Abstract
There is a continuing need to develop new techniques for the rapid and specific identification of bacterial pathogens in human body fluids especially given the increasing prevalence of drug resistant strains. Efforts to develop a surface enhanced Raman spectroscopy (SERS) based approach, which encompasses sample preparation, SERS substrates, portable Raman microscopy instrumentation and novel identification software, are described. The progress made in each of these areas in our laboratory is summarized and illustrated by a spiked infectious sample for urinary tract infection (UTI) diagnostics. SERS bacterial spectra exhibit both enhanced sensitivity and specificity allowing the development of an easy to use, portable, optical platform for pathogen detection and identification. SERS of bacterial cells is shown to offer not only reproducible molecular spectroscopic signatures for analytical applications in clinical diagnostics, but also is a new tool for studying biochemical activity in real time at the outer layers of these organisms.
Introduction
The ability to rapidly detect and identify bacterial cells in human body fluids at relatively low cost and in point-of-care settings is a continuing need for health care providers worldwide. Bacteremia, the presence of bacteria in blood, results from severe infections at sites in the body, surgical wounds, or contaminated implanted devices and may lead to the potentially fatal condition of sepsis. Owing to a ~40% mortality rate, sepsis ranks 13th among the causes of death overall in the United States.(1–3) Urinary tract infections (UTI), evidenced by the appearance of bacteria in urine, are among the most common types of infections in humans. Approximately 50% of all woman will have at least one UTI in their lifetime and a sizable percentage of those will suffer from chronic UTIs.(4, 5) In the US alone, UTIs are responsible for more than 7 million doctors office visits and over one million hospital admissions at a cost of approximately $1 billion per year. Sepsis and UTI are caused by a variety of bacterial species and strains. Furthermore, the increasing proliferation of drug resistant strains places an increasing premium on being able to indentify these microbes with strain specificity in a time frame useful for narrow spectrum antibiotic drug prescription.(6) In addition, the need for rapid bacterial identification methods for food and water safety applications has been illustrated by the several high profile incidents reported nationwide over the past few years.(7)
Traditional methods of bacterial identification are phenotypic based approaches that require a cell growth period and are consequentially slow (24 – 48 hours or longer). Furthermore, distinguishing closely related strains may be difficult via traditional methods and it is not a point of care technique. The best current methods are molecular diagnostic approaches that utilize specific primers or probes for particular gene targets, such as “real time” polymerase chain reaction methods (PCR),(8) which are increasingly finding use in clinical settings. If no culturing is required PCR time frames are typically in the 2 – 6 hour framework. However, PCR is not without some limitations or liabilities such as sample contamination, infectious mixture resolution, need for required primer sets, speed, cost and point-of-care capabilities.(9) Hence, at the very least, there is a continuing need for competing or orthogonal bacterial identification methods.
For the past few years we have been developing an optical approach for rapid, sensitive and specific bacterial diagnostics based on surface enhanced Raman spectroscopy (SERS).(10–14) SERS is a well-known spontaneous light scattering technique, discovered ~35 years ago which results in the 105 – 108 effective enhancement of the Raman scattering intensity of some molecular vibrational modes of molecules that are close (≤ ~5 nm) to nanostructured metal surfaces.(15, 16) This effect is predominantly attributed to the plasmonically enhanced local electric fields that become concentrated near nanosized structures that are coincident with the surface plasmon resonances of these nanomaterials. These plasmon resonances are usually in the NIR to visible for the most commonly employed metals, Ag or Au. Aside from the advantages of speed, ease of use and potentially cost introduced by the development of an optical approach for bacterial diagnostics, a SERS based platform also offers the advantage of portability since only a few milliwatts of laser power is required for acquisition of high quality SERS spectra. This portability is central for the application of this SERS platform to a wide variety of applications in addition to clinical bacterial diagnostics, such as forensics, food and water safety testing, and in situ studies of works of art and cultural heritage materials.
Overview of SERS platform description
The total SERS pathogen detection platform may be viewed as consisting of four key components:
Sample preparation device/procedure. Although often overlooked, sample preparation is a key ingredient for the effective use of novel pathogen detection methods. We are developing a two stage approach for bacterial enrichment from human blood.(12, 17) Detecting bacteria in blood is extremely challenging because red blood cell concentrations are ~109/mL in human blood and other particles, such as white blood cells and platelets are present at ~106/mL levels. In contrast medically relevant concentrations corresponding to bacteremic presentations are in the 102 – 103 /mL range, and the surface area of a red blood cell is about an order of magnitude larger than that of a typical bacterial cell. However, via selective lysis, centrifugation, and micro-evaporation sufficient enrichments of ~105 can be achieved allowing acquisition of SERS spectra in 100 nL volumes starting with 10 mL of infected blood. Sensitivities down to 103/mL have already been achieved in a bench top procedure and an automated device is currently being developed in our lab.(12) Required sample preparation procedures for UTI detection is simpler because the concentration of other cells in urine is normally low normally and the clinical levels of bacterial infection concentrations are much higher in urine (≥105/mL) than blood as described further below.
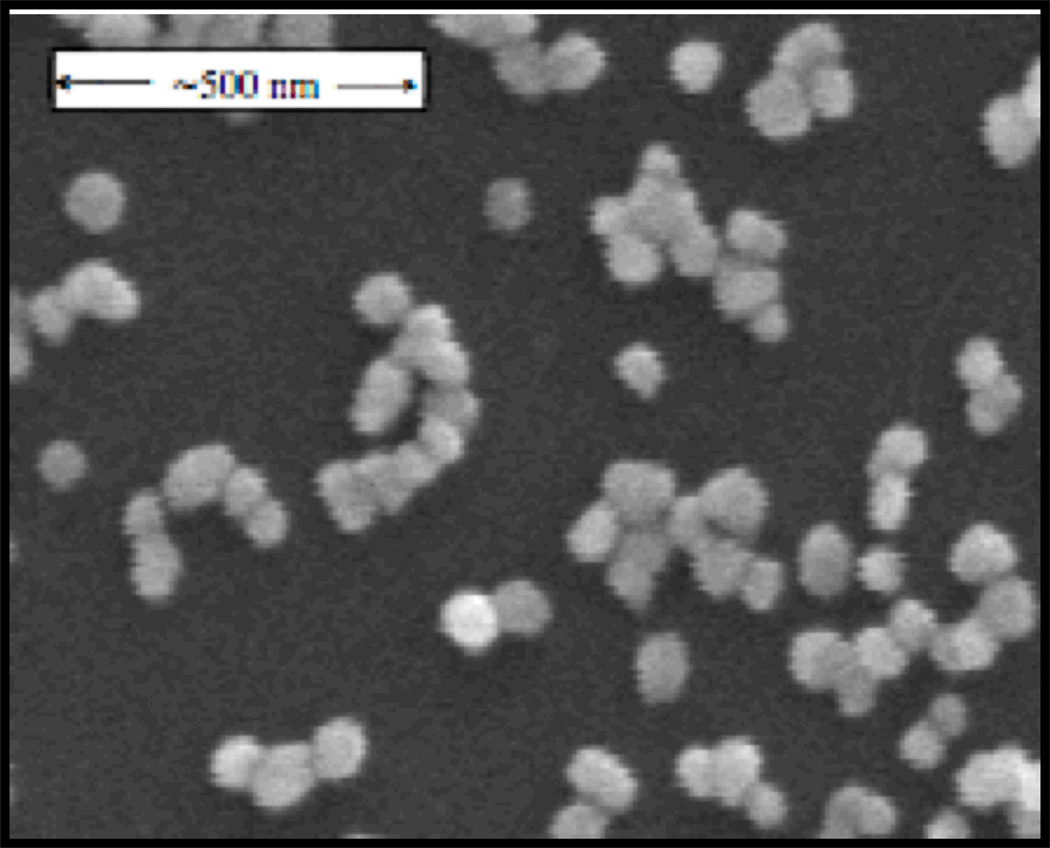
SERS substrate. Most of our SERS data has been acquired from bacterial cells placed on a substrate produced by a Au ion doped sol-gel procedure.(10) These substrates are found to yield strongly enhanced, reproducible SERS signal for vegetative bacterial cells with 785 nm excitation. As shown in Fig. 1, these substrates have clusters of 80 nm Au nanoparticles physi-adsorbed to the outer surface of a SiO2 matrix. Further improvements in SERS chip performance will only enhance the value of this technique. We have collaborated on the design and production of several engineered substrates,(18–20) however, the sol-gel Au nanoparticle “chips” have proven to provide the best signal/noise for bacterial identification purposes of those we have tested.
A portable SERS signal acquisition instrument. Although several hand held Raman devices with nearly point and click capabilities are commercially available, true microscopic capabilities are necessary for this SERS application. Thus, we designed and built several prototype high-performance (cooled CCD array, ~5 cm−1 spectral resolution, piezo-driven sample positioning stages, submicron imaging resolution) portable Raman microscopes for this purpose. This unique instrument allows positioning of the 785 nm Raman excitation on a specific cell or sample region on the SERS substrate for Raman analysis. Imaging also provides additional morphology information that can aid identification procedures. Our most recent version of the Raman microscope was built in collaboration with BioTools, Inc. (Jupiter FL).
Bacterial identification software. As described further below we have developed a novel multivariate data analysis technique that provides accurate distinction between the SERS spectra of closely related bacterial species and strains when combined with a previously developed reference library.
Figure 1.
A scanning electron micrograph image of the Au nanoparticle covered SERS substrate produced by an Au ion doped sol-gel process. Clusters of 1–15 ~80 nm Au particles are evident on the surface of the SiO2 SERS chip.
Characteristics of bacterial SERS
Following multiple cycles of centrifugation and washing with distilled water, bacterial cells are resuspended in water and placed on the SERS substrate for signal acquisition. The maximum scattering intensity of the SERS spectrum on a per bacterium, not per molecule, basis is between 104 to 105 times larger than the observed maximum signal for the normal Raman spectrum of the same bacterium.(10) This per bacterium signal enhancement factor enables signal acquisition close to the single cell level, although usually SERS spectra are acquired from about 10 – 20 bacterial cells in the sample region. This level of signal enhancement allows the development of a portable Raman instrument because excellent signal to noise scattering can be achieved with low incident laser power at 785 nm. Aside from a measure of sensitivity, the capability of obtaining spectra at the single cell level offers the possibility of resolving infectious mixtures by selectively interrogating different members of the bacterial population.
Interestingly, there is no correspondence between the vibrational bands observed in the normal Raman spectrum and those in the SERS spectrum.(10, 13) Due to the distance dependence of the SERS enhancement mechanism, only those molecules at the outer layers, or very close to the cell wall, can contribute to the bacterial SERS spectrum. In contrast, all the molecules in the Raman illuminated region contribute to the normal Raman spectrum and that volume is dominated by the cell cytoplasm. Hence the SERS and normal Raman spectrum of a given bacterial cell type are from totally different parts of the cell and have virtually no features in common.
Specificity
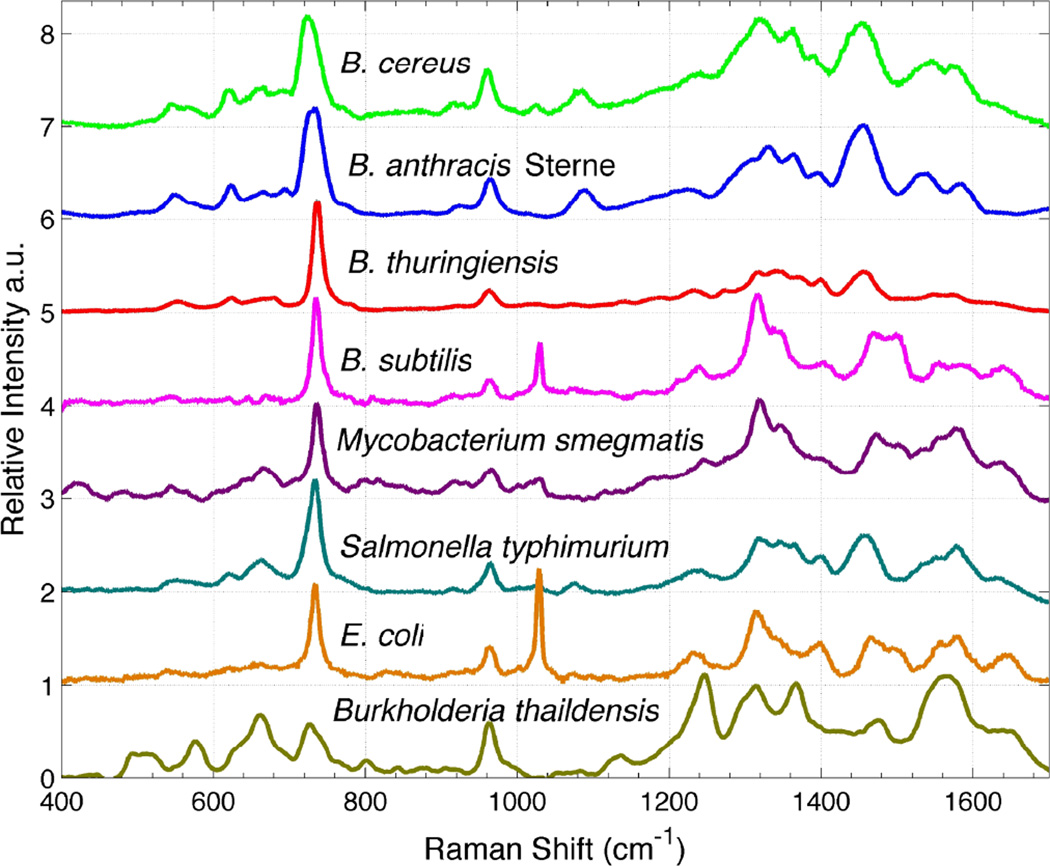
SERS spectra of 8 different lab cultured bacterial species from 5 genera are shown in Fig. 2. These spectra are acquired with 10 seconds of ~2 mW of 785 nm incident laser power and results from the Raman excitation of 10 – 20 cells typically. Spectra with excellent signal-to-noise ratios are obtained on our SERS substrates. As evident in this figure, each species exhibits a unique SERS signature. Although the spectra have been arranged top to bottom by phylogenic proximity, only moderate correlation with this lineage is observed. Furthermore, no clear distinction between Gram-positive and Gram-negative bacteria is apparent. An under-appreciated aspect of bacterial SERS spectra is that, in addition to the enhanced sensitivity that SERS provides, the spectral differences between SERS spectra of different species/strains is greater in SERS than for normal Raman.(13) In other words, SERS results in enhanced specificity as well as enhanced sensitivity for bacterial samples.
Figure 2.
SERS spectra of eight bacterial species obtained on gold aggregate coated SiO2 chips (Figure 1). An incident laser power of ~2 mW and a data accumulation time of 10 seconds were used to obtain these spectra. Spectra are offset vertically for display purposes and top-to-bottom ordered according to their phylogenetic relationship.
Software for bacterial diagnostics
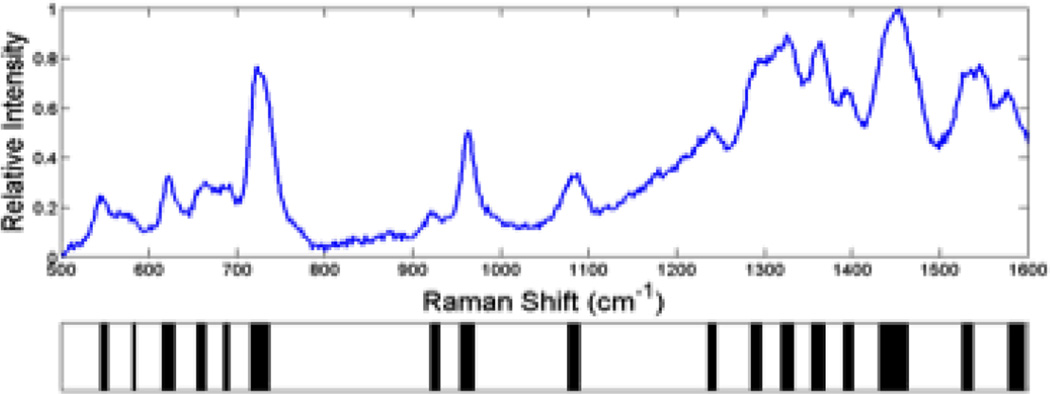
To be able to exploit these unique SERS signatures for bacterial diagnostics, we have developed a principle components analysis (PCA) based methodology for matching the acquired SERS spectrum to that of a known species/strain in a previously compiled library.(13) After Fourier filtering the SERS spectrum, a barcode is generated by using the sign of the second derivative of the spectrum, i.e. whether the spectrum has curvature up or curvature down at each wavenumber, to assign a “one” or a “zero” as a function of scattered frequency. An example of a SERS based barcode is shown in Fig. 3 for a SERS spectrum of B. cereus. These barcodes, or arrays of ones and zeroes, color-coded as black (curvature down) or white (curvature up) per wavenumber interval in this figure, are the inputs for PCA and related standard analyses such as DFA or HCA. The barcode methodology results in significantly enhanced diagnostic specificity and is the tool used for quantitative measures of reproducibility.
Figure 3.
SERS spectrum of B. cereus and the corresponding second derivative “barcode” spectrum is given below. The barcode is determined only by the sign of the second derivative (curvature up = white, curvature down = black). These barcodes are the inputs for the principal component analysis (PCA) used to identify unknown bacteria via SERS demonstrated in Figs 4 and 5.
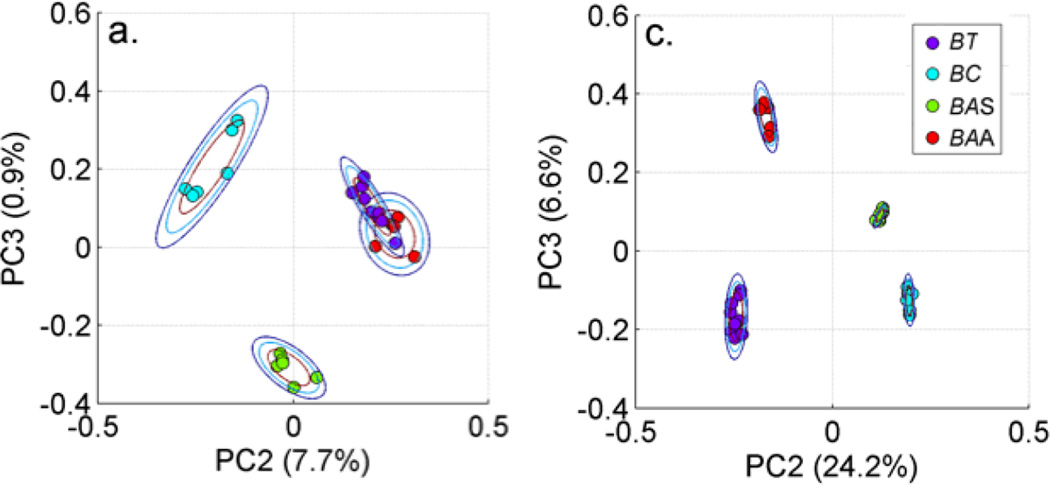
As an example of the effectiveness of this barcode methodology and how it is used for bacterial diagnostics, two PCA plots (PC2 vs. PC3) are shown in Fig. 4. The left hand panel results when six SERS spectra of four closely related Bacillus bacteria (B. thuringiensis, B. cereus, B. anthracis Sterne, B. anthracis Ames) are the input vectors to a standard PCA procedure. Each spectrum corresponds to a “dot” in this plot, which has been color coded to indicate the organism it represents and the rings correspond to 2D standard deviations. The tightness of the clusters for a given species/strain characterizes the reproducibility of the spectra and the distance between the color-coded clusters represents the specificity offered by these measurements. B. anthracis Ames and B. thurengiensis for example cannot be separated in this treatment (Fig. 4, left). However, when the input to the PCA treatment are the barcode spectra, all species/strain are well-separated, and much tighter clusters are obtained as seen in the PCA plot on the right hand side in Fig. 4. The contrast between these two panels illustrates the enhanced specificity that results from this barcoding procedure and is key to the use of this methodology for bacterial diagnostics.(13)
Figure 4.
Comparison of PCA plots (PC3 vs, PC2) for SERS spectra of four closely related Bacillus organisms (B. thuringiensis, B. cereus, B. anthracis Sterne, B. anthracis Ames). When the observed SERS spectra are the inputs to the PCA, the contour plot shown in the left hand panel results. By contrast, when the input to the PCA are the barcode representations of the observed SERS spectra, the contour plot shown in the right hand panel results. The enhancement in specificity resulting from the barcode spectra is clearly evident by this comparison.
Following acquisition of the SERS spectrum of the unknown sample, the spectrum is converted to its corresponding barcode and PCA analysis is performed with groups of barcode SERS spectra of known bacterial identity selected from a previously developed library in order to identify the unknown bacteria. The species/strain group that the unknown clusters with determines the identity of the unknown. The identification method is only limited by the library size, which can be continuously and readily expanded, and is not dependent upon specific primer sets or antibody specificity. This approach is quite general and in principle could be expanded to other pathogens such as fungi or viral particles.
Origin of SERS signals
The molecular origins of the vibrational bands usually observed in the SERS spectra of vegetative bacterial cells are usually attributed to the diversity of lipoproteins, proteins, lipids, and other cell wall components at the outer membrane layer of these bacterial cells.(21–23) The distance dependence of the SERS enhancement mechanism supports such a supposition. However, we have discovered that nearly all of the observed vibrational signatures are due to small metabolites which are concentrated at the outer region of bacterial cells.(24) In particular, the largest features in the SERS spectra are generally products in the purine degradation pathway. The large range and unique reproducible signatures found for the SERS bacterial spectra result from the large number of enzymatic variations in these metabolic pathways and the resulting different relative concentration of the many small molecules found in this near extracellular region. Thus, SERS offers not only reproducible molecular signatures for analytical detection and identification applications as shown here, but also a new tool for studying biochemical activity in real time at the outer layers of these organisms.
An example: UTI diagnosis
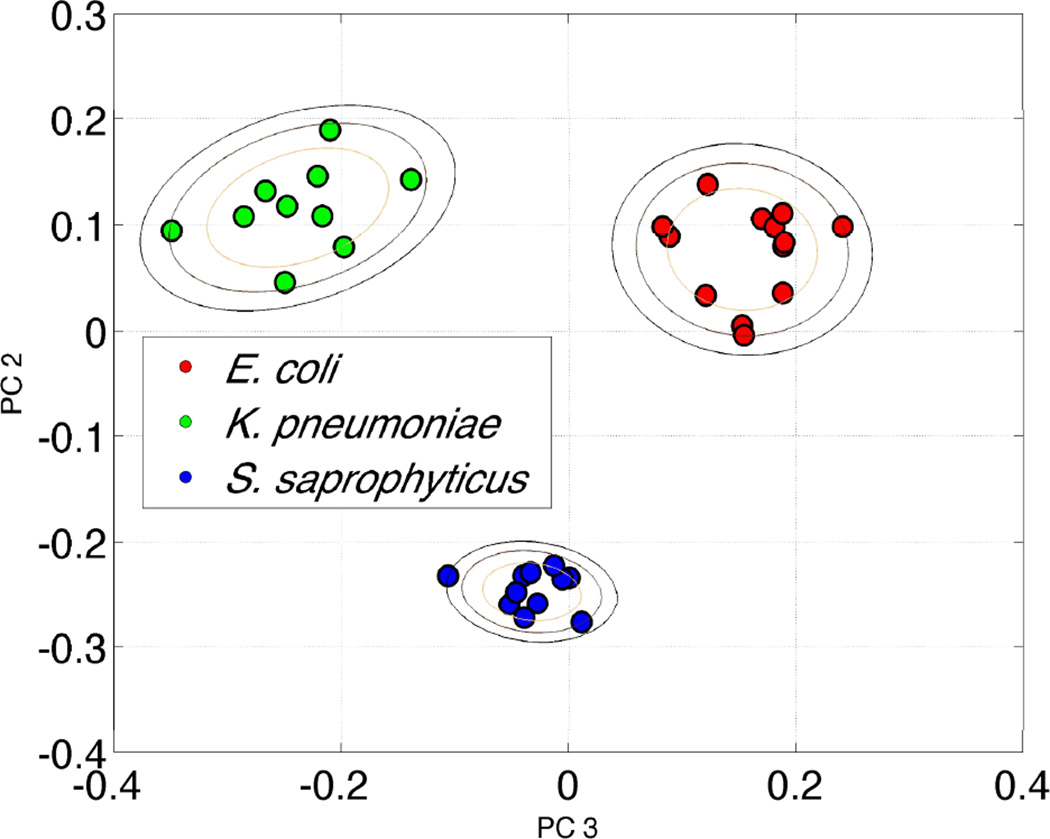
Although the bulk of our effort has been directed towards the very challenging problem of bacteremia diagnostics, efforts in our laboratory have recently expanded to develop this SERS based approach for UTI diagnostics. With relatively routine centrifugation based techniques bacterial concentration enrichments of 103 to 104 can be achieved in urine samples that have been spiked at the clinically relevant concentration of 105 cfu/mL. Fig. 5 shows the PCA plot for three different organisms known to be causative agents of UTI. These bacteria were spiked into human urine at 105 cfu/mL concentration, enriched via a centrifugation procedure and then placed on the SERS substrate for data acquisition. Distinct clusters are readily obtained for the bacterial cells after recovery from urine indicting that species-specific SERS spectral signatures can be identified by this SERS barcode procedure. This prototype, bench top UTI test took about 30 minutes to perform and the SERS based analysis illustrates the potential value of this promising methodology.
Figure 5.
A PCA plot resulting from a SERS barcode cluster analysis for three bacteria (E. coli, K. pneumoniae, and S. saphrophyticus) that were recovered from spiked urine samples at medically relevant concentrations (105/mL).
Conclusion
The elements and basis for a SERS based platform for bacterial diagnostics, which includes a sample preparation device, a portable Raman microscope, SERS substrates and software for bacterial identification, have been described. This novel optical approach is a potentially transformative diagnostic procedure for the rapid, reliable identification of bacterial species/strains in patients presenting with infectious disease symptoms. In addition, SERS provides an effective tool for learning about the chemical activity of molecules at the outer layers of these organisms and, in particular, is a new methodology for use in the recently established field of metabolomics.
References
- 1.Bearman GM, Wenzel RP. Bacteremias: a leading cause of death. Arch. Med. Res. 2005;36:646–659. doi: 10.1016/j.arcmed.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 2001;29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien JM, Jr, Ali NA, Aberegg SK, Abraham E. Sepsis. Am J Med. 2007;120:1012–1022. doi: 10.1016/j.amjmed.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 4.Schappert SM, Rechtsteiner EA. National health statistics reports. no 8. Hyattsville, MD: National Center for Health Statistics; 2008. Ambulatory medical care utilization estimates for 2006. [PubMed] [Google Scholar]
- 5.Griebling TL, Litwin MS, Saigal CS, editors. Department of Health and Human Services, Public Health Service. Washington, D.C.: National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; 2007. Urologic Diseases in America; pp. 587–619. [Google Scholar]
- 6.Yu X, Susa M, Weile J, Knabbe C, Schmid RD, Bachmann TT. Rapid and sensitive detection of fluoroquinolone-resistant Escherichia coli from urine samples using a genotyping DNA microarray. Int J Med Microbiol. 2007;297:417–429. doi: 10.1016/j.ijmm.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 7. http://en.wikipedia.org/wiki/List_of_foodborne_illness_outbreaks_in_the_United_States.
- 8.Harris KA, Hartley JC. Development of broad-range 16S rDNA PCR for use in the routine diagnostic clinical microbiology service. J Med Microbiol. 2003;52:685–691. doi: 10.1099/jmm.0.05213-0. [DOI] [PubMed] [Google Scholar]
- 9.Corless CE, Guiver M, Borrow R, Edwards-Jones V, Kaczmarski EB, Fox AJ. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J Clin Microbiol. 2000;38:1747–1752. doi: 10.1128/jcm.38.5.1747-1752.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Premasiri WR, Moir DT, Klempner MS, Krieger N, Jones G, II, Ziegler LD. Characterization of the Surface Enhanced Raman Scattering (SERS) of Bacteria. J. Phys. Chem. B. 2005;109:312–320. doi: 10.1021/jp040442n. [DOI] [PubMed] [Google Scholar]
- 11.Premasiri WR, Moir DT, Ziegler LD. Vibrational Fingerprinting of Bacterial Pathogens by Surface Enhanced Raman Scattering. SPIE. 2005;5795 [Google Scholar]
- 12.Premasiri WR, Moir DT, Klempner MS, Ziegler LD, Kneipp K, Aroca R, Kneipp H, Wentrup-Byrne E, editors. New Approaches in Biomedical Spectroscopy. New York: Oxford University Press; 2007. p. 164. [Google Scholar]
- 13.Patel IS, Premasiri WR, Moir DT, Ziegler LD. Barcoding bacterial cells: A SERS based methodology for pathogen identification. J Raman Spectrosc. 2008;39:1660–1672. doi: 10.1002/jrs.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Premasiri WR, Gebregziabher Y, Ziegler LD. On the difference between surface-enhanced raman scattering (SERS) spectra of cell growth media and whole bacterial cells. Appl Spectrosc. 2011;65:493–499. doi: 10.1366/10-06173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeanmaire DL, Van Duyne RP. Surface Raman spectroelectrochemistry: Part I. Heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode. J. Electroanal. Chm. 1977;84:1–20. [Google Scholar]
- 16.Camden JP, Dieringer JA, Zhao J, Van Duyne RP. Controlled plasmonic nanostructures for surface-enhanced spectroscopy and sensing. Acc Chem Res. 2008;41:1653–1661. doi: 10.1021/ar800041s. [DOI] [PubMed] [Google Scholar]
- 17.Zhang JY, Do J, Premasiri WR, Ziegler LD, Klapperich CM. Rapid point-of-care concentration of bacteria in a disposable microfluidic device using meniscus dragging effect. Lab Chip. 2010;10:3265–3270. doi: 10.1039/c0lc00051e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopinath A, Boriskina SV, Premasiri WR, Ziegler LD, Reinhard BM, Dal Negro L. Plasmonic Nanogalaxies: Multiscale Aperiodic Arrays for Surface-Enhanced Raman Sensing. Nano Lett. 2009;9:3922–3929. doi: 10.1021/nl902134r. [DOI] [PubMed] [Google Scholar]
- 19.Yan B, Thubagere A, Premasiri WR, Ziegler LD, Dal Negro L, Reinhard BM. Engineered SERS Substrates for Bacterial Pathogen Detection: Nanoparticle Cluster Arrays with Multiscale Signal Enhancement. NanoLett. 2009 doi: 10.1021/nn800836f. [DOI] [PubMed] [Google Scholar]
- 20.Yang L, Yan B, Premasiri WR, Ziegler LD, Dal Negro L, Reinhard BM. Engineering Nanoparticle Cluster Arrays for Bacterial Biosensing: The Role of the Building Block in Multiscale SERS Substrates. Adv. Functional Mater. 2010;20:2619–2628. [Google Scholar]
- 21.Jarvis RM, Goodacre R. Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal Chem. 2004;76:40–47. doi: 10.1021/ac034689c. [DOI] [PubMed] [Google Scholar]
- 22.Jarvis RM, Brooker A, Goodacre R. Surface-enhanced Raman scattering for the rapid discrimination of bacteria. Faraday Discuss. 2006;132:281–292. doi: 10.1039/b506413a. discussion 309-19. [DOI] [PubMed] [Google Scholar]
- 23.Liu T-T, Lin Y-H, Hung C-H, Liu T-J, Chen Y, Huang Y-C, Tsai T-H, Wang H-H, Wang D-W, Wang J-K, Wang Y-L, Lin C-H. A High Speed Detection Platform Based on Surface-Enhanced Raman Scattering for Monitoring Antibiotic-Induced Chemical Changes in Bacteria Cell Wall. PLoS ONE. 2009;4:e5470. doi: 10.1371/journal.pone.0005470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Premasiri WR, Lee JC, Theberge R, Costello C, Ziegler LD. On the molecular origin of bacterial SERS spectra. in preparation. [Google Scholar]