Abstract
The virologic determinants of progressive liver disease associated with hepatitis B virus (HBV) remain unclear. Previous investigations have associated HBV disease with specific mutations but this association may be confounded by HBV genotype, HLA haplotype of the infected individual or both. The association between non-synonymous mutations located within putative cytotoxic T-lymphocyte directed epitopes (CDE) of the HBV core region and disease states was investigated. Subjects infected with HBV were enrolled from a clinical cohort in Seoul, Korea, and HBV core gene sequences were analyzed for mutational patterns inside and outside of CDE with respect to subject demographics and HBV-related disease states. No specific mutation or pattern of mutations were associated with progressive disease states; however, individuals with cirrhosis and hepatocellular carcinoma had greater numbers of non-synonymous mutations within CDE when compared to those with chronic HBV infection who were HBeAg positive (P = 0.007 and 0.026, respectively). In conclusion, this study demonstrates that HBV disease progression is associated with viral escape mutations that are a marker of CTL activity. These data suggest that the number of non-synonymous mutations in the HBV core region may predict HBV disease progression better than any single mutation or pattern of mutations.
Keywords: hepatitis B virus, mutations, cytotoxic T-lymphocyte epitopes, cirrhosis, hepatocellular carcinoma
INTRODUCTION
Approximately two billion people have been exposed to Hepatitis B virus (HBV) (Family: Hepadnavirus, Genus: Orthohepadnaovirus), and an estimated 350 million people have chronic HBV infection representing country-specific endemic rates of 2–20% [Kao and Chen, 2002]. In addition, chronic HBV infection is the leading cause of liver cirrhosis and hepatocellular carcinoma (HCC) in the world, accounting for 563,000 deaths annually worldwide [Perz et al., 2006]. Cytotoxic T lymphocytes (CTL) play a primary role in the host's immune response against HBV infection, which are engaged by HLA Class I-restricted epitopes of HBV proteins presented on the surface of infected hepatocytes [Bertoletti et al., 1991; Gehring et al., 2007]. Epitopes within the core region of HBV are especially important targets for CTL responses [Bozkaya et al., 1996; Lee et al., 1996; Liu et al., 2003; Whalley et al., 2004; Osiowy et al., 2006; Wang et al., 2010]. Although robust CTL responses can clear HBV infection [Missale et al., 1993; Maini et al., 2000; Whalley et al., 2004], inefficient responses result in viral persistence and chronic HBV [Rehermann et al., 1996; Whalley et al., 2004; Boni et al., 2007]. Continued CTL-associated chronic inflammation over decades during chronic infection can lead to cirrhosis and HCC [Chisari, 1997; Nakamoto et al., 1998; El-Serag and Rudolph, 2007], and CTL pressure on HBV can select for mutations within the HBV genome that allow for immune escape [Bertoletti et al., 1994a,b; Khakoo et al., 2000].
Recent studies have demonstrated that HBV mutations are associated with advanced HBV disease manifested as HCC and cirrhosis, and various single amino acid changes have been implicated as being responsible [Ni et al., 2003; Liu et al., 2009]. This cross-sectional study of patients with various HBV disease states (chronic HBV, HBV with cirrhosis, and HCC) evaluates how observed non-synonymous mutations within the HBV core gene are associated with CTL-mediated immune pressure and disease status.
MATERIALS AND METHODS
Study Population
This work received approval from the institutional review boards of the respective institutions. Serum samples were collected from all patients who visited Kangnam St. Mary's Hospital, Seoul, Korea from 2003 to 2009 and diagnosed clinically with chronic HBV, cirrhosis, or HCC. All serum samples were screened initially for HBV viral DNA using PCR. Data on AST, ALT, and hepatitis B virus e antigen (HBeAg) were obtained from patient charts. Patients were divided into four groups for analysis: chronic HBV HBeAg-positive individuals, chronic HBV HBeAg-negative individuals, patients infected with HBV, cirrhosis, and infected patients with HCC.
Polymerase Chain Reaction (PCR) and Sequencing
Viral DNA was extracted from serum using QIAamp DNA Mini kit (Qiagen, Valencia, CA) following the manufacturer's protocol. The precore/core gene of HBV (GenBank Accession No. NC_003977, nt 1901–2452) was amplified via nested-PCR using primers HBC1 and HBC2 for the first round and primers HBC3 and HBC4 for the second round, as described previously [Lusida et al., 2008]. The PCR products fragments were purified using QIAquick Gel Extraction Kit (Qiagen) per manufacturer's protocol and were sequenced directly using the ABI Prism Big Dye kit version 3.0 (Applied Biosystems, Foster City, CA) on an ABI 3730XL DNA automated sequencer (Applied Biosystems).
Phylogenetic, Mutation, and CTL Analysis
The nucleotide sequences were aligned with ClustalX (ver. 1.81) software and phylogenetic trees were constructed by neighbor-joining method with other HBV reference strains from GenBank. Genetic distances between sequences were calculated by bootstrap on 1,000 repeats under a TN 69 model using Geneious Pro software (Biomatters, Auckland, New Zealand). Nucleotide alignments were translated and analyzed using Geneious Pro software (Biomatters) [Drummond et al., 2005]. The 9 HLA Class I/CTL-defined epitopes in the core region analyzed in this study have been described previously [Desmond et al., 2008]. These epitopes overlap with each other to form four specific regions within the HBV core protein (HBcAg), encompassing amino acid (aa) residues 11–27, 88–96, 107–125, and 139–151. The HLA haplo-types recognizing these epitopes are all found in the Korean population [Park et al., 1998]. To evaluate sequence changes, a consensus nucleotide sequence for all subject sequences was generated using Geneious Pro software (Biomatters), and sequences from individuals were then compared to this consensus sequence and nucleotide mutations were noted. The mutations resulting in aa changes (i.e., non-synonymous mutations) were then analyzed both within and outside CDEs.
Selection Analysis and Signature Pattern Analysis
Selective pressure on individual codons was assessed in the sequence data via single likelihood ancestor counting (SLAC) analysis [Poon et al., 2009], as implemented in the HyPhy software package on the Datamonkey server [www.datamonkey.org accessed July 26, 2010]. Briefly, likelihood-based selection analyses were performed on sequence alignments for each codon with a conservative significance threshold of P = 0.1. Next, any overlap in the codons found to be under selection was compared to the non-synonymous mutations for each sequence. Sequences were then analyzed for possible patterns of mutations that were particular to HBV clinical status using viral epidemiology signature pattern analysis (VESPA) [www.hiv.lanl.gov/content/sequence/VESPA/vespa; accessed July 26, 2010]. Briefly, a background group of aa sequences (e.g., sequences from chronic HBV infected and e antigen-positive individuals) was compared to a query group of sequences (e.g., sequences from patients with cirrhosis), and the software identified sites where the most commonly found aa differed between the background and query sequences [Korber and Myers, 1992]. As there were only 35 sequences in the baseline group of individuals with chronic HBV infection and e antigen-positive status, 35 randomly selected sequences from each group were used for the analysis comparing this group with the cirrhotic group. Likewise, as the HCC group only had 33 sequences, 33 randomly selected sequences from both the group of individuals with chronic HBV infection and e antigen-positive status and the group with HCC groups were used for analysis. Finally, the amino acid sites noted in the signature patterns for the clinical status groups, the codons noted to be under selection in the SLAC analysis, and the nucleotide sites noted to contain non-synonymous mutations were evaluated for overlap.
Statistical Analysis
Statistical analysis was performed with PASW Statistics GradPack 18 for Mac (2009). Binary logistic regression was used to compare the number of mutations in the above-defined clinical groups of: chronic HBV with e antigen-positive status, chronic HBV with e antigen-negative status, HBV infected with cirrhosis and HBV infected with HCC, with age and sex as covariates and backward likelihood ratio testing to select for the best model. Chi-squared and Fisher's exact tests were used to compare the mutation profiles of clinical groups. Bonferroni's correction was used to correct for multiple comparisons. ANOVA was used to compare means of ALT and age between groups.
RESULTS
Between 2003 and 2009, 132 patients were diagnosed clinically with chronic HBV, HBV and cirrhosis, or HBV and HCC at the Kangnam St. Mary's Hospital, Seoul, Korea and were eligible for this study. Nine were excluded from the analysis due to missing HBeAg data. Patient characteristics are summarized in Table I. All clinical groups were predominantly male and mean ages for the cirrhosis and HCC groups were greater than for the chronic HBV group. Similarly, the proportion HBeAg-negative subjects was significantly greater for the cirrhosis and HCC groups than for the chronic HBV group. All but one HBV isolate obtained was of genotype C2.
TABLE I.
Patient Characteristics
| CHB (n = 46)a | LC (n = 44) | HCC (n = 33) | P-valueb | |
|---|---|---|---|---|
| Number of male patients (%) | 35 (76.1) | 25 (58.1) | 30 (76.2) | 0.037 |
| Mean age in years (range) | 36.7 (12–68) | 49.7 (25–100) | 52.1 (35–63) | <0.001 |
| Number of HBeAg positive (%) | 35 (76.1) | 18 (40.9) | 19 (57.5) | 0.003 |
| Mean ALT (SD) | 143.8 (129.1) | 62.6 (73.2) | 102.5 (140.6) | NS |
CHB, chronic HBV infection; LC, liver cirrhosis; HCC, hepatocellular carcinoma; HBeAg, hepatitis B virus e antigen; NS, not significant P > 0.05; ALT, alanine aminotransferase; SD, standard deviation.
No significant differences between HBeAg-positive CHB and HBeAg-negative CHB groups.
P-value of comparison between groups. Proportions of male patients and percentage of HBeAg-positive individuals were compared using chi-squared test. Mean age and ALT were compared using Student's t-test.
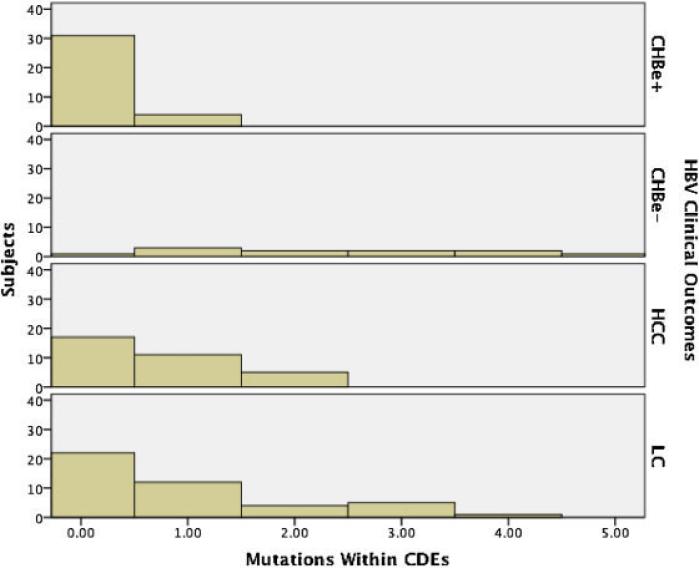
When investigating the association between potential viral escape (i.e., non-synonymous changes within the viral HBV core gene) and clinical disease state, we found a median of three non-synonymous mutations per sequence (range, 0–14). Logistic regression found that the sequences from the group of patients with chronic HBV and without e antigen, and the group with cirrhosis had significantly more non-synonymous mutations than those from the group of individuals with chronic HBV and e antigen-positive status (P = 0.002 and 0.017, respectively). No such association was found for the HCC group (P = 0.114). To evaluate more specifically viral CTL escape, we evaluated mutations inside and outside potential CTL defined epitopes (CDE) and found that overall more non-synonymous mutations were found outside CDE (median two mutations, range 0–10) versus inside CDE (median zero mutations, range 0–4) (Fig. 1). Logistic regression found that patients with cirrhosis, HCC and chronic HBV with e antigen-negative status all had significant odds ratios of having greater numbers non-synonymous mutations within CDE when compared to the control group of individuals with chronic HBV and e antigen-positive status (P = 0.007, 0.026, and <0.003, respectively) (Table II). Additionally, HBeAg-positive cases of HCC and cirrhosis had greater numbers of mutations in CDE as compared to the patients with chronic HBV and e antigen-positive status (P = 0.018 and 0.023, respectively). Notably, the odds ratios increased exponentially for each additional mutation. Taken together, this indicates that larger numbers of putative CTL viral escape mutations are associated with progressive HBV-related liver disease, irrespective of the HBeAg status.
Fig. 1.
Mutations by HBV Disease state. The average number of non-synonymous mutations within cytotoxic T-lymphocyte directed epitopes (CDEs) were higher among progressive disease states. Individuals with chronic HBV infection and e antigen present (CHBe+) had fewer mutations than those with hepatocellular carcinoma (HCC) (P = 0.026), and liver cirrhosis (LC) (P = 0.007). Additionally, individuals with chronic HBV infection and e antigen present (CHBe+) had fewer mutations than individuals with chronic HBV and e antigen absent (CHBe–) (P = <0.003).
TABLE II.
Multivariate Analyses Adjusted for Age, Sex, and Mutations Outside Cytotoxic T-Cell Defined Epitopes
| Disease state | OR fold increase per mutation | 95% CI | P-value | Other covariates (P-value) |
|---|---|---|---|---|
| HCC | 4.70 | 1.20–18.37 | 0.026 | Age (<0.001) Mutations outside CDE (0.694)a Sex (0.551)a |
| HBeAg-positive HCC | 5.70 | 1.34–24.22 | 0.018 | Age (0.003) Mutations outside CDE (0.362)a Sex (0.603)a |
| LC | 4.29 | 1.48–12.44 | 0.007 | Age (0.002) Mutations outside CDE (0.550)a Sex (0.252)a |
| HBeAg-positive LC | 4.96 | 1.24–19.77 | 0.023 | Age (0.099)a Mutations outside CDE (0.543)a Sex (0.099)a |
| HBeAg-negative CHB | 30.21 | 3.24–281.28 | 0.003 | Age (0.711)a Mutations outside CDE (0.708)a Sex (0.974)a |
| HBeAg negativeb | 1.98 | 1.32–2.96 | 0.001 | Age (0.048) Mutations outside CDE (0.505)a Sex (0.710)a |
CDE, cytotoxic T-lymphocyte directed epitopes; OR, odds ratio; HBeAg, hepatitis B virus e antigen; HCC, hepatocellular carcinoma; LC, liver cirrhosis; CHB, chronic hepatitis B.
Excluded from the final selected regression model because of lack of significance.
Comparison of all HBeAg-positive versus all HBeAg-negative subjects.
When evaluating if any single viral mutation was associated with clinical status, mutations in the HBV regions corresponding to amino acids 11–27 and 107–125 were associated significantly with the group of individuals with chronic HBV and e antigen-negative status as compared to the individuals with chronic HBV and e antigen-positive status (P < 0.001 for both). Specifically, mutations at residues 13, 21, 26, 27, 113, 116, 147, and 149 occurred greater than five times among the e antigen-negative group, although no significant associations were found for any single amino acid mutation and clinical state after Bonferroni correction. When evaluating for selection among the HBV sequences, seven positively and 29 negatively selected codon mutations were identified, but there was no enrichment in these selected sites between the group with chronic HBV and e antigen-positive group and the groups with chronic HBV and without e antigen, cirrhosis, or HCC groups (P > 0.1). There was also no pattern for multiple mutations, that is, signature, among all clinical status groups by VESPA signature pattern analysis.
DISCUSSION
Chronic inflammation of the liver during the host's immune response to chronic HBV is proposed as a mechanism for the progression of chronic HBV to cirhosis and HCC [El-Serag and Rudolph, 2007]. In addition to causing inflammation, the CTL-mediated immune response directed against the virus can produce a selective pressure on the viral genome. The virus may evade this immune response via escape mutations in the coding regions encoding CTL epitopes. Therefore, ongoing CTL immune response to HBV could be associated with continued liver inflammation and risk of disease progression and also ongoing mutagenesis and viral escape. Consistent with this hypothesis and relevant to the current investigation, a study of full-length HBV sequences from a Korean population showed that increased genetic divergence between HBV sequences was associated with HCC and cirrhosis as compared to chronic HBV [Ahn et al., 2010]. Additionally, a number of studies have demonstrated that particular mutations in the HBV genome are associated with advanced HBV disease states [21,30] and some of these mutations fall within CTL epitopes [Hosono et al., 1995; Maruyama et al., 2000; Sung et al., 2009]. All of these observations are consistent with the data presented above, but it remains unclear if these mutations cause an increased risk for disease progression or if they are markers of continued liver inflammation or both. In particular, mutations at residues 21, 113, 116, and 147 have been reported previously in association with HCC or cirrhosis, but the only statistically significant association was between a mutation at residue 21 and HCC [Ni et al., 2003]. Although mutations were found at each of these sites, no associations were identified between a mutation at a single site and clinical disease state in this study.
The relationship of e antigen status on viral mutations and the clinical state of the infected individual was also investigated. Chronic HBV without HBeAg represents an HBV infection in which actively replicating virus has lost its ability to produce HBeAg [Yim and Lok, 2006]. This is important because the lack of HBeAg expression is associated with HBV-related disease progression [Lindh et al., 1996; Hadziyannis and Vassilopoulos, 2001; Zarski et al., 2006], which may confound the associations between mutations and clinical state. Similar to the above results, previous work has shown that HBeAg seroconversion is associated with an increased frequency of mutations in CDE [Maruyama et al., 2000; Wang et al., 2010]; however, in this manuscript these observations are extended to show associations between increased number of mutations in the coding regions of CDE and disease progression states regardless of the HBeAg status in the cirrhosis and HCC groups.
A battery of statistical and selection analyses demonstrated that the total number of non-synonymous mutations within the coding region of CDEs was more important than any single mutation or group of “signature” mutations. However, most of the described associations between individual mutations and disease progression states were unable to be reproduced. This is probably explained by the targeting of different CDEs between study populations because of differing racial and ethnic groups, that is, HLA haplotypes. Thus, this work suggests that evidence of cumulative viral escape mutations is a marker of chronic CTL-induced inflammation, which is a mechanistic link between chronic HBV and disease progression [Nakamoto et al., 1998].
This study had a number of important limitations. Additional clinical data were not available, including concurrent viral loads, HBV treatment history, longitudinal disease course, and alcohol or tobacco use history, all of which could be potential confounders. The study also lacked a group of subjects in the inactive carrier phase, which would have provided a useful comparison. The homogeneity of the study population and viral isolates are both a limitation and strength for this study. A single racial group with very sparse minorities exists in Korea which is in contrast to other HBV endemic countries, such as Taiwan, which has at least four major racial groups to take into account with differing HLA profiles [Shaw et al., 1999]. In addition, Korea has a single circulating major HBV genotype, C2, in contrast to Taiwan, which has multiple circulating major genotypes [Yang et al., 2008]. As different genotypes can influence HBV disease progression [Kao et al., 2000], the homogeneity of viral genotypes reduces these effects on the analysis. However, this same homogeneity limits the extrapolation of the results of this study outside of Korea. In the future, additional studies using populations with different HLA haplotypes and HBV genotypes similar to the present study would validate further this study's findings and make them more generalizable.
In summary, current treatment guidelines suggest antiviral management based on liver inflammation markers, viral load, HBeAg status, and histology [Lok and McMahon, 2009]. This work demonstrates that the number of non-synonymous mutations in the coding regions for the HLA Class I epitopes of the HBV core region are associated strongly with clinical disease progression. The measurement of these escape mutations may be a clinically useful predictor for disease progression, and the finding of even a single such mutation, regardless of viral load, should direct the clinician to consider the use of antiviral therapy.
Acknowledgments
Grant sponsor: National Institutes of Health; Grant numbers: T32 RR023254, AI69432, AI043638, MH62512, MH083552, AI077304, AI36214, AI047745, AI74621, AI080353; Grant sponsor: James B. Pendleton Charitable Trust.
Abbreviations
- CTL
cytotoxic T lymphocytes
- CDE
CTL-defined epitopes
Footnotes
Conflict of interest: Nothing to disclose.
REFERENCES
- Ahn SH, Yuen L, Han KH, Littlejohn M, Chang HY, Damerow H, Ayres A, Heo J, Locarnini S, Revill PA. Molecular and clinical characteristics of hepatitis B virus in Korea. J Med Virol. 2010;82:1126–1134. doi: 10.1002/jmv.21844. [DOI] [PubMed] [Google Scholar]
- Bertoletti A, Ferrari C, Fiaccadori F, Penna A, Margolskee R, Schlicht HJ, Fowler P, Guilhot S, Chisari FV. HLA class I-restricted human cytotoxic T cells recognize endogenously synthesized hepatitis B virus nucleocapsid antigen. Proc Natl Acad Sci USA. 1991;88:10445–10449. doi: 10.1073/pnas.88.23.10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoletti A, Costanzo A, Chisari FV, Levrero M, Artini M, Sette A, Penna A, Giuberti T, Fiaccadori F, Ferrari C. Cytotoxic T lymphocyte response to a wild type hepatitis B virus epitope in patients chronically infected by variant viruses carrying substitutions within the epitope. J Exp Med. 1994a;180:933–943. doi: 10.1084/jem.180.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoletti A, Sette A, Chisari FV, Penna A, Levrero M, De Carli M, Fiaccadori F, Ferrari C. Natural variants of cytotoxic epitopes are T-cell receptor antagonists for antiviral cytotoxic T cells. Nature. 1994b;369:407–410. doi: 10.1038/369407a0. [DOI] [PubMed] [Google Scholar]
- Boni C, Fisicaro P, Valdatta C, Amadei B, Di Vincenzo P, Giuberti T, Laccabue D, Zerbini A, Cavalli A, Missale G, Bertoletti A, Ferrari C. Characterization of hepatitis B virus (HBV)-specific T-cell dysfunction in chronic HBV infection. J Virol. 2007;81:4215–4225. doi: 10.1128/JVI.02844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkaya H, Ayola B, Lok AS. High rate of mutations in the hepatitis B core gene during the immune clearance phase of chronic hepatitis B virus infection. Hepatology. 1996;24:32–37. doi: 10.1002/hep.510240107. [DOI] [PubMed] [Google Scholar]
- Chisari FV. Cytotoxic T cells and viral hepatitis. J Clin Invest. 1997;99:1472–1477. doi: 10.1172/JCI119308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond CP, Bartholomeusz A, Gaudieri S, Revill PA, Lewin SR. A systematic review of T-cell epitopes in hepatitis B virus: Identification, genotypic variation and relevance to antiviral therapeutics. Antivir Ther. 2008;13:161–175. [PubMed] [Google Scholar]
- Drummond J, Rowe B, Cheung L, Mayers I. The use of noninvasive mechanical ventilation for the treatment of acute exacerbations of chronic obstructive pulmonary disease in Canada. Can Respir J. 2005;12:129–133. doi: 10.1155/2005/714792. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- Gehring AJ, Sun D, Kennedy PT, Nolte-'t Hoen Lim E, Wasser SG, Selden S, Maini C, Davis MK, Nassal DM, Bertoletti MA. The level of viral antigen presented by hepatocytes influences CD8 T-cell function. J Virol. 2007;81:2940–2949. doi: 10.1128/JVI.02415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadziyannis SJ, Vassilopoulos D. Hepatitis B e antigen-negative chronic hepatitis B. Hepatology. 2001;34:617–624. doi: 10.1053/jhep.2001.27834. [DOI] [PubMed] [Google Scholar]
- Hosono S, Tai PC, Wang W, Ambrose M, Hwang DG, Yuan TT, Peng BH, Yang CS, Lee CS, Shih C. Core antigen mutations of human hepatitis B virus in hepatomas accumulate in MHC class II-restricted T cell epitopes. Virology. 1995;212:151–162. doi: 10.1006/viro.1995.1463. [DOI] [PubMed] [Google Scholar]
- Kao JH, Chen DS. Global control of hepatitis B virus infection. Lancet Infect Dis. 2002;2:395–403. doi: 10.1016/s1473-3099(02)00315-8. [DOI] [PubMed] [Google Scholar]
- Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118:554–559. doi: 10.1016/s0016-5085(00)70261-7. [DOI] [PubMed] [Google Scholar]
- Khakoo SI, Ling R, Scott I, Dodi AI, Harrison TJ, Dusheiko GM, Madrigal JA. Cytotoxic T lymphocyte responses and CTL epitope escape mutation in HBsAg, anti-HBe positive individuals. Gut. 2000;47:137–143. doi: 10.1136/gut.47.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Myers G. Signature pattern analysis: A method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8:1549–1560. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- Lee YI, Hur GM, Suh DJ, Kim SH. Novel pre-C/C gene mutants of hepatitis B virus in chronic active hepatitis: Naturally occurring escape mutants. J Gen Virol. 1996;77:1129–1138. doi: 10.1099/0022-1317-77-6-1129. [DOI] [PubMed] [Google Scholar]
- Lindh M, Horal P, Dhillon AP, Furuta Y, Norkrans G. Hepatitis B virus carriers without precore mutations in hepatitis B e antigen-negative stage show more severe liver damage. Hepatology. 1996;24:494–501. doi: 10.1002/hep.510240305. [DOI] [PubMed] [Google Scholar]
- Liu CJ, Chen PJ, Lai MY, Kao JH, Chang CF, Wu HL, Shau WY, Chen DS. A prospective study characterizing full-length hepatitis B virus genomes during acute exacerbation. Gastroenterology. 2003;124:80–90. doi: 10.1053/gast.2003.50003. [DOI] [PubMed] [Google Scholar]
- Liu S, Zhang H, Gu C, Yin J, He Y, Xie J, Cao G. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: A meta-analysis. J Natl Cancer Inst. 2009;101:1066–1082. doi: 10.1093/jnci/djp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok AS, McMahon BJ. Chronic hepatitis B: Update 2009. Hepatology. 2009;50:661–662. doi: 10.1002/hep.23190. [DOI] [PubMed] [Google Scholar]
- Lusida MI, Nugrahaputra VE, Soetjipto Handajani R, Nagano-Fujii M, Sasayama M, Utsumi T, Hotta H. Novel subgenotypes of hepatitis B virus genotypes C and D in Papua, Indonesia. J Clin Microbiol. 2008;46:2160–2166. doi: 10.1128/JCM.01681-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maini MK, Boni C, Lee CK, Larrubia JR, Reignat S, Ogg GS, King AS, Herberg J, Gilson R, Alisa A, Williams R, Vergani D, Naoumov NV, Ferrari C, Bertoletti A. The role of virus-specific CD8(+) cells in liver damage and viral control during persistent hepatitis B virus infection. J Exp Med. 2000;191:1269–1280. doi: 10.1084/jem.191.8.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Mitsui H, Maekawa H, Yamada H, Hirayama M, Iino S, Yasuda K, Koike K, Kimura S, Milich DR. Emergence of the precore mutant late in chronic hepatitis B infection correlates with the severity of liver injury and mutations in the core region. Am J Gastroenterol. 2000;95:2894–2904. doi: 10.1111/j.1572-0241.2000.03201.x. [DOI] [PubMed] [Google Scholar]
- Missale G, Redeker A, Person J, Fowler P, Guilhot S, Schlicht HJ, Ferrari C, Chisari FV. HLA-A31- and HLA-Aw68-restricted cytotoxic T cell responses to a single hepatitis B virus nucleocapsid epitope during acute viral hepatitis. J Exp Med. 1993;177:751–762. doi: 10.1084/jem.177.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto Y, Guidotti LG, Kuhlen CV, Fowler P, Chisari FV. Immune pathogenesis of hepatocellular carcinoma. J Exp Med. 1998;188:341–350. doi: 10.1084/jem.188.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni YH, Chang MH, Hsu HY, Tsuei DJ. Different hepatitis B virus core gene mutations in children with chronic infection and hepatocellular carcinoma. Gut. 2003;52:122–125. doi: 10.1136/gut.52.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osiowy C, Giles E, Tanaka Y, Mizokami M, Minuk GY. Molecular evolution of hepatitis B virus over 25 years. J Virol. 2006;80:10307–10314. doi: 10.1128/JVI.00996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Hwang YS, Park KS, Tokunaga K, Akaza T, Juji T, Kim SI. HLA haplotypes in Koreans based on 107 families. Tissue Antigens. 1998;51:347–355. doi: 10.1111/j.1399-0039.1998.tb02973.x. [DOI] [PubMed] [Google Scholar]
- Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Poon AF, Frost SD, Pond SL. Detecting signatures of selection from DNA sequences using Datamonkey. Methods Mol Biol. 2009;537:163–183. doi: 10.1007/978-1-59745-251-9_8. [DOI] [PubMed] [Google Scholar]
- Rehermann B, Lau D, Hoofnagle JH, Chisari FV. Cytotoxic T lymphocyte responsiveness after resolution of chronic hepatitis B virus infection. J Clin Invest. 1996;97:1655–1665. doi: 10.1172/JCI118592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CK, Chen LL, Lee A, Lee TD. Distribution of HLA gene and haplotype frequencies in Taiwan: A comparative study among Min-nan, Hakka, Aborigines and Mainland Chinese. Tissue Antigens. 1999;53:51–64. doi: 10.1034/j.1399-0039.1999.530106.x. [DOI] [PubMed] [Google Scholar]
- Sung FY, Jung CM, Wu CF, Lin CL, Liu CJ, Liaw YF, Tsai KS, Yu MW. Hepatitis B virus core variants modify natural course of viral infection and hepatocellular carcinoma progression. Gastroenterology. 2009;137:1687–1697. doi: 10.1053/j.gastro.2009.07.063. [DOI] [PubMed] [Google Scholar]
- Wang HY, Chien MH, Huang HP, Chang HC, Wu CC, Chen PJ, Chang MH, Chen DS. Distinct hepatitis B virus dynamics in the immunotolerant and early immunoclearance phases. J Virol. 2010;84:3454–3463. doi: 10.1128/JVI.02164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley SA, Brown D, Webster GJ, Jacobs R, Reignat S, Bertoletti A, Teo CG, Emery V, Dusheiko GM. Evolution of hepatitis B virus during primary infection in humans: Transient generation of cytotoxic T-cell mutants. Gastroenterology. 2004;127:1131–1138. doi: 10.1053/j.gastro.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Yang HI, Yeh SH, Chen PJ, Iloeje UH, Jen CL, Su J, Wang LY, Lu SN, You SL, Chen DS, Liaw YF, Chen CJ. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:1134–1143. doi: 10.1093/jnci/djn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim HJ, Lok AS. Natural history of chronic hepatitis B virus infection: What we knew in 1981 and what we know in 2005. Hepatology. 2006;43:S173–S181. doi: 10.1002/hep.20956. [DOI] [PubMed] [Google Scholar]
- Zarski JP, Marcellin P, Leroy V, Trepo C, Samuel D, Ganne-Carrie N, Barange K, Canva V, Doffoel M, Cales P. Characteristics of patients with chronic hepatitis B in France: Predominant frequency of HBe antigen negative cases. J Hepatol. 2006;45:355–360. doi: 10.1016/j.jhep.2006.03.007. [DOI] [PubMed] [Google Scholar]