Abstract
Neural crest cells form diverse derivatives that vary according to their level of origin along the body axis, with only cranial neural crest cells contributing to facial skeleton. Interestingly, the transcription factor Ets-1 is uniquely expressed in cranial but not trunk neural crest, where it functions as a direct input into neural crest specifier genes, Sox10 and FoxD3. We have isolated and interrogated a cis-regulatory element, conserved between birds and mammals, that drives reporter expression in a manner that recapitulates that of endogenous Ets-1 expression in the neural crest. Within a minimal Ets-1 enhancer region, mutation of putative binding sites for SoxE, homeobox, Ets, TFAP2 or Fox proteins results in loss or reduction of neural crest enhancer activity. Morpholino-mediated loss-of-function experiments show that Sox9, Pax7, Msx1/2, Ets-1, TFAP2A and FoxD3, all are required for enhancer activity. In contrast, mutation of a putative cMyc/E-box sequence augments reporter expression, consistent with this being a repressor binding site. Taken together, these results uncover new inputs into Ets-1, revealing critical links in the cranial neural crest gene regulatory network.
Keywords: Neural crest, Ets-1, Enhancer, TFAP2, Pax7, Sox9
Introduction
The neural crest is an embryonic stem/progenitor cell population characterized by its extensive migratory ability and multi-potency. Neural crest cells arise within the central nervous system, but subsequently emigrate to give rise to numerous derivatives that differ accordingly to their axial level of origin. For example, the cranial neural crest makes all glia and some neurons of the peripheral nervous system of the head, as well as contributing to the cornea and craniofacial skeleton. In contrast, the trunk neural crest does not contribute to cartilage or bone, and cannot do so even when transplanted to the head, but does contribute to peripheral nervous system and melanocytes of the skin. Between cranial and trunk is the vagal neural crest, which forms the enteric nervous system, and is responsible for septation of the heart and outflow tract.
One possible explanation for the apparent regional differences in the neural crest’s ability to form different derivatives is that the gene regulatory events underlying neural crest development differ according to axial level. Indeed there is evidence that the enhancers mediating expression of some key neural crest genes in the head are different from those that mediate expression in the trunk. For example, Sox10, a transcription factor critical for formation of nearly all neural crest lineages, has a different enhancer responsible for cranial than for trunk expression (Betancur et al., 2010b). Similarly, another neural crest gene, FoxD3, has a cranial-specific enhancer that mediates its initial expression in the dorsal neural tube (Simões-Costa et al., 2012). Interestingly, a common input into the cranial-specific enhancers for Sox10 and FoxD3 is the transcription factor Ets-1 (Betancur et al., 2010b; Simões-Costa et al., 2012).
Ets-1 is a winged helix-turn-helix transcription factor that is expressed in the premigratory cranial but not trunk neural crest (Tahtakran and Selleck, 2003). In fact, Ets-1 is one of the few transcription factors known to be selectively expressed in a subpopulation of the neural crest. Indeed, ectopic expression of Ets-1 at trunk levels causes changes in the character of emigrating neural crest cells, making them more cranial-like (Théveneau et al., 2007). In humans and mice, mutations in Ets-1 lead to craniofacial anomalies and septation defects of the heart (Ye et al., 2010; Gao et al., 2010).
Given the importance of Ets-1 in regulating key neural crest specifier genes Sox10 and FoxD3, identifying the upstream factors that control Ets-1 expression holds the promise of revealing novel critical inputs and links in the gene regulatory network (GRN) underlying neural crest formation. To this end, we have isolated a region adjacent to the Ets-1 coding region that is capable of driving GFP in a manner that recapitulates endogenous Ets-1 expression in the cranial neural crest. The results show that Msx1/2, Pax7, Sox9, FoxD3 and TFAP2A, are critical for activation of Ets-1 activity in the neural crest. By taking advantage of the power of cis-regulatory dissection, our findings help to expand the cranial neural crest gene regulatory network.
Materials and methods
Isolation of conserved regions
Using the UCSC Gene browser (Kent et al., 2002), we identified regions in the chicken genome in the vicinity of the Ets-1 coding region that are conserved among vertebrates. Using PCR, we amplified the conserved regions from the BAC clone CH261-52I7 (ARK Genomics) and cloned them into the vector pTK-GFP which contains a basal TK promoter driving GFP expression. We cloned conserved regions upstream and downstream of the coding region as well as conserved regions within introns. Generally, the size of the cloned fragments was less than 1 kb, though in some cases, we amplified several adjoining Conserved Regions in a larger clone.
Electroporation
The cloned Conserved Regions were electoporated into HH stage 4 (Hamburger and Hamilton, 1953) embryos ex ovo as previously described (Sauka-Spengler and Barembaum, 2008). After 18 h the GFP expression was examined in unfixed embryos. Using this procedure we were able to electroporate the entire embryo. Once a clone was identified with the requisite neural crest expression, potential transcription factor binding sites were identified using the Jaspar database (Bryne et al., 2008) and mutated using fusion PCR (Szewczyk et al., 2006). The plasmid with the mutated enhancer was co-electroporated with a plasmid containing the non-mutated enhancer driving Cherry expression. The enhancer activity of the mutant construct driving GFP was then compared to the non-mutated construct driving Cherry in the same embryo, thus controlling for electroporation efficiency. Antisense morpholino oligos (MO) (1.5 mM) (Gene-Tools, Philomath, OR) aimed at the putative regulators were co-electroporated with reporter constructs (1 mg/ml) on the right side (dorsal side up) and a control morpholino oligo (1.5 mM) and reporter construct (1 mg/ml) on the contralateral side.
In situ hybridization
Whole mount in situ hybridization was performed according to the previous protocols (Wilkinson, 1992) using the Chicken EST clone ChEST218o8 from the BBSRC database (Boardman et al. 2002).
Results
Endogenous pattern of chick Ets-1 expression in the neural crest
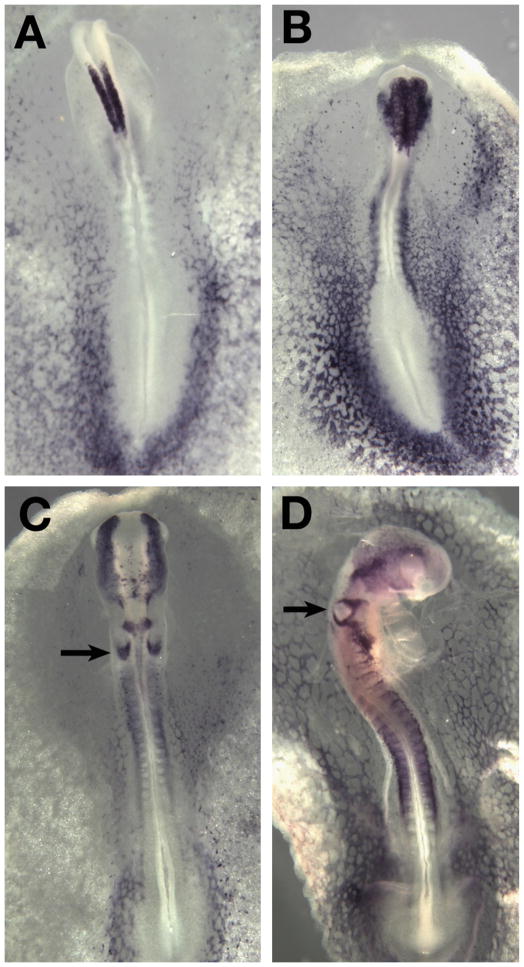
Ets-1 transcripts are expressed in the premigratory neural crest, with onset of expression at the 4 somite stage (4ss; Fig. 1A). Expression is retained on migrating cranial and vagal neural crest cells, but is absent from the trunk premigratory or migrating neural crest (Fig. 1B and C). Later, Ets-1 expression is maintained as crest cells reach their final position in the frontonasal process and branchial arches (Fig. 1D). In addition to the cranial neural crest, Ets-1 is observed in the caudal half of the otic vesicle (Fig. 1C) as well as endothelial cells of the body and extraembryonic regions, which exhibit high levels of Ets-1 expression at all stages examined (Fig. 1) (Tahtakran and Selleck, 2003).
Fig. 1.
Ets-1 is expressed in the dorsal neural tube of HH stage 8 embryos (A) and in the migrating crest of HH stage 10 (B) HH stage 12 (C) and HH stage 14 (D) chicken embryos. Arrows point to the Ets-1 expression in the otic pit.
Identification of the Ets-1 regulatory region mediating neural crest expression
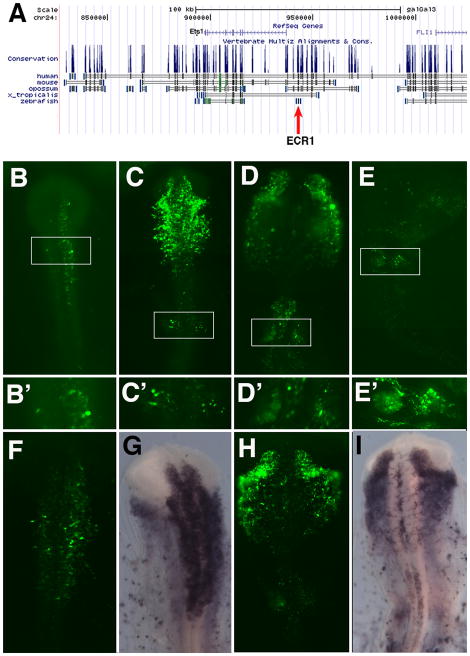
Using the UCSC gene browser, we identified regions in the vicinity of the Ets-1 coding region that are conserved between birds and mammals. Conserved fragments were isolated by PCR and cloned into an EGFP reporter vector upstream of a thymidine kinase (TK) basal promoter (Uchikawa et al., 2003). Using an ex ovo electroporation technique (Sauka-Spengler and Barembaum, 2008), the embryonic ectoderm of HH stage 4 embryos was electroporated with the reporter construct together with a ubiquitously expressed pCIG H2B-RFP to assess the efficiency of transfection. Embryos were cultured to HH stages 9–14 (Hamburger and Hamilton, 1953), and analyzed for GFP and RFP expression. The RFP was expressed throughout the rostrocaudal axis of the embryos (data not shown). Of the regions tested, which included regions on either side of the coding region as well as introns, only one conserved region mediated GFP activity at the stages examined. This 694 bb region, which we refer to as Ets Conserved Region (ECR) 1 (Fig. 2A), drove expression of GFP in chicken embryos (Fig. 2B–E) in a pattern that reflects endogenous Ets-1 expression in the cranial neural crest and otic placode. GFP expression was first observed at the 6 somite stage (HH stage 9) in premigratory crest cells in the dorsal neural tube (Fig. 2B). Later, GFP was detected in emigrating cranial neural crest departing the neural tube (HH stage 10, Fig. 2C) and migrating at HH stage 12 (Fig. 2D) toward their final positions (HH stage 14, Fig. 2E). At later stages, GFP was expressed throughout the head mesenchyme. The ECR1 region also mediated GFP expression in the neural crest cells at the hindbrain level, including the rhombomere (r) 4 and r6 streams and the branchial arches, the latter corresponding to the migrating vagal crest stream. However, no GFP expression was seen in the trunk neural crest, even though a co-electroporated construct expressing RFP ubiquitously showed expression throughout the embryo (data not shown). These results show that ECR1 enhancer mediates expression in the cranial neural crest similar to that of endogenous Ets-1.
Fig. 2.
A. Using the UCSC genome browser, we identified conserved regions near the Ets-1 gene. Conserved regions were cloned into pTK vector and tested by electroporation for neural crest enhancer activity (A). One of these, ECR1 (red arrow) had enhancer activity. ECR1 drives GFP expression in the neural crest of HH stage 9 (B,B′), HH stage 10 (C,C′), HH stage 12 (D,D′) and HH stage 14 (E,E′) embryos. The rectangles in B, C, D, and E correspond to higher magnification images of the midbrain (B′), otic placode (C′), otic pit (D′) and otic vesicle (E′) respectively. Expression of GFP by ECR1 in HH stage 9 embryos (F) recapitulates the Ets-1 expression as seen by in situ hybridization (G). Similarly, expression of GFP by ECR1 in HH stage 11 embryos (H) recapitulates the Ets-1 expression as seen by in situ hybridization (I).
In addition to neural crest, expression of GFP was detected in the otic placode (Fig. 2C), otic pit (Fig. 2D) and otic vesicle (Fig. 2E). In situ hybridization of embryos electroporated with ECR1 showed that the endogenous Ets-1 expression overlapped with that of GFP expression driven by the ECR1 enhancer in HH stage 10 (Fig. 2F and G) and HH stage 11 embryos (Fig. 2H and I). The ECR1 driven GFP expression also reproduced endogenous Ets-1 expression in the otic placode (Fig. 2H and I). However, it did not drive expression of GFP in the head mesoderm or endothelial cells despite the fact that these regions also express Ets-1. Thus the ECR1 enhancer recapitulates endogenous Ets-1 expression specifically in the neural crest and otic placode.
A conserved 288 bp element is the minimal essential core regulatory element of enhancer ECR1
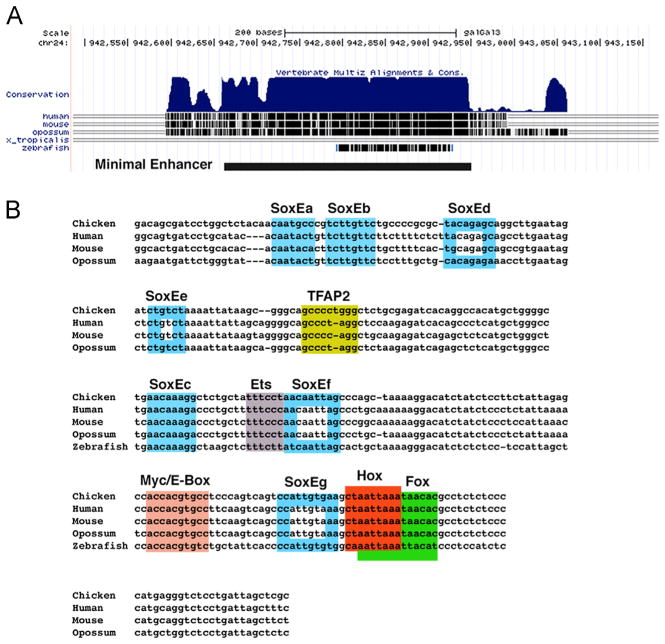
To narrow down the region necessary for enhancer activity, we progressively deleted the ends of the ECR1 enhancer to define the minimal required sequence to mediate reporter activity. A 288 bp fragment retained the GFP expression pattern of the full length ECR1 enhancer construct, albeit at a somewhat reduced level compared to the ECR1 Cherry construct. Enhancer activity was eliminated when the conserved sequence at either end was removed, indicating that the minimum enhancer region was 288 bp (Fig. 3A).
Fig. 3.
Conservation of ECR1 using UCSC gene browser (A). The conserved regions are required for ECR1 enhancer activity. The minimal enhancer is depicted as a solid lie at the bottom. There are a number of transcription factor binding consensus sites conserved in the ECR1 region (B) as determined using the JASPAR database. The open rectangles correspond to SoxE consensus sequences whose mutation did alter enhancer activity.
As a first step in identifying potential inputs that mediate Ets-1 expression in the neural crest, we explored potential transcription factor binding sites within the minimal ECR1 enhancer using the JASPAR database (Bryne et al., 2008) (Fig. 3B). This analysis revealed consensus binding sequences for SoxE genes, TFAP2A, Ets genes, cMyc/E-box sequence, a homeobox binding site, and a Fox transcription factor binding site. Interestingly, members of each of these transcription factor families have been identified as functioning within the neural crest GRN.
Dissection of ECR1 reveals eight critical binding sites for enhancer activity
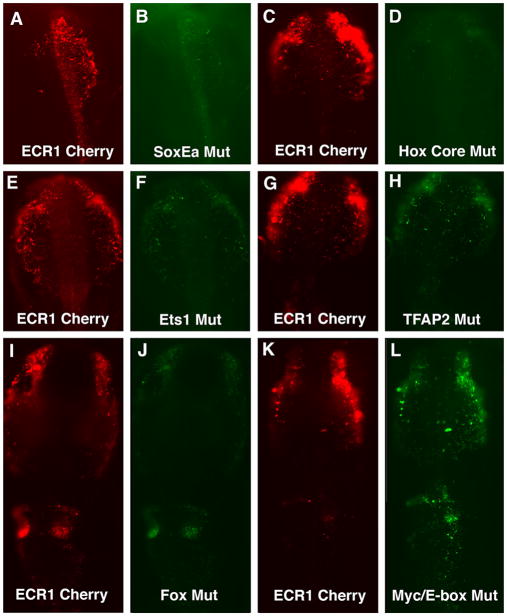
To test the functional relevance of these putative binding motifs, we turned to mutational analysis. Constructs were created with mutations of each of the putative binding motifs to determine if these conserved sites are required for ECR1 enhancer activity. To this end, the endogenous consensus sequences were replaced with heterologous sequence from GFP. Mutant constructs were then co-electroprated along with a ECR1-pTK-Cherry control constructs to determine the differences between the GFP expression driven by the mutant ECR1 enhancer compared with Cherry fluorescent protein expression driven by the wild type ECR1 enhancer construct.
Seven SoxE sequences were tested in this way. Of these, mutation of three sites (SoxEa shown in Fig. 4A and B; SoxEb and SoxEc are not shown) resulted in significant reduction of the fluorescence signal compared to the intact ECR1 control, suggesting that they were important for mediating enhancer activity in the cranial neural crest. Similarly replacement of the homeobox consensus sequence eliminated enhancer activity (Fig. 4C and D). Mutation of the Ets (Fig. 4E and F) or TFAP2 (Fig. 4G and H) consensus sequences decreased enhancer activity, albeit less than removal of either Hox or Sox sites. Mutation of the Fox sequence also reduced activity in the neural crest compared to the wild type enhancer. However, reporter activity in the otic pit was unchanged, suggesting differential regulation in the neural crest and ear (Fig. 4I and J)
Fig. 4.
Putative transcription binding sites were mutated using fusion PCR and cloned into pTK vector. The constructs were co-electroporated with a pTK-ECR1 construct expressing Cherry. Mutations in the SoxE sites (B) reduced enhancer activity versus intact ECR1 driving Cherry expression (A). Mutations in the core Hox2 sequence (D) also reduced enhancer activity compared to control (C ). Mutations in the consensus Ets-1 sequence (F) decrease enhancer activity compared to controls (E). Similarly, the control Cherry construct (G) had higher enhancer activity compared to the TFAP2 consensus site mutation construct (H). Mutations in a Fox consensus sequence reduced neural crest GFP expression but not otic expression (I), compared to control (J). Mutations in a putative cMyc/E-box binding sequence increased enhancer activity in the vagal neural crest only (K) compared to controls (L).
In contrast to the above mutations all of which reduced enhancer-mediated reporter activity, mutation of cMyc/E-box consensus sequence resulted in a very different change in reporter expression (the E-box consensus sequence is part of the longer cMyc consensus sequence). While no detectable differences were noted in reporter expression in the cranial neural crest compared with the wild type enhancer, reporter expression was expanded in the hindbrain region (Fig. 4K and L). These results suggest that the Myc/E-box site is bound by a repressor of Ets-1 activity at hindbrain levels. However, we never detected enhancer activity in the trunk neural crest either with the Myc/E-box mutant or with any other mutant we tested.
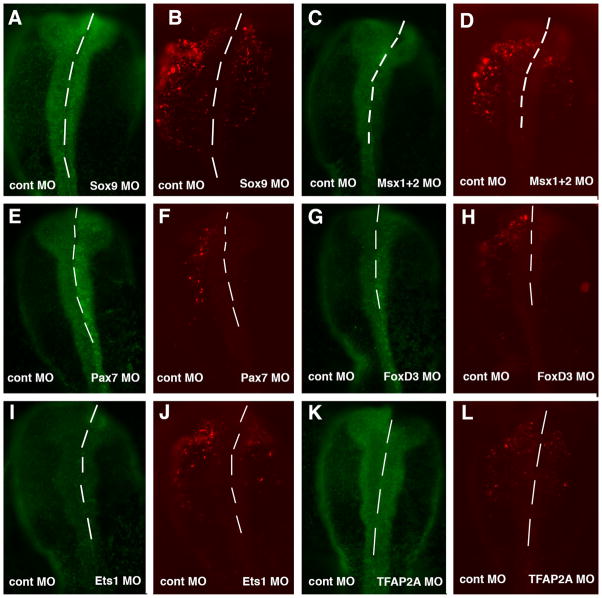
Knockdown of potential regulators reveals transcriptional inputs into ECR1
By identifying the sequences in the ECR1 enhancer that are required for enhancer activity, we identified several transcription factor families that that may be involved in mediating Ets-1 expression in the cranial neural crest. To determine which transcription factors are necessary for enhancer activity, we took a candidate approach to determine specific factors that are required for reporter activity by co-electroporating antisense morpholino oligos (MO) against specific factors together with the wild type ECR1 reporter construct.
As potential transcriptional inputs into the homeobox consensus sequences, we tested the effects of knock-down of Msx1, Msx2 and Pax7, all of which are neural plate border specifier genes (Meulemans and Bronner-Fraser, 2004). The results show that a combination of Msx1 and Msx2 MO significantly abrogated enhancer mediated reporter activity in the neural crest compared to the control MO (Fig. 5C and D). Similarly, electroporation of Pax7 MO resulted in a reduction in enhancer activity compared to the control MO side (Fig. 5E and F). These results suggest that a combination of neural plate border genes is important for activation of Ets-1 expression in the neural crest.
Fig. 5.
Morpholino oligos (MO) to transcription factors (right side) and control (left side) were co-electroporated with ECR1-pTK Cherry. MO to Sox9 (A) reduced ECR1 enhancer activity (B). A combination of Msx1 and Msx2 MO (C) reduced enhancer activity in the neural crest (D) compared to the control MO. The Pax7 MO electroplated side (E) had reduced enhancer activity compared to the control MO side (F). FoxD3 MO (G) also reduced the ECR1 enhancer activity compared to the control MO (H). Similarly, the Ets-1 MO (I) reduced enhancer activity versus the control MO (J). The MO to TFAP2A (K) reduced enhancer activity compared to the control MO side (L).
Three SoxE sites appeared to be essential for ECR1 activity. Sox9, a member of the SoxE gene family of transcription factors, is known to be important for neural crest development (Cheung and Briscoe, 2003; Mori-Akiyama et al., 2003). By electroporating a Sox9 MO on the right side and a control MO on the left side of the same embryo, we noted that Sox9 MO caused a reduction of ECR1 enhancer activity (Fig. 5A and B). In contrast, MO to its paralog, Sox10, had little effect on its own and did not enhance the knock-down when co-electroporated with Sox9 MO (data not shown). This suggests that Sox9, but not Sox10, is required for enhancer activity. This is interesting since both Sox9 and Ets-1 are direct inputs into Sox10 (Betancur et al., 2010B). To test if Sox9 was sufficient to activate the ECR1 enhancer, we over-expressed Sox9 plus the enhancer and found that it was able to ectopically induce its activity (Supplementary Fig. 1A–D).
Finally, we tested putative inputs into the Fox, Ets, TFAP2, and cMyc/E-Box sites. One Fox family member well-known to be expressed in the neural crest is FoxD3. FoxD3 expression in the neural folds slightly precedes that of Ets-1 (Théveneau et al., 2007)). To test whether FoxD3 is a required input into ECR1 activity, we electroporated a translation-blocking FoxD3 MO together with ECR1 enhancer constructs. The results show that the FoxD3 MO reduces ECR1 enhancer activity in the neural crest compared to the control MO side (Fig. 5G and H). However, there was no change in reporter expression in the ear, suggesting differential regulation of the same enhancer in these two tissues. Curiously, FoxD3 has been shown to be a repressor in neural crest (Simões-Costa et al., 2012). Thus the effects with the FoxD3 MO may be indirect. Also we cannot exclude the possibility that some other Fox family member, not yet identified, may be responsible for activation of this site. Alternatively, FoxD3 may act as an activator in this case, though we are unaware of any evidence that FoxD3 can act in this way.
Additional ECR1 inputs observed in this study include Ets-1 itself and TFAP2A. Our results reveal that a translation blocking Ets-1 MO reduced ECR1 enhancer activity compared to the control MO side (Fig. 5I and J). This suggests that Ets-1 autoregulates its own expression. In addition, a TFAP2A MO resulted in a reduction in enhancer activity in the neural crest compared to a control MO (Fig. 5K and L). In contrast, we saw no effect using either a cMyc MO or a Snail2 MO (data not shown).
To test specificity of the morpholino, we co-electroporated embryos with TFAP2-MO together with the mutated ECR1 construct lacking the TFAP2 binding site. The results show no effect on change in the small amount of residual Cherry expression (Supplementary Fig. 2A and B). Similarly, Ets-1 morpholino did not alter reporter expression of the construct in which the Ets binding site was mutated, nor were there changes in immunostaining (Supplementary Fig. 2D and C). Interesting, individual TFAP2 or Ets-1 morpholinos did not alter endogenous Ets-1 mRNA expression (Supplementary Fig. 3A and B), nor did MO to FoxD3, cMyc, or Sox9 (Supplementary Fig. 3C–E).
Taken together, these results show that numerous inputs are required for mediating enhancer activity in the neural crest. Notably, Sox9, Msx1, Msx2, FoxD3, Ets-1 and TFAP2A all are required for activation of Ets-1 expression in the cranial neural crest.
Discussion
Recent studies have formulated a gene regulatory network (GRN) that explains many aspects of cranial neural crest development (Meulemans and Bronner-Fraser, 2004; Sauka-Spengler and Bronner-Fraser, 2008; Betancur et al. 2010A). Neural crest cells are first induced in a swathe of epiblast between the neural and non-neural ectoderm at the gastrula stage. This induction is mediated by the combined action of signaling molecules including BMPs, FGFs ad WNTs (Stuhlmiller and Garcia-Castro, 2012). These signals activate a set of genes, referred to as neural border specifier genes that maintain the cells in a state competent to form neural crest. Transcription factors expressed in the neural borders, including Msx1, Id, cMyc, AP2a, Zic1 as well as Pax7, then act to induce the expression of the neural crest specifier genes, including Snail2, Sox9, Sox10, FoxD3, cMyb and Ets-1, that imbue cranial neural crest cells with the ability to undergo an epithelial-to-mesenchymal transition, to migrate extensively and ultimately differentiate into multiple derivatives.
The Ets-1 transcription factor is an important input into the neural crest GRN at cranial levels. Recent work has shown that Ets-1 directly regulates cranial neural crest enhancers that mediate expression of Sox10 and the FoxD3 (Betancur et al. 2010B; Simões-Costa et al., 2012). To understand regulation of Ets-1 itself, we have identified an Ets-1 minimal enhancer, ECR1, that mediates reporter expression in the cranial neural crest and otic placode and examined the inputs required for this expression. The ECR1 enhancer closely recapitulates the endogenous pattern of Ets-1 expression in the neural crest. We detect GFP activity in the premigratory neural crest as early as 6 somite stage. GFP expression remains evident on the neural crest continuously throughout the migratory phase as well as later, when the neural crest have reached their destination in the head and the branchial arches at HH stage 14, the last stage examined.
In addition to its expression in the neural crest, Ets-1 is well known for its expression in the endothelial, immune and hematopoetic systems (Dittmer, 2003). Previously, an Ets-1 regulatory region that controls expression in erythropoetic cells has been identified. Its inputs include Pea3, AP1, TFAP2A, Oct, and Ets-1 itself (Jorcyk et al., 1991; Oka et al., 1991; Chen and Wright, 1993). In mice, the first intron was shown to be necessary for expression in endothelial cells as well as erythropoetic cells. Although a 5′ flanking region was reported to drive expression in the branchial arches, it was not examined further (Jorcyk et al., 1997).
In contrast to cranial neural crest and otic expression, we did not observe reporter expression mediated by ECR1 or other conserved regions in other cell types that express Ets-1, like endothelial cells or the head mesoderm. There are several possible explanations for this. Perhaps the endothelial enhancer is not conserved between chicken and mammals, or its conservation was below the level of detection using the UCSD genome browser. Also, the enhancers controlling endothelial and head mesoderm Ets-1 expression may be located outside the regions we examined. Another possibility is that there may be multiple enhancers whose coordinate activation is required to recapitulate expression in these other cell types. The results from the mouse transgenic studies support the latter possibility (Jorcyk et al., 1997).
We tested a number of conserved putative consensus transcription factor binding sites within the ECR1 enhancer. A conserved homeobox consensus recognition sequence is required for cranial neural crest enhancer activity and represents a potential binding site for several neural plate border specifier genes, including Pax7, Msx1 and Msx2 (Meulemans and Bronner-Fraser, 2004). Consistent with this possibility, our results show that knock-down of Pax7, or Msx1/2 reduces reporter activity suggesting that both of these factors are required for expression mediated by the ECR1 enhancer. A Fox gene consensus binding sequence overlaps with the homeobox consensus sequence that is important for ECR1 enhancer activity and morpholino knockdown shows that FoxD3 appears to mediate expression through this binding site.
Of the seven conserved SoxE transcription factor consensus binding sites within ECR1, three were individually necessary for proper reporter activity. We were unable to discern any differences between the active site sequences and the inactive site sequences. Sox9 is the most likely input into the SoxE binding sites in the ECR1 enhancer. Not only is it expressed early during neural crest development, but it also has been shown to be important for regulation of Sox10 enhancer activity in the cranial neural crest (Betancur et al., 2010B) and its expression precedes that of Sox10 and Ets-1. Because Ets-1 is expressed prior to onset of Sox10 transcription, the lack of obvious effect of knocking down Sox10 on Ets-1 enhancer activity is not unexpected. TFAP2A is also required for ECR1 enhancer activity as well as for the activity of an additional Sox10 enhancer discovered in the mouse (Werner et al., 2007; Wahlbuhl et al., 2012).
Interestingly, our results show that mutation of a conserved cMyc/E-box consensus sequence actually increases enhancer activity in the hindbrain neural crest. Thus, this is site is likely to represent a repressor binding site. In contrast, we failed to detect any conserved neural crest recognition sites involved in trunk neural crest development. A possible explanation for the lack trunk expression is the rostrally limited expression of one or more of the upstream factors. Alternatively, there may be other yet to be identified sites necessary for neural crest enhancer activity orwhich may contain repressors of trunk neural crest expression. It is not surprising that expression of a particular gene would be mediated by a combination of activation and repression, with the latter limiting or reducing the level of gene expression in particular locations and times.
These data show that many of the same transcription factors that directly regulate other neural crest specifiers (e.g. Sox9, Pax7, FoxD3) also regulate Ets-1, which in turn regulates other neural crest specifiers such as Sox10 and FoxD3. Thus the neural crest specifier genes in the neural crest GRN appear not only to receive an initial signal (such as from Pax7, Msx1, Msx2, and Sox9), but also act to reinforce that signal, both by acting on other specifiers (FoxD3 and Sox10) and by feeding back upon themselves. A summary of known direct and proposed inputs into the neural crest gene regulatory network influencing expression of Ets-1 and other neural crest specifier genes is shown in Fig. 6. As illustrated, there is abundant feedback that could act to amplify the initial signal, ensuring that the neural crest cells remain specified by this reinforcement.
Fig. 6.

Schematic of direct interactions in the cranial neural crest on neural crest specifier genes (solid lines) as previously described (Betancur et al., 2010B; Simões-Costa et al., 2012) and the proposed interactions on Ets-1 as determined by morpholino knock-down of putative inputs and effects on Ets-1 enhancer expression (dashed lines).
Supplementary Material
Acknowledgments
We would like to thank Tatjana Sauka-Spengler for her valuable advice and Marcos Simões-Costa for his comments in writing this paper. This work was supported by USPHS P01 HD037105 and DE16459.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2013.08.009.
References
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010a;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc Natl Acad Sci U S A. 2010b;107:3570–3575. doi: 10.1073/pnas.0906596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman PE, Sanz-Ezquerro J, Overton IM, Burt DW, Bosch E, Fong WT, Tickle C, Brown WRA, Wilson SA, Hubbard SJ. A comprehensive collection of chicken cDNAs. Curr Biol. 2002;12:1965–1969. doi: 10.1016/s0960-9822(02)01296-4. [DOI] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JH, Wright CD. PEA3, Oct 1 and Oct 2 positively regulate the human ETS1 promoter. Oncogene. 1993;8:3375–3383. [PubMed] [Google Scholar]
- Cheung M, Briscoe J. Neural crest development is regulated by the transcription factor Sox9. Development. 2003;130:5681–5693. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- Dittmer J. The biology of the Ets-1 proto-oncogene. Mol Cancer. 2003;2:229. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Kim GH, Mackinnon AC, Flagg AE, Bassett B, Earley JU, Svensson EC. Ets1 is required for proper migration and differentiation of the cardiac neural crest. Development. 2010;137:1543–1551. doi: 10.1242/dev.047696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger H, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1953;88:49–92. [PubMed] [Google Scholar]
- Jorcyk CL, Watson DK, Mavrothalassitis GJ, Papas TS. The human ETS1 gene: genomic structure, promoter characterization and alternative splicing. Oncogene. 1991;6:523–532. [PubMed] [Google Scholar]
- Jorcyk CL, Garrett LJ, Maroulakou IG, Watson DK, Green JE. Multiple regulatory regions control the expression of Ets-1 in the developing mouse: vascular expression conferred by intron I. Cell Mol Biol. 1997;43:211–225. [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Mori-Akiyama Y, Akiyama H, Rowitch DH, de Crombrugghe B. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc Natl Acad Sci U S A. 2003;100:9360–9365. doi: 10.1073/pnas.1631288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Rairkar A, Chen JH. Structural and functional analysis of the regulatory sequences of the ets-1 gene. Oncogene. 1991;6:2077–2083. [PubMed] [Google Scholar]
- Sauka-Spengler T, Barembaum M. Gain- and loss-of-function approaches in the chick embryo. Methods Cell Biol. 2008;87:237–256. doi: 10.1016/S0091-679X(08)00212-4. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Simões-Costa MS, McKeown SJ, Tan-Cabugao J, Sauka-Spengler T, Bronner ME. Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is encrypted in the genome. PLoS Genet. 2012;8:e1003142. doi: 10.1371/journal.pgen.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlmiller TJ, Garcia-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 2012;69:3715–3737. doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Tahtakran SA, Selleck MA. Ets-1 expression is associated with cranial neural crest migration and vasculogenesis in the chick embryo. Gene Expr Patterns. 2003;3:455–458. doi: 10.1016/s1567-133x(03)00065-6. [DOI] [PubMed] [Google Scholar]
- Théveneau E, Duband JL, Altabef M. Ets-1 confers cranial features on neural crest delamination. PLoS One. 2007;2:e1142. doi: 10.1371/journal.pone.0001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell. 2003;4:509–519. doi: 10.1016/s1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Wahlbuhl M, Reiprich S, Vogl MR, Bösl MR, Wegner M. Transcription factor Sox10 orchestrates activity of a neural crest-specific enhancer in the vicinity of its gene. Nucleic Acids Res. 2012;40:88–101. doi: 10.1093/nar/gkr734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Hammer A, Wahlbuhl M, Bösl MR, Wegner M. Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res. 2007;35:6526–6538. doi: 10.1093/nar/gkm727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG. Wholemount in situ hybridization of vertebrate embryos. In: Wilkinson DG, editor. In Situ Hybridization: A Practical Approach. IRL Press; Oxford: 1992. pp. 75–83. [Google Scholar]
- Ye M, Coldren C, Liang X, Mattina T, Goldmuntz E, Benson DW, Ivy D, Perryman MB, Garrett-Sinha LA, Grossfeld P. Deletion of ETS-1, a gene in the Jacobsen syndrome critical region, causes ventricular septal defects and abnormal ventricular morphology in mice. Hum Mol Genet. 2010;19:648–656. doi: 10.1093/hmg/ddp532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.