Biol. 1999;287:1011–1022. doi: 10.1006/jmbi.1999.2649. [DOI] [PubMed] [Google Scholar]
- Chang C, Yu YM, Dai SM, Law SK, Gill SS. High-level cryIVD and cytA gene expression in Bacillus thuringiensis does not require the 20-kilodalton protein, and the coexpressed gene products are synergistic in their toxicity to mosquitoes. Appl Environ Microbiol. 1993;59:815–821. doi: 10.1128/aem.59.3.815-821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles JF, de Barjac H. [Action of crystals of Bacillus thuringiensis var. israelensis on the midgut of Aedes aegypti L larvae, studied by electron microscopy] Ann Microbiol (Paris) 1983;134A:197–218. [PubMed] [Google Scholar]
- Chen J, Aimanova KG, Fernandez LE, Bravo A, Soberon M, Gill SS. Aedes aegypti cadherin serves as a putative receptor of the Cry11Aa toxin from Bacillus thuringiensis subsp israelensis. Biochem J. 2009a;424:191–200. doi: 10.1042/BJ20090730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Aimanova KG, Pan S, Gill SS. Identification and characterization of Aedes aegypti aminopeptidase N as a putative receptor of Bacillus thuringiensis Cry11A toxin. Insect Biochem Mol Biol. 2009b;39:688–696. doi: 10.1016/j.ibmb.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott CN, Ellar DJ. Comparative toxicity of Bacillus thuringiensis varisraelensis crystal proteins in vivo and in vitro. J Gen Microbiol. 1988;134:2551–2558. doi: 10.1099/00221287-134-9-2551. [DOI] [PubMed] [Google Scholar]
- Cowles EA, Yunovitz H, Charles JF, Gill SS. Comparison of toxin overlay and solid-phase binding assays to identify diverse CryIA(c) toxin-binding proteins in Heliothis virescens midgut. Appl Environ Microbiol. 1995;61:2738–2744. doi: 10.1128/aem.61.7.2738-2744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Chen S, Bartsch U, Schachner M. Generation of affinity matured scFv antibodies against mouse neural cell adhesion molecule L1 by phage display. Biochem Biophys Res Commun. 2003;301:60–70. doi: 10.1016/s0006-291x(02)02933-9. [DOI] [PubMed] [Google Scholar]
- Fernandez LE, Aimanova KG, Gill SS, Bravo A, Soberon M. A GPI-anchored alkaline phosphatase is a functional midgut receptor of Cry11Aa toxin in Aedes aegypti larvae. Biochem J. 2006;394:77–84. doi: 10.1042/BJ20051517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Cowles EA, Francis V. Identification, isolation, and cloning of a Bacillus thuringiensis CryIAc toxin-binding protein from the midgut of the lepidopteran insect Heliothis virescens. J Biol Chem. 1995;270:27277–27282. doi: 10.1074/jbc.270.45.27277. [DOI] [PubMed] [Google Scholar]
- Gill SS, Cowles EA, Pietrantonio PV. The mode of action of Bacillus thuringiensis endotoxins. Annual review of entomology. 1992;37:615–636. doi: 10.1146/annurev.en.37.010192.003151. [DOI] [PubMed] [Google Scholar]
- Hooper NM. Families of zinc metalloproteases. FEBS letters. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- Hua G, Zhang R, Bayyareddy K, Adang MJ. Anopheles gambiae alkaline phosphatase is a functional receptor of Bacillus thuringiensis jegathasan Cry11Ba toxin. Biochemistry. 2009;48:9785–93. doi: 10.1021/bi9014538. [DOI] [PubMed] [Google Scholar]
- Jenkins JL, Lee MK, Valaitis AP, Curtiss A, Dean DH. Bivalent sequential binding model of a Bacillus thuringiensis toxin to gypsy moth aminopeptidase N receptor. J Biol Chem. 2000;275:14423–14431. doi: 10.1074/jbc.275.19.14423. [DOI] [PubMed] [Google Scholar]
- Kang’ethe W, Aimanova KG, Pullikuth AK, Gill SS. NHE8 mediates amiloride-sensitive Na+/H+ exchange across mosquito Malpighian tubules and catalyzes Na+ and K+ transport in reconstituted proteoliposomes. Am J Physiol Renal Physiol. 2007;292:F1501–1512. doi: 10.1152/ajprenal.00487.2005. [DOI] [PubMed] [Google Scholar]
- Knight PJ, Knowles BH, Ellar DJ. Molecular cloning of an insect aminopeptidase N that serves as a receptor for Bacillus thuringiensis CryIA(c) toxin. J Biol Chem. 1995;270:17765–17770. doi: 10.1074/jbc.270.30.17765. [DOI] [PubMed] [Google Scholar]
- Lereclus D, Agaisse H, Gominet M, Chaufaux J. Overproduction of encapsulated insecticidal crystal proteins in a Bacillus thuringiensis spo0A mutant. Biotechnology (N Y) 1995;13:67–71. doi: 10.1038/nbt0195-67. [DOI] [PubMed] [Google Scholar]
- Masson L, Lu YJ, Mazza A, Brousseau R, Adang MJ. The CryIA(c) receptor purified from Manduca sexta displays multiple specificities. J Biol Chem. 1995;270:20309–20315. doi: 10.1074/jbc.270.35.20309. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Yaoi K, Nagino Y, Hara H, Kitami M, Atsumi S, Miura N, Sato R. Aminopeptidase N isoforms from the midgut of Bombyx mori and Plutella xylostella -- their classification and the factors that determine their binding specificity to Bacillus thuringiensis Cry1A toxin. FEBS Lett. 2002;519:215–220. doi: 10.1016/s0014-5793(02)02708-4. [DOI] [PubMed] [Google Scholar]
- Nielsen-Leroux C, Charles JF. Binding of Bacillus sphaericus binary toxin to a specific receptor on midgut brush-border membranes from mosquito larvae. Eur J Biochem. 1992;210:585–590. doi: 10.1111/j.1432-1033.1992.tb17458.x. [DOI] [PubMed] [Google Scholar]
- Pardo-Lopez L, Soberon M, Bravo A. Bacillus thuringiensis insecticidal three-domain Cry toxins: mode of action, insect resistance and consequences for crop protection. FEMS Microbiol Rev. 2013;37:3–22. doi: 10.1111/j.1574-6976.2012.00341.x. [DOI] [PubMed] [Google Scholar]
- Perez C, Fernandez LE, Sun J, Folch JL, Gill SS, Soberon M, Bravo A. Bacillus thuringiensis subsp israelensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane-bound receptor. Proc Natl Acad Sci USA. 2005;102:18303–18308. doi: 10.1073/pnas.0505494102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigott CR, Ellar DJ. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol Mol Biol Rev. 2007;71:255–281. doi: 10.1128/MMBR.00034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal R, Sivakumar S, Agrawal N, Malhotra P, Bhatnagar RK. Silencing of midgut aminopeptidase N of Spodoptera litura by dsRNA establishes its role as BT toxin receptor. J Biol Chem. 2002;107:1. doi: 10.1074/jbc.C200523200. [DOI] [PubMed] [Google Scholar]
- Ravoahangimalala O, Charles JF, Schoeller-Raccaud J. Immunological localization of Bacillus thuringiensis serovar israelensis toxins in midgut cells of intoxicated Anopheles gambiae larvae (Diptera: Culicidae) Res Microbiol. 1993;144:271–278. doi: 10.1016/0923-2508(93)90011-p. [DOI] [PubMed] [Google Scholar]
- Saengwiman S, Aroonkesorn A, Dedvisitsakul P, Sakdee S, Leetachewa S, Angsuthanasombat C, Pootanakit K. In vivo identification of Bacillus thuringiensis Cry4Ba toxin receptors by RNA interference knockdown of glycosylphosphatidylinositol-linked aminopeptidase N transcripts in Aedes aegypti larvae. Biochemical and biophysical research communications. 2011;407:708–713. doi: 10.1016/j.bbrc.2011.03.085. [DOI] [PubMed] [Google Scholar]
- Sivakumar S, Rajagopal R, Venkatesh GR, Srivastava A, Bhatnagar RK. Knockdown of aminopeptidase-N from Helicoverpa armigera larvae and in transfected Sf21 cells by RNA interference reveals its functional interaction with Bacillus thuringiensis insecticidal protein Cry1Ac. J Biol Chem. 2007;282:7312–7319. doi: 10.1074/jbc.M607442200. [DOI] [PubMed] [Google Scholar]
- Soberon M, Gill SS, Bravo A. Signaling versus punching hole: How do Bacillus thuringiensis toxins kill insect midgut cells? Cell Mol Life Sci. 2009;66:1337–1349. doi: 10.1007/s00018-008-8330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiewsiri K, Wang P. Differential alteration of two aminopeptidases N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc Natl Acad Sci USA. 2011;108:14037–14042. doi: 10.1073/pnas.1102555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rie J, Jansens S, Hofte H, Degheele D, Van Mellaert H. Specificity of Bacillus thuringiensis delta-endotoxins. Importance of specific receptors on the brush border membrane of the mid-gut of target insects. Eur J Biochem. 1989;186:239–247. doi: 10.1111/j.1432-1033.1989.tb15201.x. [DOI] [PubMed] [Google Scholar]
- Yaoi K, Kadotani T, Kuwana H, Shinkawa A, Takahashi T, Iwahana H, Sato R. Aminopeptidase N from Bombyx mori as a candidate for the receptor of Bacillus thuringiensis Cry1Aa toxin. Eur J Biochem. 1997;246:652–657. doi: 10.1111/j.1432-1033.1997.t01-1-00652.x. [DOI] [PubMed] [Google Scholar]
- Yaoi K, Nakanishi K, Kadotani T, Imamura M, Koizumi N, Iwahana H, Sato R. Bacillus thuringiensis Cry1Aa toxin-binding region of Bombyx mori aminopeptidase N. FEBS letters. 1999;463:221–224. doi: 10.1016/s0014-5793(99)01626-9. [DOI] [PubMed] [Google Scholar]
- Zalunin IA, Chaika S, Dronina MA, Revina LP. [Cytopathological effect of Bacillus thuringiensis israelensis endotoxins on the intestines of Aedes aegypti mosquito larvae] Parazitologiia. 2002;36:337–344. [PubMed] [Google Scholar]
- Zhang R, Hua G, Andacht TM, Adang MJ. A 106-kDa aminopeptidase is a putative receptor for Bacillus thuringiensis Cry11Ba toxin in the mosquito Anopheles gambiae. Biochemistry. 2008;47:11263–11272. doi: 10.1021/bi801181g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Hua G, Urbauer JL, Adang MJ. Synergistic and Inhibitory Effects of Aminopeptidase Peptides on Bacillus thuringiensis Cry11Ba Toxicity in the Mosquito Anopheles gambiae. Biochemistry. 2010;49:8512–8919. doi: 10.1021/bi1009908. [DOI] [PubMed] [Google Scholar]
- Zhang S, Cheng H, Gao Y, Wang G, Liang G, Wu K. Mutation of an aminopeptidase N gene is associated with Helicoverpa armigera resistance to Bacillus thuringiensis Cry1Ac toxin. Insect Biochem Mol Biol. 2009;39:421–9. doi: 10.1016/j.ibmb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Zhang X, Candas M, Griko NB, Taussig R, Bulla LA., Jr A mechanism of cell death involving an adenylyl cyclase/PKA signaling pathway is induced by the Cry1Ab toxin of Bacillus thuringiensis. Proc Natl Acad Sci USA. 2006;103:9897–9902. doi: 10.1073/pnas.0604017103. [DOI] [PMC free article] [PubMed] [Google Scholar]